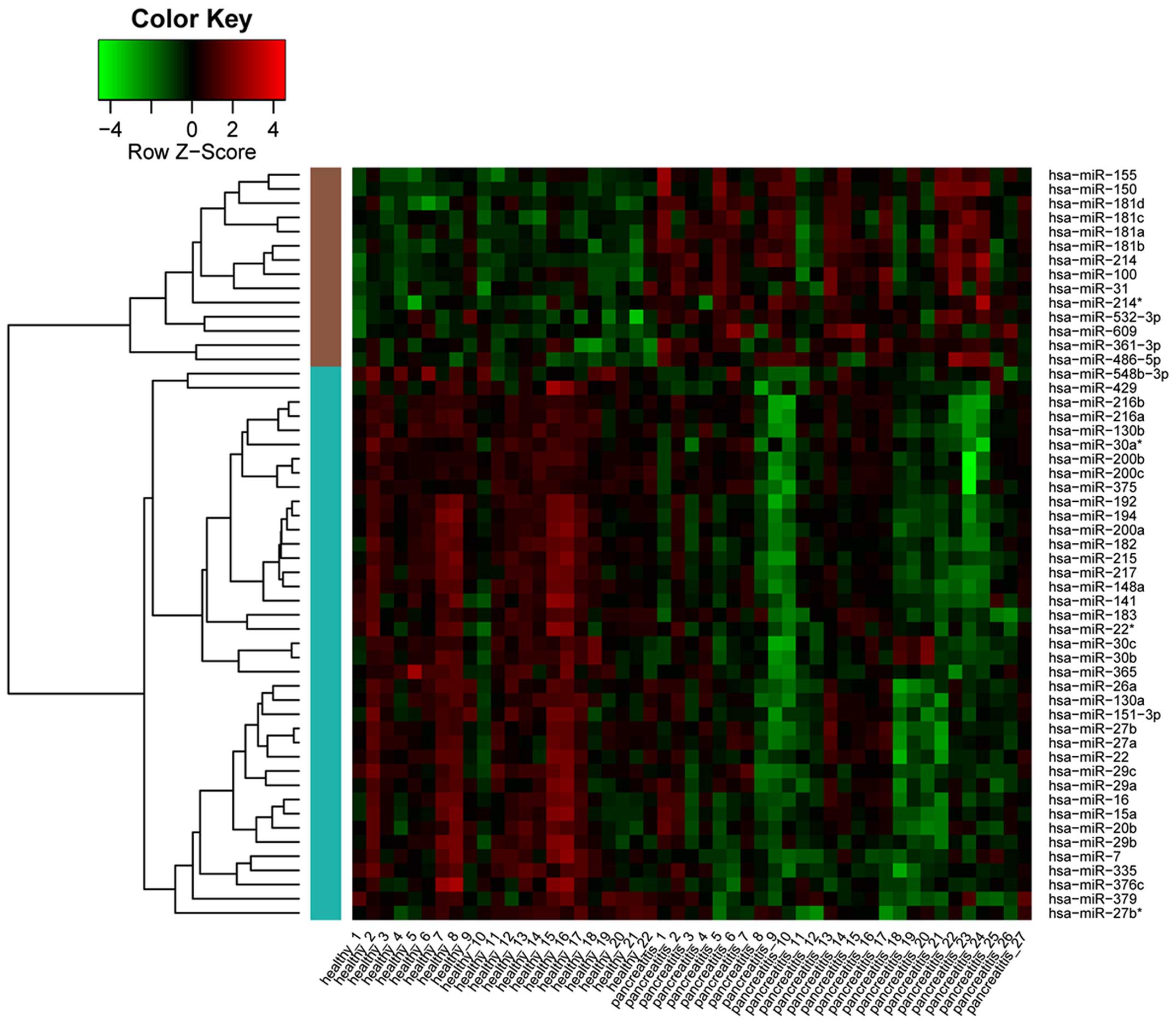
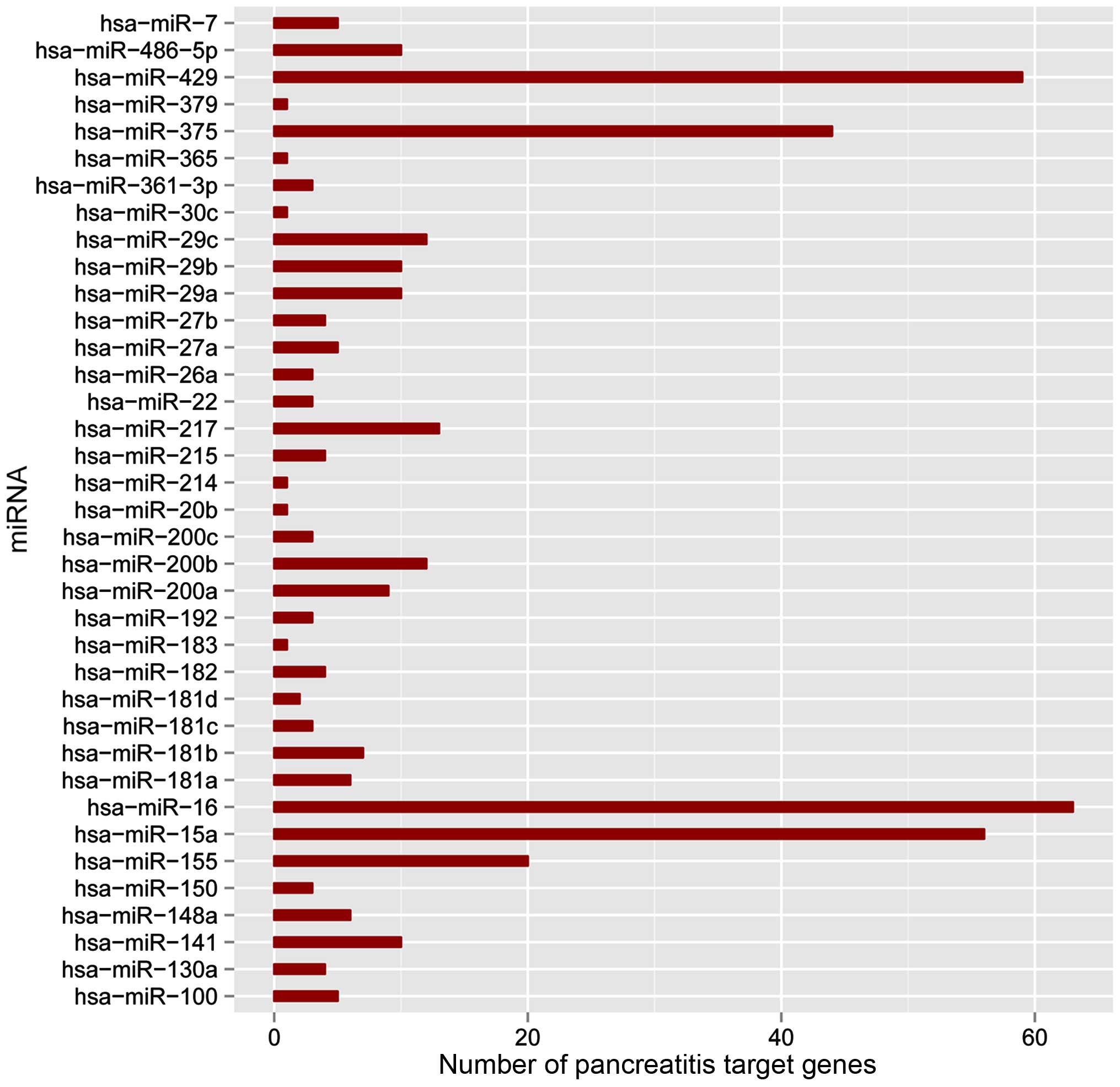
|
1
|
Apte MV, Pirola RC and Wilson JS:
Pancreas: Alcoholic pancreatitis-it's the alcohol, stupid. Nat Rev
Gastroenterol Hepatol. 6:321–322. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yadav D, Hawes RH, Brand RE, Anderson MA,
Money ME, Banks PA, Bishop MD, Baillie J, Sherman S, DiSario J, et
al: Alcohol consumption, cigarette smoking, and the risk of
recurrent acute and chronic pancreatitis. Arch Intern Med.
169:1035–1045. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hofmeyr S, Meyer C and Warren BL: Serum
lipase should be the laboratory test of choice for suspected acute
pancreatitis. S Afr J Surg. 52:72–75. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Turner B: Acute pancreatitis: Symptoms,
diagnosis and management. Nurs Times. 99:38–40. 2003.PubMed/NCBI
|
|
5
|
Banks PA and Freeman ML: Practice
Parameters Committee of the American College of Gastroenterology:
Practice guidelines in acute pancreatitis. Am J Gastroenterol.
