Introduction
Down syndrome (DS), or trisomy 21, results from the
presence of all or part of an extra chromosome (Chr) 21 and is a
high-incidence birth defect that is often associated with physical
disorders (1–3) that have no explicit mechanism.
Incorrect partitioning of sister chromatids to the daughter cells
may induce aneuploidy, including trisomy 21 (4,5).
Centromeres, the evident structures that are located in the center
of chromosomes are responsible for chromosome segregation. A
previous study indicated that the highly conserved centromere
protein B (CENP-B) and its 17 base pair (bp) binding site (CENP-B
box) are required for de novo mammalian artificial
chromosome formation (6). In
vivo analyses with cultured human cells suggested that the
presence of the CENP-B box is essential for the formation of
functional centromere components (7,8). In
addition, CENP-B binding to the CENP-B boxes influences nucleosome
positioning in centromeric regions and this binding is required for
accurate chromosome segregation (9–11).
Therefore, this suggests that nondisjunction of Chr21 may be
induced by a defective functioning of the centromere with a CENP-B
box mutation.
To determine whether the mutation or absence of the
CENP-B box is involved with extra Chr21 formation in DS, the CENP-B
boxes in Chr21 α-satellite DNA (α-satDNA) of 127 DS children were
investigated using polymerase chain reaction (PCR) and direct
sequencing. The observations of the present study provide an
insight into the molecular association between the CENP-B box and
α-satDNA in Chr21, which may be a major contributor to the
occurrence of trisomy 21.
Materials and methods
Patient recruitment
A total of 127 Chinese children with DS were
recruited from the Children's Hospital of Chongqing Medical
University (Chongqing, China). Cytogenetic analysis demonstrated
the karyotypes were 47, XY, +21 or 47, XX, +21 in all the patients.
In addition, 100 healthy children with normal karyotypes were also
enrolled as the control group. The distribution of age and
residential placement did not differ between controls and DS
