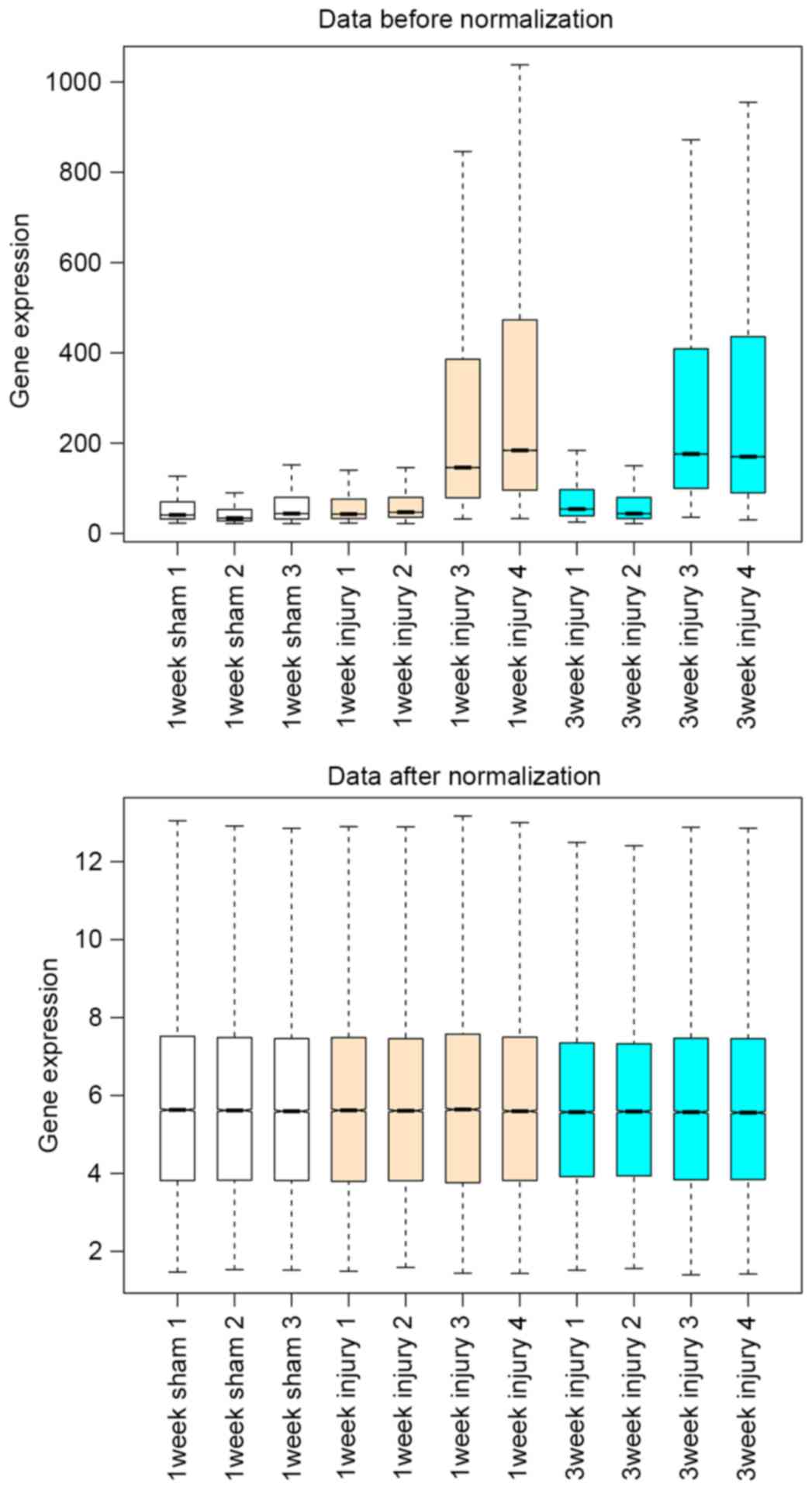
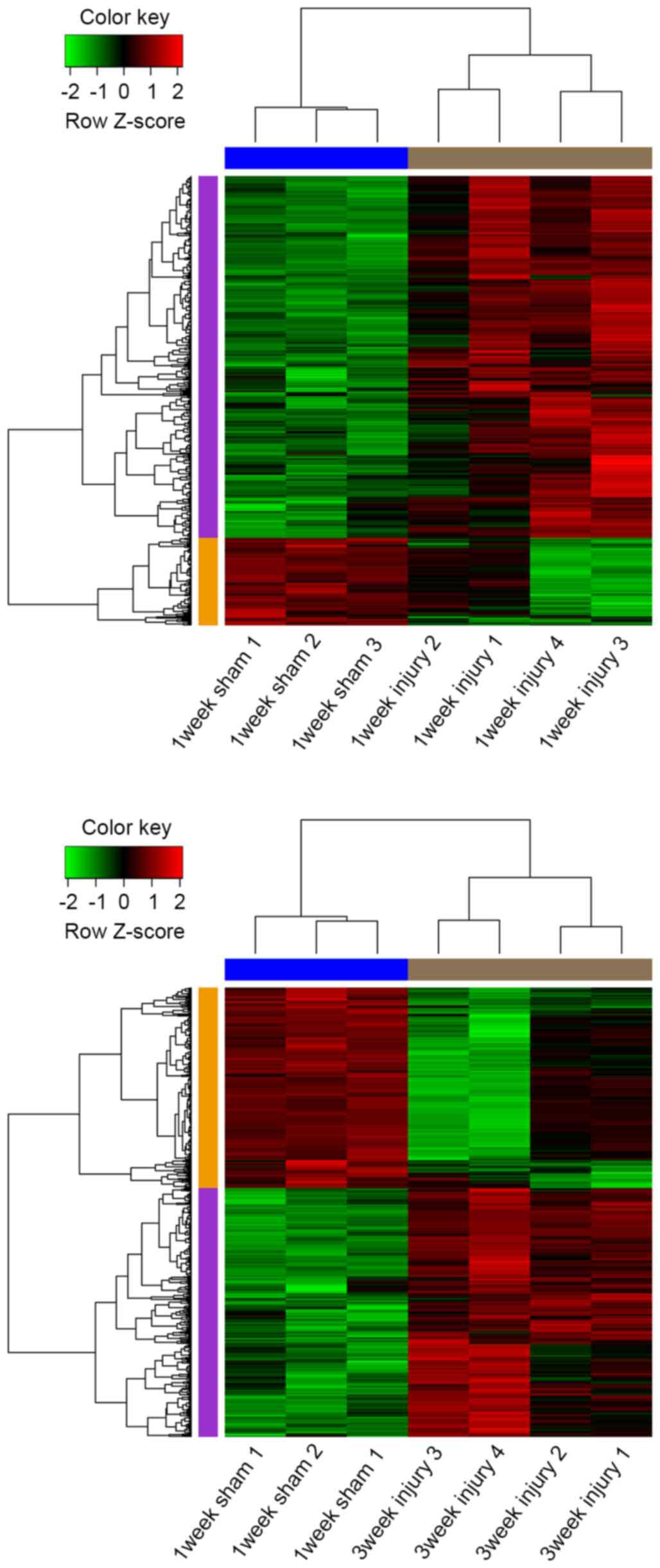
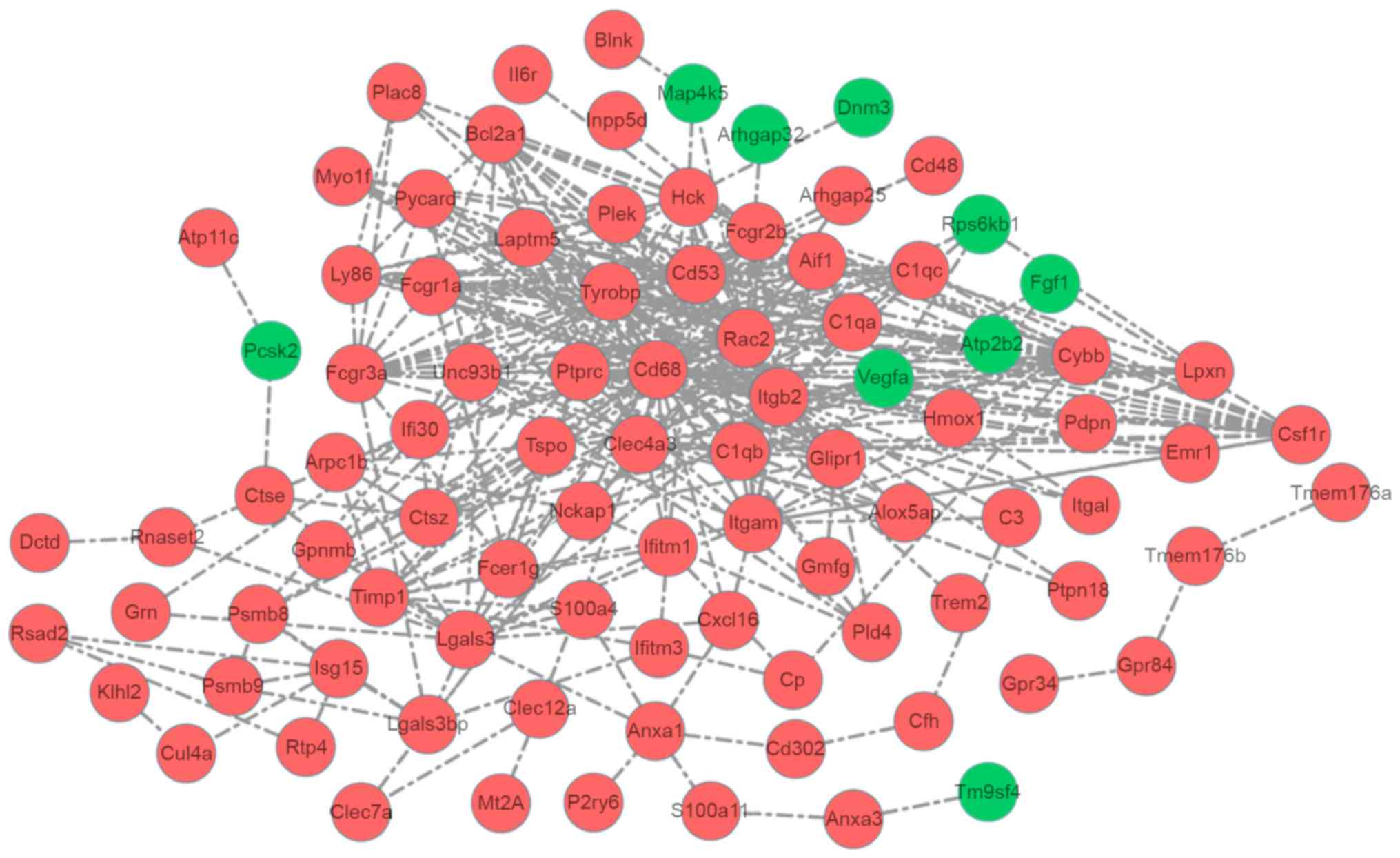
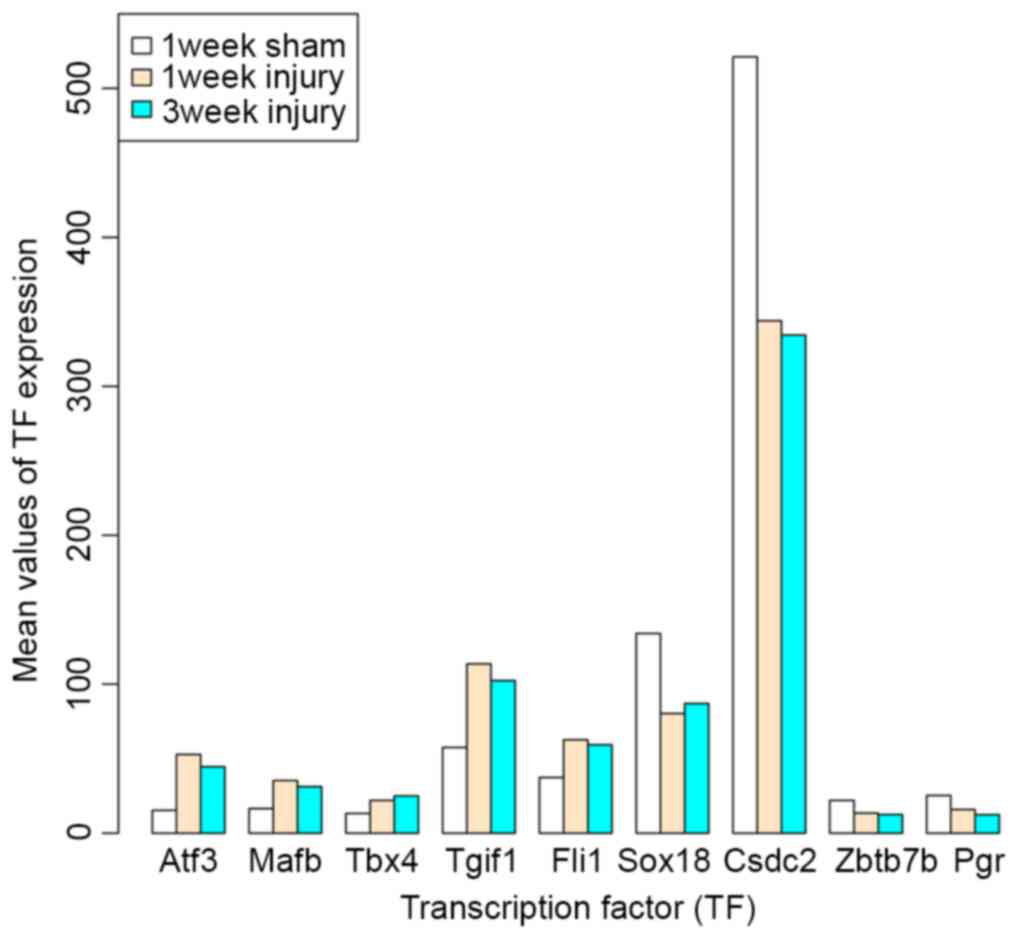
|
1
|
Jain NB, Ayers GD, Peterson EN, Harris MB,
Morse L, O'Connor KC and Garshick E: Traumatic spinal cord injury
in the United States, 1993–2012. Jama. 313:2236–2243. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen MJ, Kress B, Han X, Moll K, Peng W,
Ji RR and Nedergaard M: Astrocytic CX43 hemichannels and gap
junctions play a crucial role in development of chronic neuropathic
pain following spinal cord injury. Glia. 60:1660–1670. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen M, Ni Y, Liu Y, Xia X, Cao J, Wang C,
Mao X, Zhang W, Chen C, Chen X and Wang Y: Spatiotemporal
expression of EAPP modulates neuronal apoptosis and reactive
astrogliosis after spinal cord injury. J Cell Biochem.
116:1381–1390. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhu P, Hata R, Nakata K, Cao F, Samukawa
