Introduction
Allergic rhinitis (AR) is a symptomatic disorder of
the nose induced after exposure to allergens via IgE-mediated
hypersensitivity reactions, which are characterized by the cardinal
symptoms of watery rhinorrhea, nasal obstruction, nasal itching and
sneezing (1). It represents one of
the greatest health problems in modern societies (2). A conservative estimate revealed that
AR occurs in over 500 million people around the world (3). The prevalence is about 10–30% in
adults and nearly 40% in children (4). This makes AR become one of the most
common allergic diseases in the world, with increasing prevalence
and often far-reaching consequences for quality of life.
Allergen-specific immunotherapy (SIT) is the most available
treatment for AR. It can alter the natural course of allergic
disease by preventing new sensitization/onset and providing
long-term remission after discontinuation of treatment (5). However, the conventional SIT,
subcutaneous injection, requires frequent hospital visits and is
painful, resulting in a low patient compliance. Furthermore, it may
cause some adverse events such as anaphylaxis (6). It is urgent to make a deeper
understanding of the pathogenesis of AR and find new therapeutic
methods.
Recent studies showed that genetic factors played
important roles in the development of AR. There was ample evidence
suggesting that AR was a complex multifactorial disorder including
both genetic and environmental factors (7,8).
Several genes and pathways had been reported to be associated with
AR. One of the important biological signals involved in the
pathogenesis of AR was histamine, which was released after relevant
antigenic stimulation of sensitized subjects, initiating the early
phase of allergic reaction (9).
Moreover, one research demonstrated that thymic stromal
lymphopoietin (TSLP) gene SNP rs1837253 was associated with
reduced odds for AR in boys with asthma (10). In addition, apolipoprotein A-IV was
also reported to be associated with the pathogenesis of AR, and
could be served as a candidate target molecule for the treatment of
seasonal AR (3). However, the
precise molecular mechanism of AR is still not well understood.
In this study, differentially expressed genes (DEGs)
in AR samples compared with normal samples were identified through
bioinformatics methods. The construction of the differential
co-expression network and the protein-protein interaction (PPI)
network might provide us a better understanding of the pathogenesis
of AR. Our study might provide references for the diagnosis and
therapy of AR.
Materials and methods
Microarray data
The gene expression profile of GSE43523 was
downloaded from National Center of Biotechnology Information (NCBI)
Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) database. The
dataset contained 7 nasal epithelial cells of seasonal AR samples
and 5 nasal epithelial cells of non-allergic normal samples. The
expression profile was detected based on GPL6883 Illumina
HumanRef-8 v3.0 expression beadchip platform.
Data processing and identification of
DEGs
The raw data were background corrected, quantile
normalized and log2 transformed using the preprocessCore package in
R (11). Affymetrix probe
IDs were converted to official gene symbol. If multiple probes
corresponded to one given gene, the mean expression value of those
probes was defined as the gene expression value. DEGs in rhinitis
samples compared with normal samples were identified via the limma
package of R (12).
Bonferroni and Hochberg method was used for the correction of
P-value. The threshold was P<0.05 and |log2 (fold change)|>
0.58. The threshold was |log2 (fold change)|>0.58 that mean gene
expression quantity in rhinitis samples change >1.5-fold
compared with normal samples. Besides, hierarchical clustering
analysis of rhinitis samples and normal samples based on the DEGs
were performed.
Pathway enrichment analysis
To further explore the biological functions and
involved pathways of the DEGs, Kyoto Encyclopedia of Genes and
Genomes (KEGG) pathway enrichment analysis was performed based on
the webGestal database with the threshold of P<0.05 (13).
Construction of the differential
co-expression network
DCGL was an R package for identifying differentially
co-expressed genes and links from gene expression microarray data
(14). It could examine the
expression correlation based on the exact co-expression changes of
gene pairs between two conditions (15). In this study, the co-expression
values for each pair of the DEGs in rhinitis samples and normal
samples were calculated via the DCGL package in R. Gene
pairs with different signs of co-expression values in two types of
samples were selected. Then, the differential co-expression pairs
were identified according to the criterion: Absolute value of the
difference of co-expression values in two samples >1.5. The
differential co-expression network of the DEGs was constructed
based on these pairs.
Construction of the PPI network
Search Tool for the Retrieval of Interacting Genes
(STRING) (http://string-db.org/) (16) was an online database for predicting
functional interactions between proteins (17). In this study, PPI pairs of the DEGs
were selected based on the STRING database with the threshold of
combined score >0.4. The PPI network of the DEGs was constructed
based on these pairs.