patients. Peripheral blood samples were collected from all
participants. The study was approved by the Ethics Committee of
Chongqing Medical University. Informed consent was obtained from
all individual participants included in the study.
PCR amplification and sequencing
Genomic DNA was extracted from peripheral blood
samples with TIANamp Blood DNA kit (Tiangen Biotech Co., Ltd.,
Beijing, China). α-satDNA (GenBank accession no. D29750.1) in the
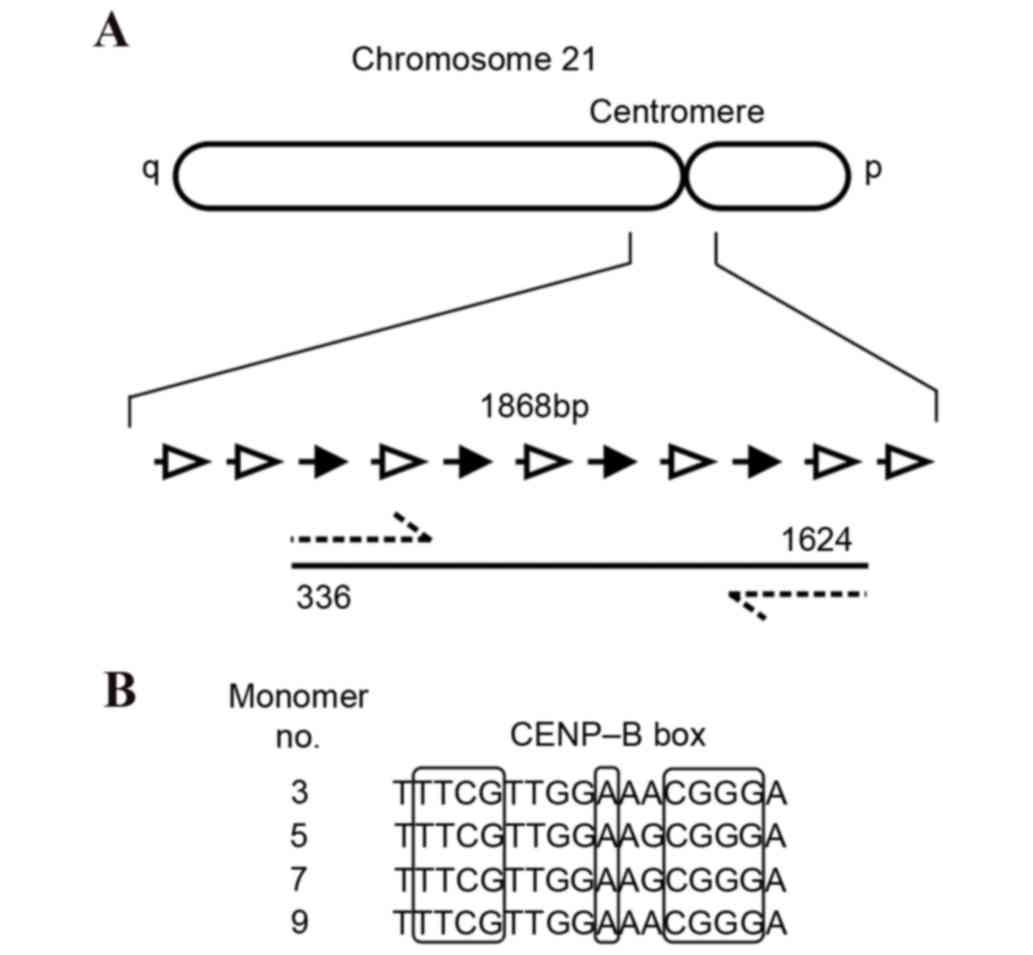
centromeric region of Chr21 was amplified as two overlapping
fragments, with each fragment containing two CENP-B boxes (Fig. 1). Oligonucleotide primer sequences
were as follows: α-satellite part A, forward
5′-GGAATATCGTCATACAAAAT-3′, reverse 5′-TATCAATGGCAAAGTTCA-3′; and
α-satellite part B, forward 5′-GTTGAACTTTGCCATTGA-3′, and reverse,
5′-GCTCTAAGAAAGCGAATG-3′. PCR was performed using the Takara
Taq™ PCR kit (Takara Biotechnology, Co., Ltd., Dalian,
China), according to the manufacturer's protocol, using 200 ng
genomic DNA and 20 nM primers (1 µl). PCR thermocycling conditions
were as follows: 94°C for 4 min (initial denaturation), followed by
30 cycles at 95°C for 30 sec (denaturation), 46°C/43°C for 30 sec
(primer annealing) for part A/B, respectively, and 72°C for 45 sec
(PCR product elongation), a final extension step was at 72°C for 5
min. PCR products were run on 1.5% agarose gels to detect possible
large sequence rearrangements. Following the removal of
unincorporated primers and excess dNTPs by exonuclease I and shrimp
alkaline phosphatase, the amplified fragments were directly
sequenced on an ABI-3100 Genetic Analyzer (Applied Biosystems;
Thermo Fisher Scientific, Inc., Waltham, MA, USA). Each sample was
compared with NCBI RefSeq sequence of α-satDNA.
Statistical analysis
Statistical analysis was used to summarize the
clinical characteristics of the subjects. Associations between
polymorphisms of DS and control group were examined using the
Chi-square test. P<0.05 was considered to indicate a
statistically significant difference. All data were analyzed using
SPSS software, version 16.0 (SPSS, Inc., Chicago, IL, USA).