K, Fujita H and Sakanaka M: Intravenous infusion of ginsenoside Rb1
ameliorates compressive spinal cord injury through upregulation of
Bcl-xL and VEGF. Int J Neurol Neurother. 2:12015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
De Biase A, Knoblach SM, Di Giovanni S,
Fan C, Molon A, Hoffman EP and Faden AI: Gene expression profiling
of experimental traumatic spinal cord injury as a function of
distance from impact site and injury severity. Physiol Genom.
22:368–381. 2005. View Article : Google Scholar
|
|
6
|
Byrnes KR, Garay J, Di Giovanni S, De
Biase A, Knoblach SM, Hoffman EP, Movsesyan V and Faden AI:
Expression of two temporally distinct microglia-related gene
clusters after spinal cord injury. Glia. 53:420–433. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sharp J, Frame J, Siegenthaler M, Nistor G
and Keirstead HS: Human embryonic stem cell-derived oligodendrocyte
progenitor cell transplants improve recovery after cervical spinal
cord injury. Stem Cells. 28:152–163. 2010.PubMed/NCBI
|
|
8
|
Shin HY, Kim H, Kwon MJ, Hwang DH, Lee K
and Kim BG: Molecular and cellular changes in the lumbar spinal
cord following thoracic injury: Regulation by treadmill locomotor
training. PLoS One. 9:e882152014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: Affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Smyth GK: Limma: Linear models for
microarray dataBioinformatics and computational biology solutions
using R and Bioconductor. Springer; pp. 397–420. 2005, View Article : Google Scholar
|
|
11
|
Obayashi T and Kinoshita K: Rank of
correlation coefficient as a comparable measure for biological
significance of gene coexpression. DNA Res. 16:249–260. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kohl M, Wiese S and Warscheid B:
Cytoscape: Software for visualization and analysis of biological
networksData Mining in Proteomics. Springer; pp. 291–303. 2011
|
|
13
|
Maere S, Heymans K and Kuiper M: BiNGO: A
Cytoscape plugin to assess overrepresentation of gene ontology
categories in biological networks. Bioinformatics. 21:3448–3449.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang HM, Liu T, Liu CJ, Song S, Zhang X,
Liu W, Jia H, Xue Y and Guo AY: Animal TFDB 2.0: A resource for
expression, prediction and functional study of animal transcription
factors. Nucleic Acids Res. 43(Database Issue): D76–D81. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang Y, Hwang CK, D'Souza UM, Lee SH, Junn
E and Mouradian MM: Three-amino acid extension loop homeodomain
proteins Meis2 and TGIF differentially regulate transcription. J
Biol Chem. 275:20734–20741. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang J, Wah IY, Pooh RK and Choy KW:
Molecular genetics in fetal neurology. Semin Fetal Neonatal Med.