Results
The DEGs
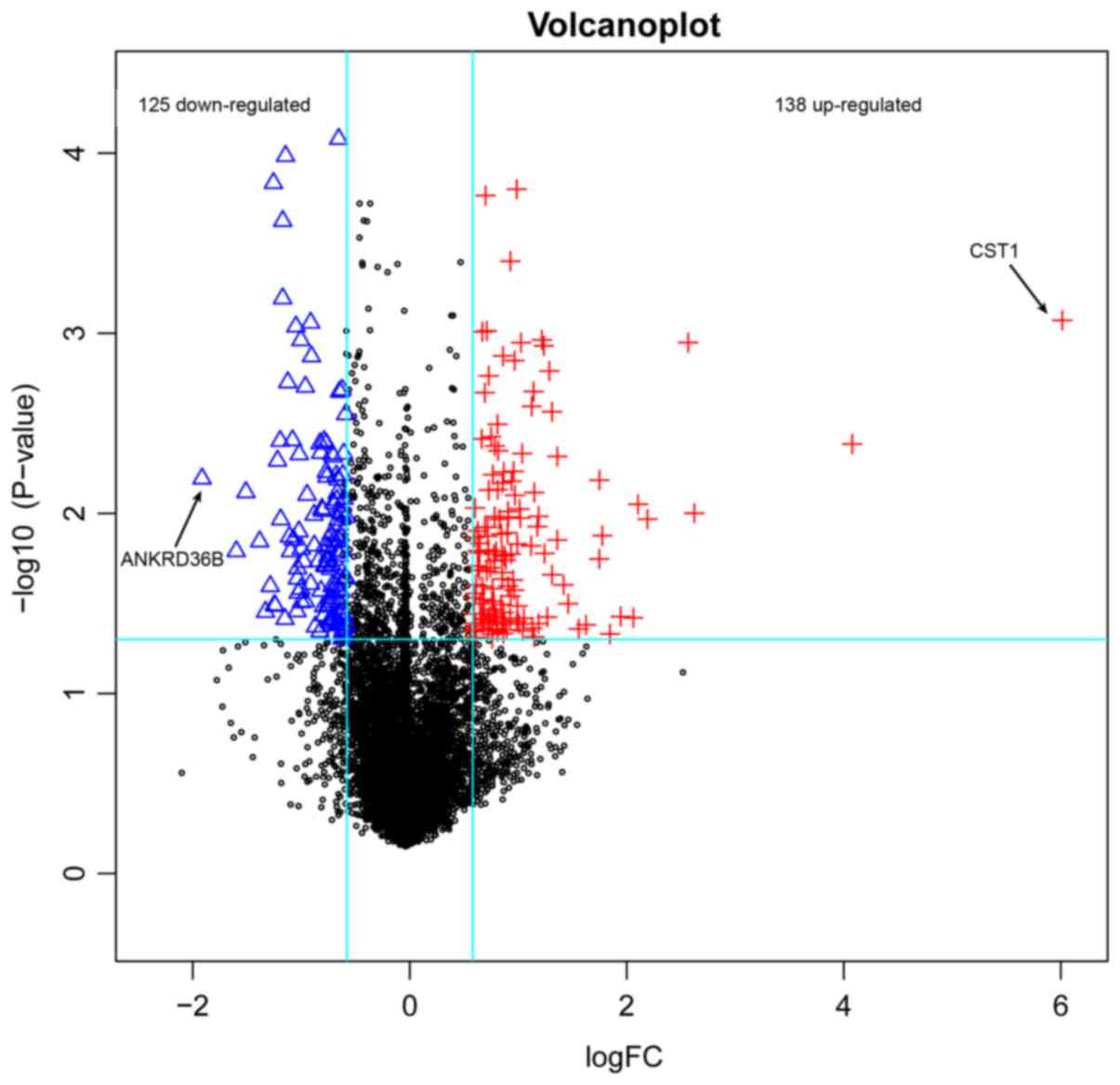
A total of 263 DEGs were identified in rhinitis
samples compared with normal samples, including 125 downregulated
ones and 138 upregulated ones (Fig.