Results
Gel electrophoresis
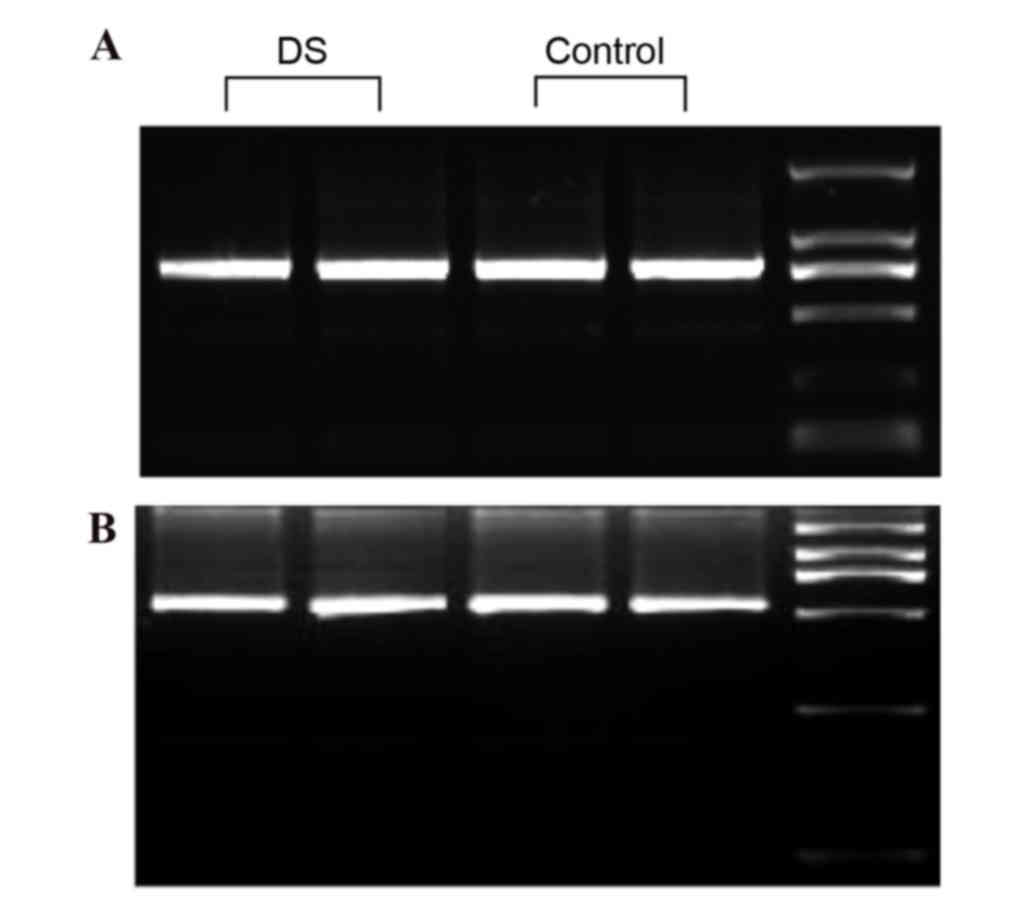
The gel electrophoresis indicated that the 773 and
535 bp PCR fragments from α-satDNA were amplified correctly. There
was no observation of a mispairing present in the agarose gels.
There was no visible difference in gel electrophoresis between
amplified fragments of DS and healthy children (Fig. 2). To examine whether the two
fragments exhibited a point mutation, direct sequencing was
performed following gel electrophoresis.
CENP-B boxes in Chr21 are highly
conserved
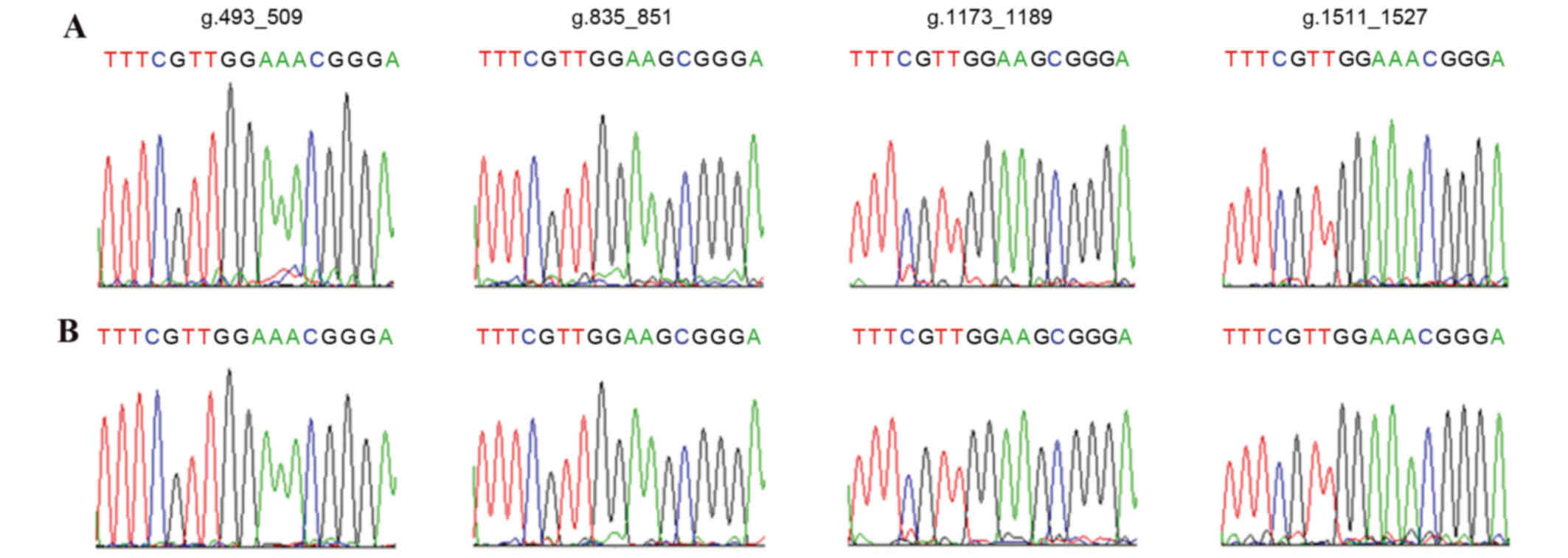
The affected patients with DS and healthy children
underwent DNA analysis by sequencing of PCR products, of four
CENP-B boxes that are respectively located at the g.493_509,
g.835_851, g.1173_1189 and g.1511_1527 sites of α-satDNA in Chr21.
