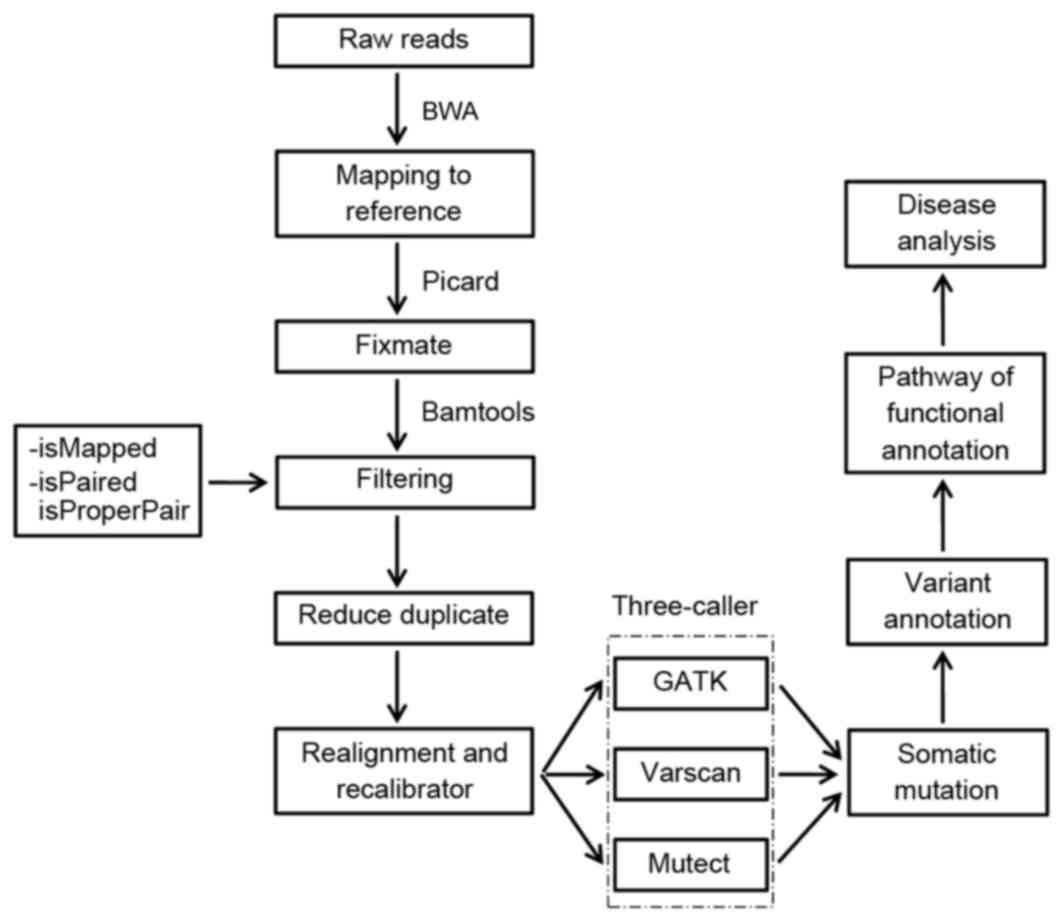
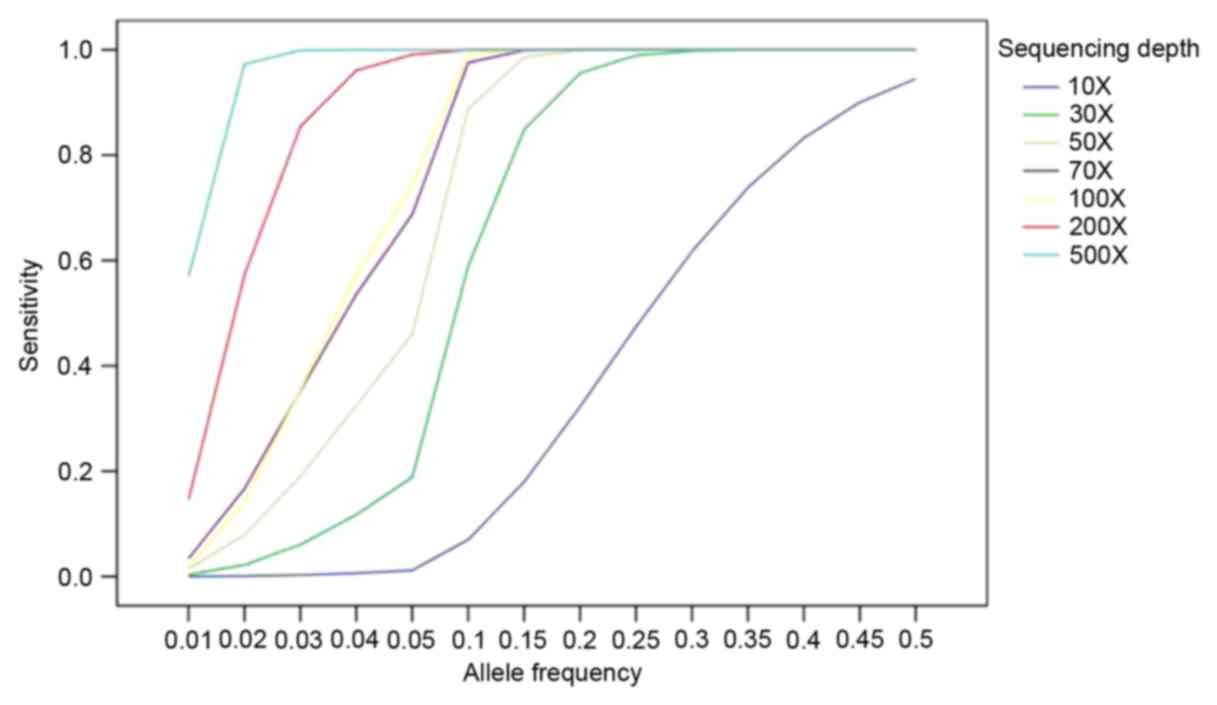
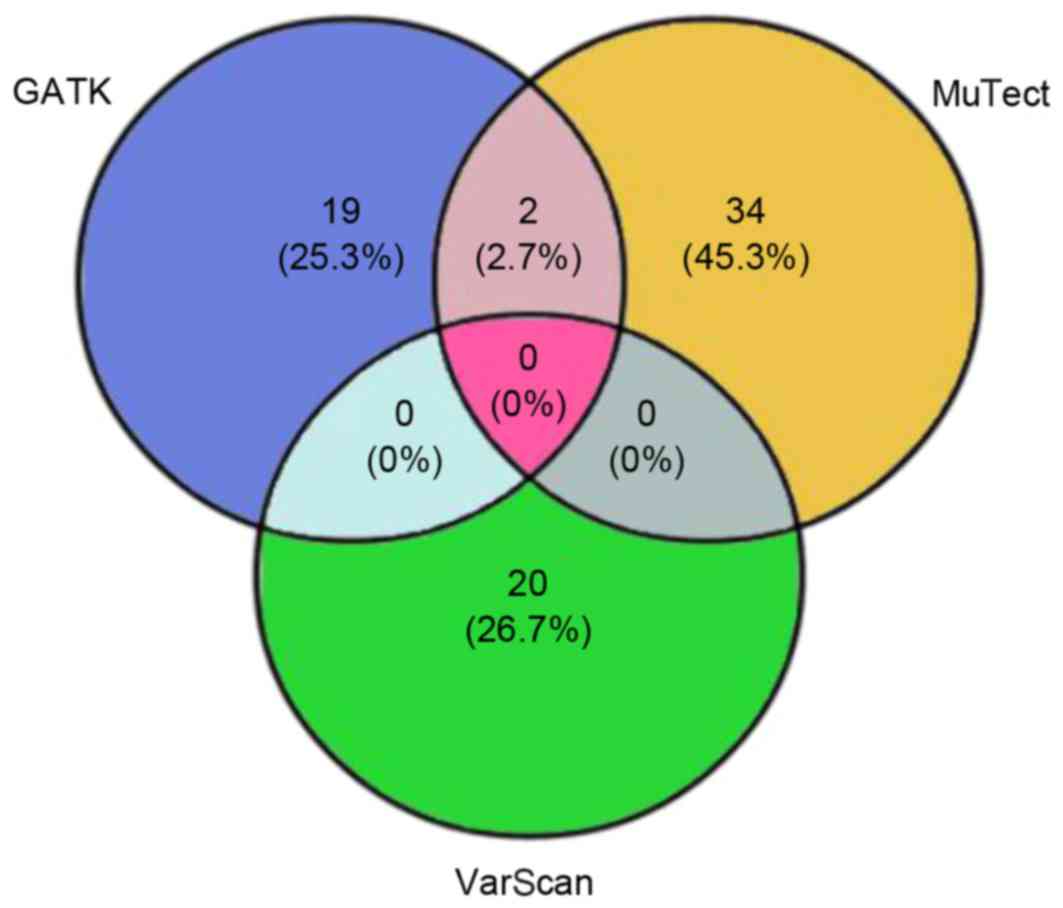
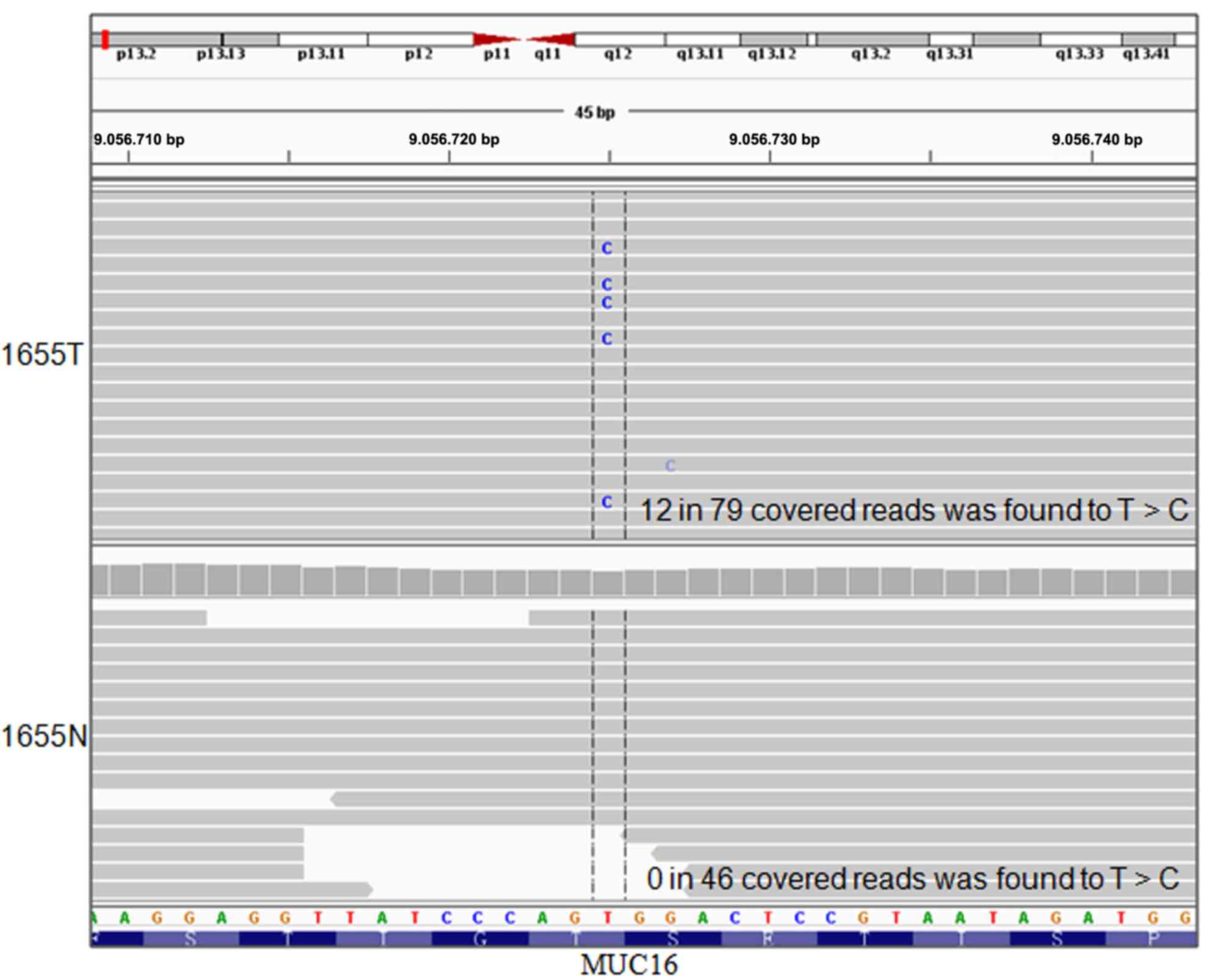
|
1
|
Lengauer C, Kinzler KW and Vogelstein B:
Genetic instabilities in human cancers. Nature. 396:643–649. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bass AJ, Lawrence MS, Brace LE, Ramos AH,
Drier Y, Cibulskis K, Sougnez C, Voet D, Saksena G, Sivachenko A,
et al: Genomic sequencing of colorectal adenocarcinomas identifies
a recurrent VTI1A-TCF7L2 fusion. Nat Genet. 43:964–968. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chapman MA, Lawrence MS, Keats JJ,
Cibulskis K, Sougnez C, Schinzel AC, Harview CL, Brunet JP, Ahmann
GJ, Adli M, et al: Initial genome sequencing and analysis of
multiple myeloma. Nature. 471:467–472. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jia D, Dong R, Jing Y, Xu D, Wang Q, Chen
L, Li Q, Huang Y, Zhang Y, Zhang Z, et al: Exome sequencing of
hepatoblastoma reveals novel mutations and cancer genes in the Wnt
pathway and ubiquitin ligase complex. Hepatology. 60:1686–1696.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Biesecker LG and Green RC: Diagnostic
clinical genome and exome sequencing. N Engl J Med.
371:11702014.PubMed/NCBI
|
|
6
|
Cancer Genome Atlas Research Network.