1). The top 20 DEGs according to P-value were listed in
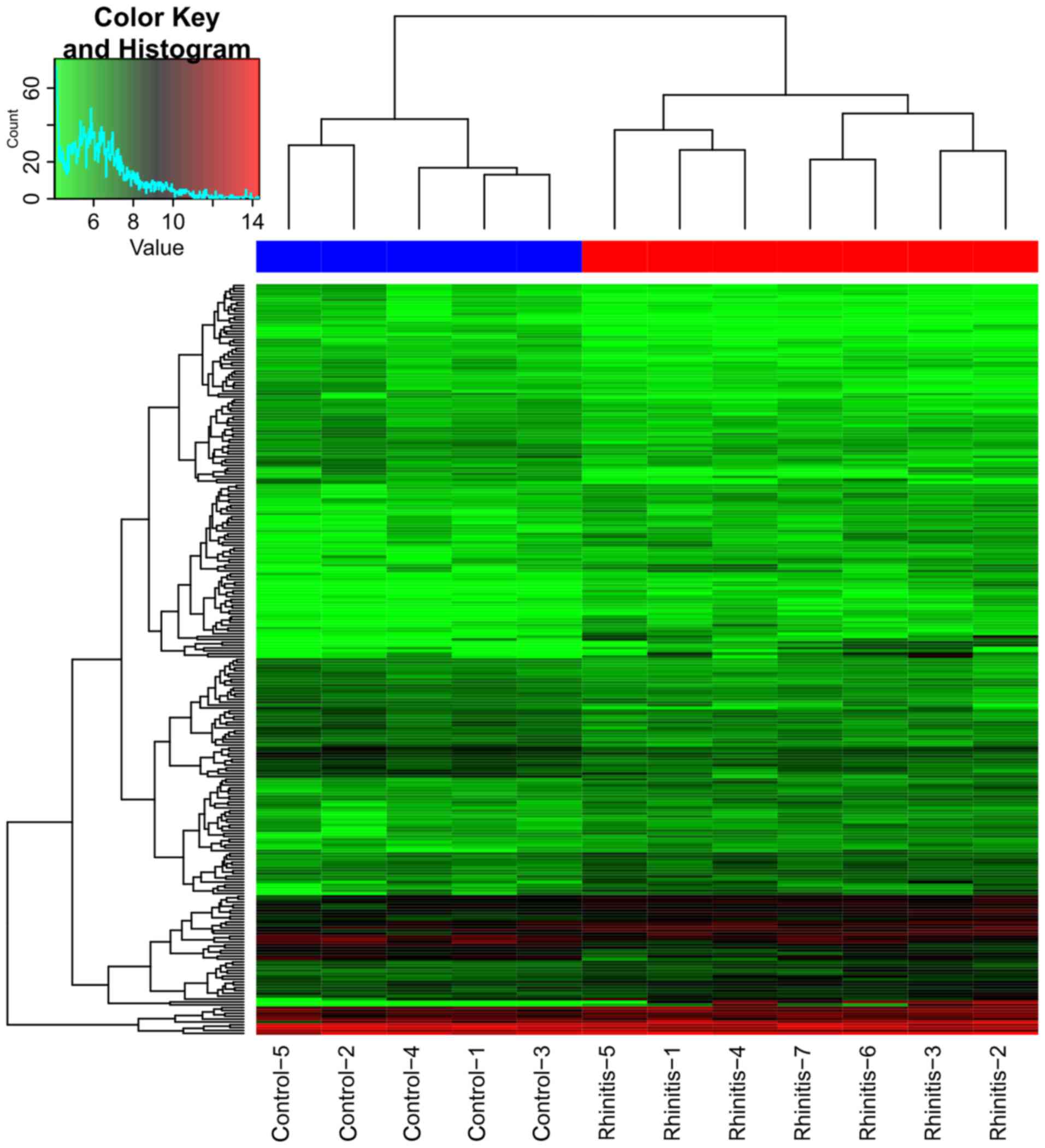
Table I. Clustering analysis
revealed a clearly distinct expression of all DEGs between rhinitis
samples and normal samples (Fig.
2). From the heatmap (Fig. 2),
we found that the gene expression of 5 nasal epithelial cells of
non-allergic normal samples was distinguished from the 7 nasal
epithelial cells of seasonal AR samples.
 | Table I.The top 20 DEGs in AR samples
compared with normal samples. |
Table I.
The top 20 DEGs in AR samples
compared with normal samples.