No mutation or absence of CENP-B boxes in the 127 patients with DS
and 100 healthy children was observed. All four CENP-B boxes in
Chr21 were demonstrated to be highly conserved (Fig. 3).
α-satDNA sequence is highly
variable
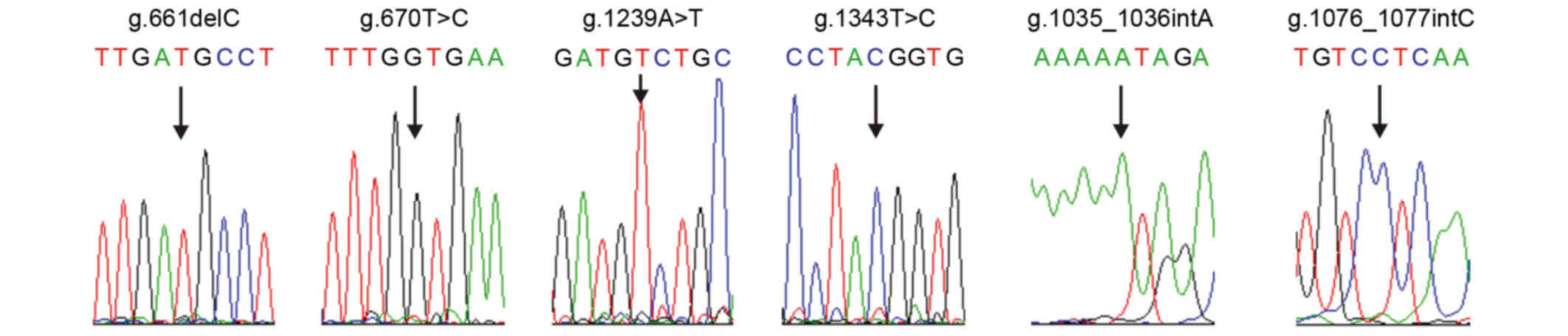
The direct sequencing of α-satDNA indicated
numerous underlying nucleotide alterations. The variations were
point nucleotide deletions, substitutions, insertions and
polymorphisms. The novel identified mutations were as follows:
g.661delC, g.670T>G, g.1239A>T, g.1343T>C, g.1035_1036insA
and g.1076_1077insC (Fig. 4). All
the mutations were observed to be present in DS and control groups,
however statistical analysis suggested that there was no
significant difference between their occurrence in the two groups
(P>0.05; Table I). Furthermore,
8 polymorphism sites were observed in the majority of samples and
to the best of our knowledge, these had not previously been
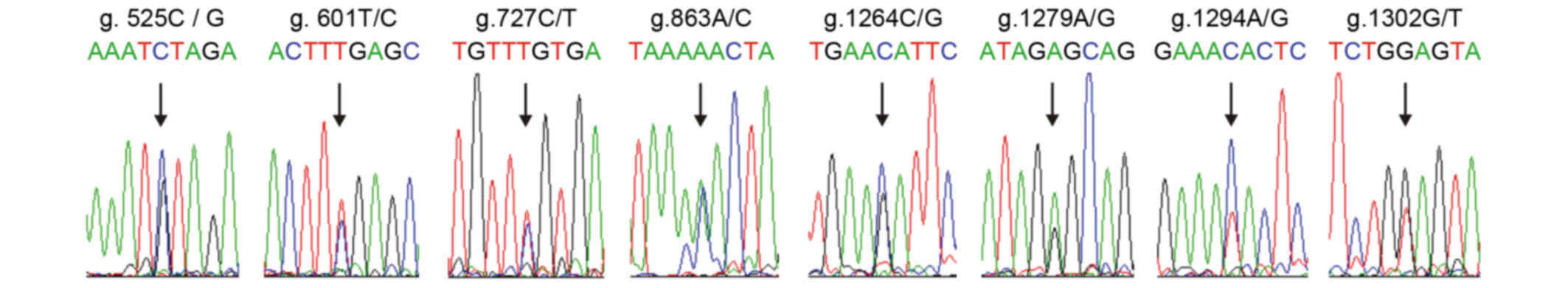
reported. However, occurrence of the g.525C/G (P=0.01), g.601T/C
(P=0.00000002), g.1279A/G (P=0.002), g.1294C/T (P=0.0006) and
g.1302 G/T (P=0.004) sites in the DS group demonstrated a
significant difference compared with the control group (P<0.05)
However, no differences in g.727C/T (P=0.36), g.863A/C (P=0.207)
and g.1264C/G (P=0.073) sites were observed between the two groups
(Fig. 5; Table I).
 | Table I.Spectrum of α-satDNA variants detected
in DS and control children. |
Table I.
Spectrum of α-satDNA variants detected
in DS and control children.
| Mutation
characteristics | DS (n=127) | Control (n=100) | P-value |
|---|
| g.661delC | 127 (100%) | 100 (100%) | 0.904 |
| g.670T>G | 127 (100%) | 100 (100%) | 0.904 |
| g.1239A>T | 124 (97.6%) | 100 (100%) | 0.122 |
| g.1343T>C | 124 (97.6%) | 100 (100%) | 0.122 |
| g.1035_1036insA | 125 (98.4%) | 97 (97%) | 0.468 |
| g.1076_1077insC | 125 (98.4%) | 97 (97%) | 0.468 |
| g.525C/G | 125 (98.4%) | 91 (91%) | 0.01a |