101:2379–2400. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lipshutz GS and Wilkinson AH:
Pancreas-kidney and pancreas transplantation for the treatment of
diabetes mellitus. Endocrinol Metab Clin North Am. 36:1015–1038.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tien YW, Lee PH, Yang CY, Ho MC and Chiu
YF: Risk factors of massive bleeding related to pancreatic leak
after pancreaticoduodenectomy. J Am Coll Surg. 201:554–559. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Marik PE and Zaloga GP: Meta-analysis of
parenteral nutrition versus enteral nutrition in patients with
acute pancreatitis. BMJ. 328:14072004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
GBD 2013 Mortality and Causes of Death
Collaborators, . Global, regional, and national age-sex specific
all-cause and cause-specific mortality for 240 causes of death,
1990–2013: A systematic analysis for the Global Burden of Disease
Study 2013. Lancet. 385:117–171. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lund E, Güttinger S, Calado A, Dahlberg JE
and Kutay U: Nuclear export of microRNA precursors. Science.
303:95–98. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bauer AS, Keller A, Costello E, Greenhalf
W, Bier M, Borries A, Beier M, Neoptolemos J, Büchler M, Werner J,
et al: Diagnosis of pancreatic ductal adenocarcinoma and chronic
pancreatitis by measurement of microRNA abundance in blood and
tissue. PLoS One. 7:e341512012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bloomston M, Frankel WL, Petrocca F,
Volinia S, Alder H, Hagan JP, Liu CG, Bhatt D, Taccioli C and Croce
CM: MicroRNA expression patterns to differentiate pancreatic
adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA.
297:1901–1908. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yu S, Lu Z, Liu C, Meng Y, Ma Y, Zhao W,
Liu J, Yu J and Chen J: miRNA-96 suppresses KRAS and functions as a
tumor suppressor gene in pancreatic cancer. Cancer Res.
70:6015–6025. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mees ST, Mardin WA, Sielker S, Willscher
E, Senninger N, Schleicher C, Colombo-Benkmann M and Haier J:
Involvement of CD40 targeting miR-224 and miR-486 on the
progression of pancreatic ductal adenocarcinomas. Ann Surg Oncol.
16:2339–2350. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liffers ST, Munding JB, Vogt M, Kuhlmann
JD, Verdoodt B, Nambiar S, Maghnouj A, Mirmohammadsadegh A, Hahn SA
and Tannapfel A: MicroRNA-148a is down-regulated in human
pancreatic ductal adenocarcinomas and regulates cell survival by
targeting CDC25B. Lab Invest. 91:1472–1479. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ohuchida K, Mizumoto K, Lin C, Yamaguchi
H, Ohtsuka T, Sato N, Toma H, Nakamura M, Nagai E, Hashizume M and
Tanaka M: MicroRNA-10a is overexpressed in human pancreatic cancer
and involved in its invasiveness partially via suppression of the
HOXA1 gene. Ann Surg Oncol. 19:2394–2402. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huber W, Von Heydebreck A, Sültmann H,
Poustka A and Vingron M: Variance stabilization applied to
microarray data calibration and to the quantification of
differential expression. Bioinformatics. 18:(Suppl 1). S96–S104.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Smyth G: Limma: Linear models for
microarray data. Bioinformatics and Computational Biology Solutions
Using R and Bioconductor. Gentleman R, Carey V, Dudoit S, Irizarry
R and Huber W: Springer Science & Business Media. (New York,
NY). 397–420. 2005. View Article : Google Scholar
|
|
20
|
Benjamini Y: Discovering the false
discovery rate. J Royal Stat Soc: Series B (Statistical
Methodology). 72:405–416. 2010. View Article : Google Scholar
|
|
21
|
Xiao F, Zuo Z, Cai G, Kang S, Gao X and Li
T: miRecords: An integrated resource for microRNA-target
interactions. Nucleic Acids Res. 37:(Database issue). D105–D110.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Dweep H, Sticht C, Pandey P and Gretz N:
miRWalk-database: Prediction of possible miRNA binding sites by
‘walking’ the genes of three genomes. J Biomed Inform. 44:839–847.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tweedie S, Ashburner M, Falls K, Leyland
P, McQuilton P, Marygold S, Millburn G, Osumi-Sutherland D,
Schroeder A, Seal R, et al: FlyBase: Enhancing Drosophila gene
ontology annotations. Nucleic Acids Res. 37:(Database issue).
D555–D559. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Davis AP, Grondin CJ, Lennon-Hopkins K,
Saraceni-Richards C, Sciaky D, King BL, Wiegers TC and Mattingly
CJ: The Comparative Toxicogenomics Database's 10th year
anniversary: Update 2015. Nucleic Acids Res. 43:(Database issue).