| Gene name | P-value | LogFC |
|---|
| ST3GAL5 |
0.83×10−4 | −0.655 |
| NR1D2 |
1.04×10−4 | −1.144 |
| AKR1B1 |
1.46×10−4 | −1.255 |
| HIST1H2BD |
1.58×10−4 |
0.985 |
| TMEM125 |
1.72×10−4 |
0.698 |
| MAP3K2 |
2.38×10−4 | −1.170 |
| AGR2 |
3.98×10−4 |
0.927 |
| RNF217 |
6.40×10−4 | −1.171 |
| CST1 |
8.47×10−4 |
6.013 |
| LIN54 |
8.74×10−4 | −0.913 |
| ZNF750 |
9.18×10−4 | −1.049 |
| DHCR24 |
9.70×10−4 |
0.711 |
| SLC39A11 |
9.82×10−4 |
0.669 |
| ATP2C2 |
10.88×10−4 |
1.218 |
| CAMK2G |
10.93×10−4 | −1.003 |
| CLC |
11.26×10−4 |
2.565 |
| SDPR |
11.28×10−4 |
1.025 |
| IL20RB |
11.74×10−4 |
1.237 |
| FAM46B |
13.36×10−4 |
0.860 |
| ANKRD13C |
13.44×10−4 | −0.906 |
The enriched pathways
Seven enriched KEGG pathways of the DEGs were
obtained in the webGestal database (Table II). The top 4 pathways were
fructose and mannose metabolism, riboflavin metabolism,
renin-angiotensin system (RAS), amino sugar and nucleotide sugar
metabolism respectively.
 | Table II.The enriched KEGG pathways of the
DEGs. |
Table II.
The enriched KEGG pathways of the
DEGs.
| Category | Pathway name | P-value |
|---|
| KEGG pathway | Fructose and
mannose metabolism | 0.001 |
| KEGG pathway | Riboflavin
metabolism | 0.008 |
| KEGG pathway | Renin-angiotensin
system | 0.020 |
| KEGG pathway | Amino sugar and
nucleotide sugar metabolism | 0.027 |
| KEGG pathway | Steroid
biosynthesis | 0.024 |
| KEGG pathway | Ribosome | 0.027 |
| KEGG pathway | Metabolic
pathways | 0.032 |
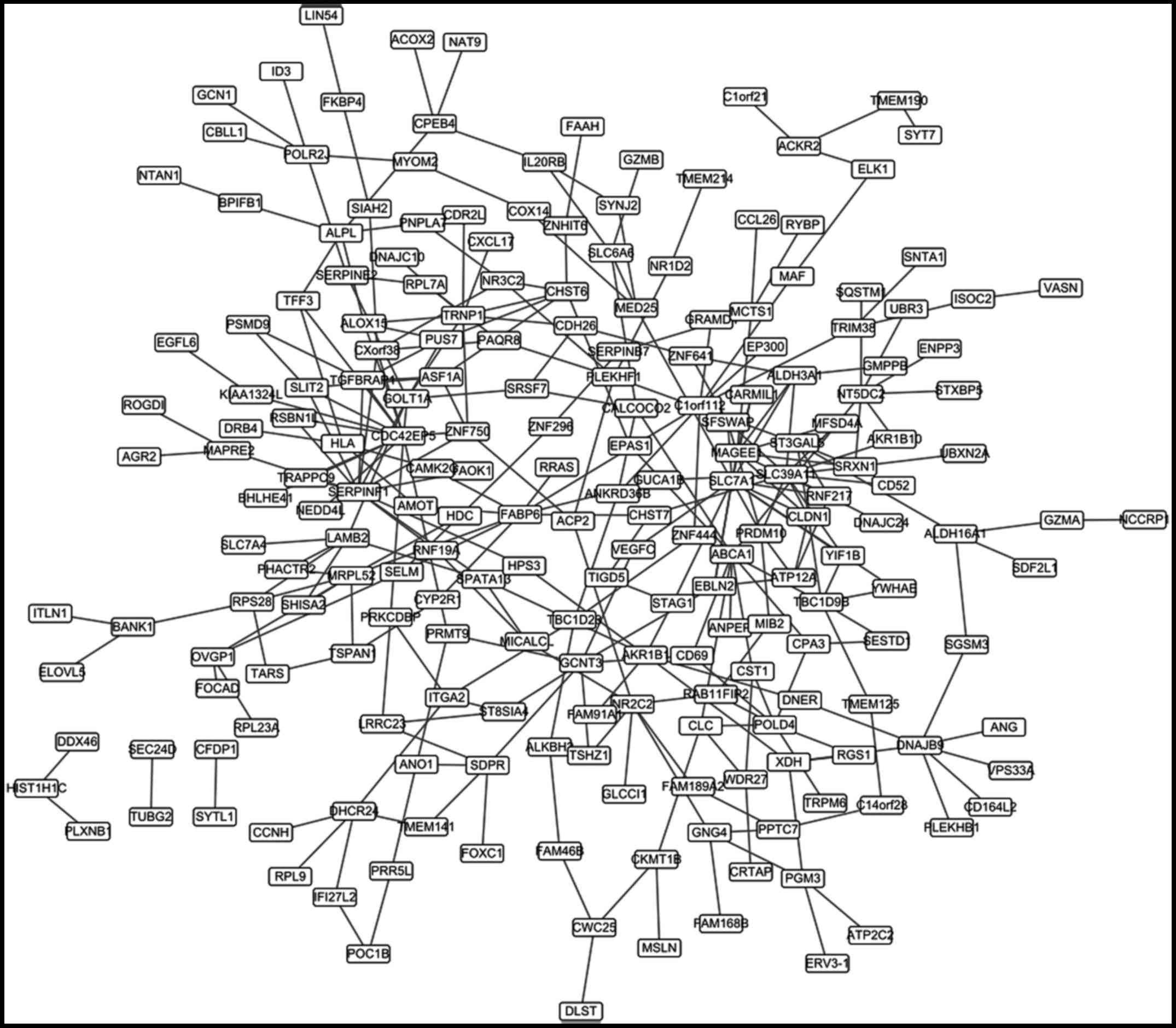
The differential co-expression
network
A total of 308 differential co-expression gene pairs
were obtained. The co-expression network based on these pairs was
constructed (Fig. 3), which
contained 212 nodes. The top 20 nodes according to the degree were
listed in Table III.