| g.601T/C | 93 (73.2%) | 100 (100%) |
0.00000002a |
| g.727C/T | 120 (94.5%) | 97 (97%) | 0.36 |
| g.863A/C | 125 (98.4%) | 100 (100%) | 0.207 |
| g.1264C/G | 123 (96.9%) | 100 (100%) | 0.073 |
| g.1279A/G | 101 (79.5%) | 94 (94%) | 0.002a |
| g.1294C/T | 105 (82.7%) | 97 (97%) | 0.0006a |
| g.1302G/T | 103 (81.1%) | 94 (94%) | 0.004a |
Discussion
DS is a common chromosomal disorder in human
aneuploidy, and a termination of pregnancy occurs for the majority
DS fetuses identified via Down's screening and amniocentesis. The
extra Chr21 results from a meiotic nondisjunction event, with 88%
from nondisjunction in the maternal gamete and 8% in the paternal
gamete. But the underlying molecular genetics mechanism linking
nondisjunction to Chr21 remains to be fully elucidated. Therefore,
it is necessary to determine the genotypes of DS to provide a
methodology for antenatal examination and gene diagnosis for
trisomy 21 in noninvasive prenatal testing.
The current study identified that the CENP-B boxes
are highly conserved in DS and healthy children. Human Chr21
contains a large repetitive sequence and consists of 11 tandem
repeats of an AT-rich 171 bp alphoid monomer unit. This 11mer
repeat construct is a 1.3 Mbp higher order α-satDNA repeat on the
centromeric area. Each monomer has a unique 17 bp sequence, however
only five have the CENP-B binding activity known as the CENP-B box
(12,13). In the human artificial chromosome
(HAC), CENP-B box mutations induced no HAC formation and diminution
of CENP-A assembly (6,11,14).
CpG methylation and point mutation of the CENP-B box reduced CENP-B
binding activity (15–17). Therefore, the CENP-B box is
required for de novo centromere chromatin assembly on human
α-satDNA. The present study identified the methylation status of
CpG dinucleotides in CG islands; however, the results did not show
any difference in the DNA methylation status between DS and healthy
people as previously described (18).