D914–D920. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Saito R, Smoot ME, Ono K, Ruscheinski J,
Wang PL, Lotia S, Pico AR, Bader GD and Ideker T: A travel guide to
Cytoscape plugins. Nat Methods. 9:1069–1076. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Habbe N, Koorstra JB, Mendell JT,
Offerhaus GJ, Ryu JK, Feldmann G, Mullendore ME, Goggins MG, Hong
SM and Maitra A: MicroRNA miR-155 is a biomarker of early
pancreatic neoplasia. Cancer Biol Ther. 8:340–346. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Greither T, Grochola LF, Udelnow A,
Lautenschläger C, Würl P and Taubert H: Elevated expression of
microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated
with poorer survival. Int J Cancer. 126:73–80. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang XJ, Ye H, Zeng CW, He B, Zhang H and
Chen YQ: Dysregulation of miR-15a and miR-214 in human pancreatic
cancer. J Hematol Oncol. 3:462010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ebert MP, Hernberg S, Fei G, Sokolowski A,
Schulz HU, Lippert H and Malfertheiner P: Induction and expression
of cyclin D3 in human pancreatic cancer. J Cancer Res Clin Oncol.
127:449–454. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Radulovich N, Pham NA, Strumpf D, Leung L,
Xie W, Jurisica I and Tsao MS: Differential roles of cyclin D1 and
D3 in pancreatic ductal adenocarcinoma. Mol Cancer. 9:242010.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Biliran H Jr, Wang Y, Banerjee S, Xu H,
Heng H, Thakur A, Bollig A, Sarkar FH and Liao JD: Overexpression
of cyclin D1 promotes tumor cell growth and confers resistance to
cisplatin-mediated apoptosis in an elastase-myc
transgene-expressing pancreatic tumor cell line. Clin Cancer Res.
11:6075–6086. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Qiao Q, Ramadani M, Gansauge S, Gansauge
F, Leder G and Beger HG: Reduced membranous and ectopic cytoplasmic
expression of beta-catenin correlate with cyclin D1 overexpression
and poor prognosis in pancreatic cancer. Int J Cancer. 95:194–197.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Handra-Luca A, Hammel P, Sauvanet A,
Ruszniewski P and Couvelard A: Tumoral epithelial and stromal
expression of SMAD proteins in pancreatic ductal adenocarcinomas. J
Hepatobiliary Pancreat Sci. 20:294–302. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Duan W, Li R, Ma J, Lei J, Xu Q, Jiang Z,
Nan L, Li X, Wang Z, Huo X, et al: Overexpression of Nodal induces
a metastatic phenotype in pancreatic cancer cells via the Smad2/3
pathway. Oncotarget. 6:1490–1506. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pan X, Wang X, Wang Z, Zhu Q and Han Y:
Expression of Smad2 and TGFβ 1 in human pancreatic carcinoma
tissues and its significance. Prog Mod Biomed. 6:0322012.
|
|
38
|
Basu A, Alder H, Khiyami A, Leahy P, Croce
CM and Haldar S: MicroRNA-375 and MicroRNA-221 Potential noncoding
RNAs associated with antiproliferative activity of Benzyl
isothiocyanate in pancreatic cancer. Genes Cancer. 2:108–119. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhou J, Song S, Cen J, Zhu D, Li D and
Zhang Z: MicroRNA-375 is downregulated in pancreatic cancer and
inhibits cell proliferation in vitro. Oncol Res. 20:197–203. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Altomare DA, Tanno S, De Rienzo A,
Klein-Szanto AJ, Tanno S, Skele KL, Hoffman JP and Testa JR:
Frequent activation of AKT2 kinase in human pancreatic carcinomas.
J Cell Biochem. 87:470–476. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Roy SK, Srivastava RK and Shankar S:
Inhibition of PI3K/AKT and MAPK/ERK pathways causes activation of
FOXO transcription factor, leading to cell cycle arrest and
apoptosis in pancreatic cancer. J Mol Signal. 5:102010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Tomita T: Cyclin-dependent kinase (cdk6)
and p16 in pancreatic endocrine neoplasms. Pathology. 36:566–570.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Tang LH, Contractor T, Clausen R, Klimstra
DS, Du YC, Allen PJ, Brennan MF, Levine AJ and Harris CR:
Attenuation of the retinoblastoma pathway in pancreatic
neuroendocrine tumors due to increased cdk4/cdk6. Clin Cancer Res.
18:4612–4620. 2012. View Article : Google Scholar : PubMed/NCBI
|