 | Table III.The top 20 nodes in the PPI network
with high degree. |
Table III.
The top 20 nodes in the PPI network
with high degree.
| Gene | Degree |
|---|
| PRDM10 | 22 |
| EP300 | 12 |
| ITGA2 | 11 |
| RRAS | 7 |
| VASN | 6 |
| ATP12A | 5 |
| SERPINE2 | 5 |
| SRSF7 | 5 |
| ABCA1 | 4 |
| ALDH3A1 | 4 |
| ASF1A | 4 |
| EDF1 | 4 |
| FKBP4 | 4 |
| GZMB | 4 |
| HPS3 | 4 |
| MAF | 4 |
| MUC2 | 4 |
| SQSTM1 | 4 |
| TYRO3 | 4 |
| ALDH16A1 | 3 |
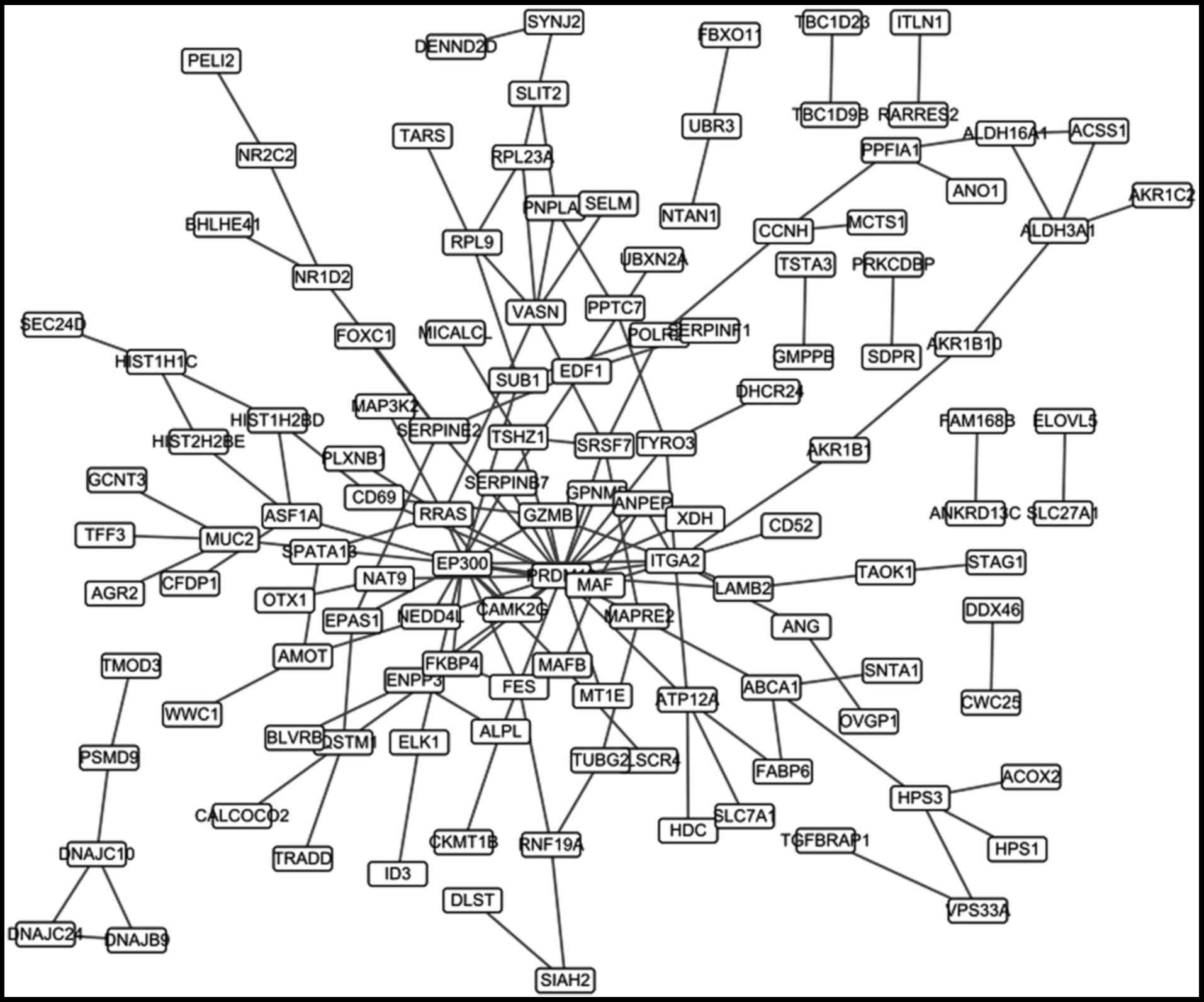
The PPI network
148 PPI pairs were identified by the STRING
database. A PPI network was constructed based on these pairs and
contained 125 genes (Fig. 4). The
top 20 nodes of the PPI network were listed in Table IV.
 | Table IV.The top 20 nodes in the differential
co-expression network with high degree. |
Table IV.
The top 20 nodes in the differential
co-expression network with high degree.
| Gene | Degree |
|---|
| CDC42EP5 | 14 |
| SERPINF1 | 13 |
| SLC39A11 | 13 |
| SLC7A1 | 12 |
| MAGEE1 | 10 |
| FABP6 | 9 |
| ABCA1 | 8 |
| TRNP1 | 8 |
| C1orf112 | 7 |
| DNAJB9 | 7 |
| GCNT3 | 7 |
| GOLT1A | 7 |
| HLA | 7 |
| MRPL52 | 7 |
| NR2C2 | 7 |
| NT5DC2 | 7 |
| POLD4 | 7 |
| AKR1B1 | 6 |
| CHST6 | 6 |
| CLDN1 | 6 |
Discussion
AR significantly affects the quality of the
patient's daily life. Despite the development of various of
pharmacological methods, avoidance of the allergen is usually not
possible and symptom relief is often limited (18). The precise pathogenesis of AR is
still not well understood. In this study, DEGs in AR samples
compared with normal samples were identified via bioinformatics
methods. Then the DEGs were further analyzed by the construction of
differential co-expression network and PPI network to a better