A cohort of 127 patients diagnosed with DS and 100
healthy children in Chongqing were investigated via chromosome
screening. The spectrum of α-satDNA variants in Chr21 was analyzed
by direct DNA sequencing. The present study identified 14 novel
variations that, to the best of our knowledge, have not previously
been reported. A total of 6 mutations were identified in the
Chinese population: g.661delC, g.670T>G, g.1239A>T,
g.1343T>C, g.1035_1036insA and g.1076_1077insC. The present
study revealed the distribution and frequency of novel variations
among the population in Chongqing. For human Chr21, α-satDNA
appears to be the major constituent of functional centromeres
(19), as it is associated with
the existence of functional centromere components on the endogenous
human chromosome and the ability to induce de novo
centromere assembly. In the present study, the CENP-B box and
α-satDNA sequence were required for de novo mammalian
artificial chromosome formation and assembly of functional
centromere components, including CENP-A, CENP-C, and CENP-E
(20,21). These results demonstrated the
presence of a variable in α-satDNA, other than the CENP-B box,
which may be important for de novo centromere formation
function (8,16,22,23).
In addition, the present study reported eight single nucleotide
polymorphisms (SNPs) of α-satDNA in Chr21. Three SNPs, including
g.727C/T, g.863A/C and g.1264C/G did not demonstrate any difference
in occurrence between DS and healthy children. It was therefore
hypothesized that the variants identified in the present study may
be due to ethnic disparities, as suggested by previous studies
(18,24) and consequently, these variants may
act as DNA markers for distinguishing ethnicities. However, five
SNPs including g.525C/G, g.601T/C, g.1279A/G, g.1294C/T and g.1302
G/T were observed to be significantly different between the DS and
control group, and may therefore act as potential molecular
diagnostic markers of DS.
The present study exhibited various limitations.
Firstly, the study was conducted on a small sample size and larger
sample sizes are required to confirm these associations in the
future. Furthermore, the patients were from the same region of
China, therefore further studies should include individuals from
different regions of China and of different ethnicities.
In conclusion, the present study investigated the
α-satDNA variants present in Chr21 and any differences in genotype
between DS and healthy children. The results demonstrated that
CENP-B box mutations were not present and the box appears to be
highly conserved in DS patients and therefore may not be
responsible for Chr21 nondisjunction events. Furthermore, 14 novel
variations were identified in this investigation and the differing
mutation spectrum of α-satDNA between DS and healthy individuals
may contribute to complex DS phenotypes and act as potential
dinucleotide markers for diagnosing trisomy 21.
Acknowledgements
The authors would like to thank Dr Lin Zou and Dr
Cui Song (Center of Clinical Molecular Medicine, Children's
Hospital of Chongqing Medical University; Chongqing, China) for
case collection. The present study was supported by the National
Natural Science Foundation of China (grant no. 30771202).
Glossary
Abbreviations
Abbreviations:
|
α-satDNA
|
α-satellite DNA
|
|
CENP-B
|
centromere protein B
|
|
Chr21
|
chromosome 21
|
|
DS
|
Down syndrome
|
|
HAC
|
human artificial chromosome
|
|
PCR
|
polymerase chain reaction
|
References
|
1
|
Shott SR, Joseph A and Heithaus D: Hearing
loss in children with Down syndrome. Int J Pediatr
Otorhinolaryngol. 61:199–205. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nespoli L, Burgio GR, Ugazio AG and
Maccario R: Immunological features of Down's syndrome: A review. J
Intellect Disabil Res. 37:543–551. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Karlsson B, Gustafsson J, Hedov G,
Ivarsson SA and Annerén G: Thyroid dysfunction in Down's syndrome:
Relation to age and thyroid autoimmunity. Arch Dis Child.