17:341–346. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kerr TC, Cuykendall TN, Luettjohann LC and
Houston DW: Maternal Tgif1 regulates nodal gene expression in
Xenopus. Dev Dyn. 237:2862–2873. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sha L, Kitchen R, Porteous D, Blackwood D,
Muir W and Pickard B: SOX11 target genes: Implications for
neurogenesis and neuropsychiatric illness. Acta Neuropsychiatrica.
24:16–25. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ramsey SA, Klemm SL, Zak DE, Kennedy KA,
Thorsson V, Li B, Gilchrist M, Gold ES, Johnson CD, Litvak V, et
al: Uncovering a macrophage transcriptional program by integrating
evidence from motif scanning and expression dynamics. PLoS Comput
Biol. 21:e10000212008. View Article : Google Scholar
|
|
20
|
Didsbury J, Weber RF, Bokoch GM, Evans T
and Snyderman R: Rac, a novel ras-related family of proteins that
are botulinum toxin substrates. J Biol Chem. 264:16378–16382.
1989.PubMed/NCBI
|
|
21
|
Iversen PO, Hjeltnes N, Holm B, Flatebo T,
Strom-Gundersen I, Ronning W, Stanghelle J and Benestad HB:
Depressed immunity and impaired proliferation of hematopoietic
progenitor cells in patients with complete spinal cord injury.
Blood. 96:2081–2083. 2000.PubMed/NCBI
|
|
22
|
Numano F, Inoue A, Enomoto M, Shinomiya K,
Okawa A and Okabe S: Critical involvement of Rho GTPase activity in
the efficient transplantation of neural stem cells into the injured
spinal cord. Mol Brain. 2:372009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tian Y and Autieri MV: Cytokine expression
and AIF-1-mediated activation of Rac2 in vascular smooth muscle
cells: A role for Rac2 in VSMC activation. Am J Physiol Cell
Physiol. 292:C841–C849. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hall A and Lalli G: Rho and Ras GTPases in
axon growth, guidance, and branching. Cold Spring Harb Perspect
Biol. 2:a0018182010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lanier LL, Corliss BC, Wu J, Leong C and
Phillips JH: Immunoreceptor DAP12 bearing a tyrosine-based
activation motif is involved in activating NK cells. Nature.
391:703–707. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gingras MC, Lapillonne H and Margolin JF:
TREM-1, MDL-1 and DAP12 expression is associated with a mature
stage of myeloid development. Mol Immunol. 38:817–824. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jones KJ: Gonadal steroids as promoting
factors in axonal regeneration. Brain Res Bull. 30:491–498. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pakulski C: Neuroprotective properties of
sex hormones. Anestezjol Intens Ter. 43:113–118. 2011.(In Polish).
PubMed/NCBI
|
|
29
|
Gonzalez SL, Labombarda F, González
Deniselle MC, Guennoun R, Schumacher M and De Nicola AF:
Progesterone up-regulates neuronal brain-derived neurotrophic
factor expression in the injured spinal cord. Neuroscience.
125:605–614. 2004. View Article : Google Scholar : PubMed/NCBI
|