understanding of the molecular mechanism of AR.
The gene expression profile of GSE43523 contained 7
nasal epithelial cells of seasonal AR samples and 5 nasal
epithelial cells of non-allergic normal samples. Small sample size
was a limitation of the study but the results of our analysis was
reliable to a certain extent. The small samples of studies have
been recognized by many researchers. For example, Zhu et al
identified endometrial cancer prognosis markers which the tissue
samples for the microarray study consisted of 7 control samples,
3-G1 samples, 8-G2 samples and 2-G3 samples (19). A total of 263 DEGs were identified
in rhinitis samples compared with normal samples which were further
analyzed by the construction of differential co-expression network
and PPI network to a better understanding of the molecular
mechanism of AR. Individual analysis of the 263 DEGs was nοt
performed as this was not considered necessary.
The enriched KEGG pathways of the DEGs were fructose
and mannose metabolism, riboflavin metabolism, RAS, amino sugar and
nucleotide sugar metabolism, steroid biosynthesis, ribosome and
metabolic pathways. Many of these pathways were associated with the
pathogenesis of AR. Angiotensin was a peptide hormone that caused
vasoconstriction and a subsequent increase in blood pressure. It
was part of the RAS, which was a major target for drugs that
lowered blood pressure (20).
Angiotensin converting enzyme (ACE), which contained 26 exons and
25 introns, was reported to be an important AR susceptibility gene
(21). ACE was essential in
converting angiotensin I into angiotensin II, which was an mainly
effector molecule in the RAS and acted as pro-inflammatory
modulator in the augmentation of immune responses (22,23).