79:242–245. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wittmann T, Hyman A and Desai A: The
spindle: A dynamic assembly of microtubules and motors. Nat Cell
Biol. 3:E28–E34. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Walczak CE and Heald R: Mechanisms of
mitotic spindle assembly and function. Int Rev Cytol. 265:111–158.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ohzeki J, Nakano M, Okada T and Masumoto
H: CENP-B box is required for de novo centromere chromatin assembly
on human alphoid DNA. J Cell Biol. 159:765–775. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Trowell HE, Nagy A, Vissel B and Choo KH:
Long-range analyses of the centromeric regions of human chromosomes
13, 14 and 21: Identification of a narrow domain containing two key
centromeric DNA elements. Hum Mol Genet. 2:1639–1649. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Okada T, Ohzeki J, Nakano M, Yoda K,
Brinkley WR, Larionov V and Masumoto H: CENP-B controls centromere
formation depending on the chromatin context. Cell. 131:1287–1300.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tanaka Y, Tachiwana H, Yoda K, Masumoto H,
Okazaki T, Kurumizaka H and Yokoyama S: Human centromere protein B
induces translational positioning of nucleosomes on alpha-satellite
sequences. J Biol Chem. 280:41609–41618. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hasson D, Panchenko T, Salimian KJ, Salman
MU, Sekulic N, Alonso A, Warburton PE and Black BE: The octamer is
the major form of CENP-A nucleosomes at human centromeres. Nat
Struct Mol Biol. 20:687–695. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Okamoto Y, Nakano M, Ohzeki J, Larionov V
and Masumoto H: A minimal CENP-A core is required for nucleation
and maintenance of a functional human centromere. EMBO J.
26:1279–1291. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ikeno M, Masumoto H and Okazaki T:
Distribution of CENP-B boxes reflected in CREST centromere
antigenic sites on long-range alpha-satellite DNA arrays of human
chromosome 21. Hum Mol Genet. 3:1245–1257. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Alkan C, Ventura M, Archidiacono N, Rocchi
M, Sahinalp SC and Eichler EE: Organization and evolution of
primate centromeric DNA from whole-genome shotgun sequence data.
PLoS Comput Biol. 3:1807–1820. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Basu J, Stromberg G, Compitello G, Willard
HF and Van Bokkelen G: Rapid creation of BAC-based human artificial
chromosome vectors by transposition with synthetic alpha-satellite
arrays. Nucleic Acids Res. 33:587–596. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Earnshaw WC, Sullivan KF, Machlin PS,
Cooke CA, Kaiser DA, Pollard TD, Rothfield NF and Cleveland DW:
Molecular cloning of cDNA for CENP-B, the major human centromere
autoantigen. J Cell Biol. 104:817–829. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Masumoto H, Masukata H, Muro Y, Nozaki N
and Okazaki T: A human centromere antigen (CENP-B) interacts with a
short specific sequence in alphoid DNA, a human centromeric
satellite. J Cell Biol. 109:1963–1973. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tanaka Y, Kurumizaka H and Yokoyama S: CpG
methylation of the CENP-B box reduces human CENP-B binding. FEBS J.
272:282–289. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xia YY, Ding YB, Liu XQ, Chen XM, Cheng
SQ, Li LB, Ma MF, He JL and Wang YX: Allelic methylation status of
CpG islands on chromosome 21q in patients with Trisomy 21. Mol Med
Rep. 9:1681–1688. 2014.PubMed/NCBI
|
|
19
|
Ugarković D and Plohl M: Variation in
satellite DNA profile-causes and effects. EMBO J. 21:5955–5959.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sullivan LL, Boivin CD, Mravinac B, Song
IY and Sullivan BA: Genomic size of CENP-A domain is proportional
to total alpha satellite array size at human centromeres and
expands in cancer cells. Chromosome Res. 19:457–470. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Irvine DV, Amor DJ, Perry J, Sirvent N,
Pedeutour F, Choo KH and Saffery R: Chromosome size and origin as
determinants of the level of CENP-A incorporation into human
centromeres. Chromosome Res. 12:805–815. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Meštrović N, Pavlek M, Car A,
Castagnone-Sereno P, Abad P and Plohl M: Conserved DNA motifs,
including the CENP-B box-like, are possible promoters of satellite
DNA array rearrangements in nematodes. PLoS One. 8:e673282013.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fachinetti D, Folco HD, Nechemia-Arbely Y,
Valente LP, Nguyen K, Wong AJ, Zhu Q, Holland AJ, Desai A, Jansen
LE and Cleveland DW: A two-step mechanism for epigenetic
specification of centromere identity and function. Nat Cell Biol.
15:1056–1066. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xia YY, Ding YB, Liu XQ, Chen XM, Cheng
SQ, Li LB, Ma MF, He JL and Wang YX: Racial/ethnic disparities in
human DNA methylation. Biochim Biophys Acta. 1846:258–262.
2014.PubMed/NCBI
|