Hammerman PS, Lawrence MS, Voet D, Jing R, Cibulskis K, Sivachenko
A, Stojanov P, McKenna A, Lander ES, et al: Comprehensive genomic
characterization of squamous cell lung cancers. Nature.
489:519–525. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cancer Genome Atlas Network. Muzny DM,
Bainbridge MN, Chang K, Dinh HH, Drummond JA, Fowler G, Kovar CL,
Lewis LR, Morgan MB, et al: Comprehensive molecular
characterization of human colon and rectal cancer. Nature.
487:330–337. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cancer Genome Atlas Network. Koboldt DC,
Fulton RS, McLellan MD, Schmidt H, Kalicki-Veizer J, McMichael JF,
Fulton LL, Dooling DJ, Ding L, et al: Comprehensive molecular
portraits of human breast tumours. Nature. 490:61–70. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Litchfield K, Summersgill B, Yost S,
Sultana R, Labreche K, Dudakia D, Renwick A, Seal S, Al-Saadi R,
Broderick P, et al: Whole-exome sequencing reveals the mutational
spectrum of testicular germ cell tumours. Nat Commun. 6:59732015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang Z: Genomic landscape of liver
cancer. Nat Genet. 44:1075–1077. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kan Z, Zheng H, Liu X, Li S, Barber TD,
Gong Z, Gao H, Hao K, Willard MD, Xu J, et al: Whole-genome
sequencing identifies recurrent mutations in hepatocellular
carcinoma. Genome Res. 23:1422–1433. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang Q, Jia P, Li F, Chen H, Ji H, Hucks
D, Dahlman KB, Pao W and Zhao Z: Detecting somatic point mutations
in cancer genome sequencing data: A comparison of mutation callers.
Genome Med. 5:912013. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gerlinger M, Rowan AJ, Horswell S, Larkin
J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A,
Tarpey P, et al: Intratumor heterogeneity and branched evolution
revealed by multiregion sequencing. N Engl J Med. 366:883–892.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cibulskis K, Lawrence MS, Carter SL,
Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES
and Getz G: Sensitive detection of somatic point mutations in
impure and heterogeneous cancer samples. Nat Biotechnol.
31:213–219. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Koboldt DC, Zhang Q, Larson DE, Shen D,
McLellan MD, Lin L, Miller CA, Mardis ER, Ding L and Wilson RK:
Varscan 2: Somatic mutation and copy number alteration discovery in
cancer by exome sequencing. Genome Res. 22:568–576. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
McKenna A, Hanna M, Banks E, Sivachenko A,
Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly
M and DePristo MA: The genome analysis toolkit: A mapreduce
framework for analyzing next-generation DNA sequencing data. Genome
Res. 20:1297–1303. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mardis ER: Next-generation DNA sequencing
methods. Annu Rev Genomics Hum Genet. 9:387–402. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Metzker ML: Sequencing technologies-the
next generation. Nat Rev Genet. 11:31–46. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dohm JC, Lottaz C, Borodina T and
Himmelbauer H: Substantial biases inultra-short read data sets from
high-throughput DNA sequencing. Nucleic Acids Res. 36:e1052008.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nielsen R, Paul JS, Albrechtsen A and Song
YS: Genotype and SNP calling from next-generation sequencing data.
Nat Rev Genet. 12:443–451. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li H and Durbin R: Fast and accurate
long-read alignment with burrows-wheeler transform. Bioinformatics.
26:589–595. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li H, Handsaker B, Wysoker A, Fennell T,
Ruan J, Homer N, Marth G, Abecasis G and Durbin R: 1000 Genome
Project Data Processing Subgroup: The sequence alignment/map format
and SAMtools. Bioinformatics. 25:2078–2079. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ramos AH, Lichtenstein L, Gupta M,
Lawrence MS, Pugh TJ, Saksena G, Meyerson M and Getz G: Oncotator:
Cancer variant annotation tool. Hum Mutat. 36:E2423–E2429. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Robinson JT, Thorvaldsdóttir H, Winckler
W, Guttman M, Lander ES, Getz G and Mesirov JP: Integrative
genomics viewer. Nat Biotechnol. 29:24–26. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
da Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Totoki Y, Tatsuno K, Yamamoto S, Arai Y,
Hosoda F, Ishikawa S, Tsutsumi S, Sonoda K, Totsuka H, Shirakihara
T, et al: High-resolution characterization of a hepatocellular
carcinoma genome. Nat Genet. 43:464–469. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lawrence MS, Stojanov P, Polak P, Kryukov
GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH,
Roberts SA, et al: Mutational heterogeneity in cancer and the
search for new cancer-associated genes. Nature. 499:214–218. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kwon SM, Cho H, Choi JH, Jee BA, Jo Y and
Woo HG: Perspectives of integrative cancer genomics in next
generation sequencing era. Genomics Inform. 10:69–73. 2012.
View Article : Google Scholar : PubMed/NCBI
|