The insertion or deletion polymorphism of ACE was a risk factor for
AR and verified by large number and representative population
(24). A steroid was an organic
compound with four rings arranged in a specific configuration
(25). Steroid were generally
considered the most effective medications for the management of
inflammatory diseases including asthma and AR (26). The corticosteroids could be given
locally into the nose and bronchi without risk of systemic adverse
effects. The introduction of potent corticosteroids in the
treatment of AR had been a major therapeutic advance, and had
emphasized the importance of pharmacological and morphological
aspects of AR (27). Topical
steroid treatment of AR could decrease nasal fluid TH2 cytokines,
eosinophils, eosinophil cationic protein and IgE (28). Metabolic pathways also played
critical roles in the development of AR. Kinin metabolism in human
nasal secretions during experimentally could induce AR (29). In addition, one study demonstrated
that serum tryptophan metabolism could be served as a biomarker in
patients with AR (30). The
metabolism of vitamin D was also reported to be different in AR
patients (31).
The top 5 genes in the differential co-expression
network according to the degree were CDC42EP5,
SERPINF1, SLC39A11, SLC7A1 and MAGEE1,
respectively. While the top 5 genes in the PPI network were
PRDM10, EP300, ITGA2, RRAS and VASN,
respectively. Shi and his team found that FOS, JUN, and CEBPD may
play crucial roles during the process of seasonal allergic rhinitis
(SAR) by the microarray data GSE50101 (32). The different results with our study
may be caused by distinction between AR and SAR. AR was an
inflammatory diseases, and many of the above genes were reported to
be associated with the pathogenesis of AR or inflammatory diseases
(33). SLC39A11 encoded a
type of human zinc transporter, which was one of the critical
regulators that maintained intracellular zinc concentrations, and
played a role in regulating cell survival during inflammation
(34,35). Zinc was an essential micronutrient
and cytoprotectant involved in the host response to inflammatory
stress. Zip protein was demonstrated to be an essential zinc
importer at the onset of inflammation for facilitating
cytoprotection (36). Zinc was
also an antioxidant and had anti-inflammatory actions. Zinc could
induce A20 which inhibited nuclear transcription factor κB (NF-κB)
activation resulting in decreased generation of inflammatory
cytokines (37). Zinc deficiency
was confirmed in patients with AR (38). CDC42EP5 was a member of
CDC42 effector protein family. It could bind to CDC42 and regulate
its function negatively (39).
CDC42 was a Rho-family GTPase. It had been implicated in several
signal transduction pathways, including NF-κB activation,
activation of the c-Jun N-terminal MAP kinase and stimulation of
the NF-κB (40,41). CDC42 could be activated by the
inflammatory cytokines TNFα and IL-1, which was associated with the
development of inflammatory diseases (42). In addition, CDC42 signaling was
identified as a mediator of chronic inflammation associated with
endothelial senescence. Inhibition of CDC42 or NF-κB signaling
would attenuate the sustained upregulation of pro-inflammatory
genes in human endothelial cells. CDC42 pathway was critically
involved in senescence-associated inflammation and could be served
as a therapeutic target for chronic inflammation in patients with
inflammatory diseases (43).
However, the directly relationship between CDC42 and AR had not
been reported. PRDM10 was a member of the PRDM family, which
has emerged as prime regulators of many types of tissue
differentiation and disease pathogenesis (44–46).
Studies demonstrated that PRDM family played critical roles in the
development of inflammatory diseases. For example, PRDM1
genetic variants could be used to prognose, diagnose, and treat
inflammatory disease (47).
PRDM11 mutation was associated with inflammatory response in
mice (48). However, the direct
relationship between PRDM10 and AR were still not well
understood.
Bioinformatics methods could help us identify
significant genes and pathways related to the pathogenesis of AR.
Steroid biosynthesis pathway and metabolic pathways might play an
important role in the development of AR. Genes such as CDC42EP5,
SLC39A11 and PRDM10 might be also associated with the
pathogenesis of AR. However, further studies were still needed to
confirm our results and explore the specific regulatory mechanism
of these genes and pathways.
Acknowledgements
We would like to thank all the members of our group
for their enthusiastic participation in this study.
References
|
1
|
Min YG: The pathophysiology, diagnosis and
treatment of allergic rhinitis. Allergy Asthma Immunol Res.
2:65–76. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Meltzer EO, Nathan R, Derebery J, Stang
PE, Campbell UB, Yeh WS, Corrao M and Stanford R: Sleep, quality of
life, and productivity impact of nasal symptoms in the United
States: Findings from the Burden of Rhinitis in America survey.
Allergy Asthma Proc. 30:pp. 244–254. 2009; View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Makino Y, Noguchi E, Takahashi N,
Matsumoto Y, Kubo S, Yamada T, Imoto Y, Ito Y, Osawa Y, Shibasaki
M, et al: Apolipoprotein A-IV is a candidate target molecule for
the treatment of seasonal allergic rhinitis. J Allergy Clin
Immunol. 126:1163–1169.e5. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nathan RA: The burden of allergic
rhinitis. Allergy Asthma Proc. 28:pp. 3–9. 2007; View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Passalacqua G and Durham SR; Global
Allergy and Asthma European Network, : Allergic rhinitis and its
impact on asthma update: Allergen immunotherapy. J Allergy Clin
Immunol. 119:881–891. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Calderon MA, Alves B, Jacobson M, Hurwitz
B, Sheikh A and Durham S: Allergen injection immunotherapy for
seasonal allergic rhinitis. Cochrane Database Syst Rev.
24:CD0019362007.
|
|
7
|
Andiappan AK, Wang de Y, Anantharaman R,
Parate PN, Suri BK, Low HQ, Li Y, Zhao W, Castagnoli P, Liu J and
Chew FT: Genome-wide association study for atopy and allergic
rhinitis in a singapore chinese population. PLoS One. 6:e197192011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dávila I, Mullol J, Ferrer M, Bartra J,
del Cuvillo A, Montoro J, Jáuregui I, Sastre J and Valero A:
Genetic aspects of allergic rhinitis. J Investig Allergol Clin
Immunol. 19 Suppl 1:S25–S31. 2009.
|
|
9
|
Hanuskova E and Plevkova J: The role of
histamine H4 receptors as a potential targets in allergic rhinitis
and asthma. Open J Mol Int Physiol. 3:6–14. 2013. View Article : Google Scholar
|
|
10
|
Bunyavanich S, Melen E, Wilk JB, Granada
M, Soto-Quiros ME, Avila L, Lasky-Su J, Hunninghake GM, Wickman M,
Pershagen G, et al: Thymic stromal lymphopoietin (TSLP) is
associated with allergic rhinitis in children with asthma. Clin Mol
Allergy. 9:12011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Thompson JA, Tan J and Greene CS:
Cross-platform normalization of microarray and RNA-seq data for
machine learning applications. Peerj. 4:e16212016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Diboun I, Wernisch L, Orengo CA and
Koltzenburg M: Microarray analysis after RNA amplification can
detect pronounced differences in gene expression using limma. BMC
Genomics. 7:2522006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhao Z, Guo AY, van den Oord EJ, Aliev F,
Jia P, Edenberg HJ, Riley BP, Dick DM, Bettinger JC, Davies AG, et
al: Multi-species data integration and gene ranking enrich
significant results in an alcoholism genome-wide association study.
BMC Genomics. 13 Suppl 8:S162012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang J, Yu H, Liu BH, Zhao Z, Liu L, Ma
LX, Li YX and Li YY: DCGL v2.0: An R package for unveiling
differential regulation from differential co-expression. PLoS One.
8:e797292013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yu H, Liu BH, Ye ZQ, Li C, Li YX and Li
YY: Link-based quantitative methods to identify differentially
coexpressed genes and gene pairs. BMC Bioinformatics. 12:3152011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Szklarczyk D, Franceschini A, Kuhn M,
Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork
P, et al: The STRING database in 2011: Functional interaction
networks of proteins, globally integrated and scored. Nucleic Acids
Res. 39(Database Issue): D561–568. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C
and Jensen LJ: STRING v9.1: Protein-protein interaction networks,
with increased coverage and integration. Nucleic Acids Res.
41(Database Issue): D808–D815. 2013.PubMed/NCBI
|
|
18
|
Brehmer D: Endonasal phototherapy with
Rhinolight for the treatment of allergic rhinitis. Expert Rev Med
Devices. 7:21–26. 2014. View Article : Google Scholar
|
|
19
|
Zhu XL, Ai ZH, Wang J, Xu YL and Teng YC:
Weighted gene co-expression network analysis in identification of
endometrial cancer prognosis markers. Asian Pac J Cancer Prev.
13:4607–4611. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Blood Pressure Lowering Treatment
Trialists' Collaboration, ; Turnbull F, Neal B, Pfeffer M, Kostis
J, Algert C, Woodward M, Chalmers J, Zanchetti A and MacMahon S:
Blood pressure-dependent and independent effects of agents that
inhibit the renin-angiotensin system. J Hypertens. 25:951–958.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bernstein KE, Ong FS, Blackwell WL, Shah
KH, Giani JF, Gonzalez-Villalobos RA, Shen XZ, Fuchs S and Touyz
RM: A modern understanding of the traditional and nontraditional
biological functions of angiotensin-converting enzyme. Pharmacol
Rev. 65:1–46. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Okwan-Duodu D, Datta V, Shen XZ, Goodridge
HS, Bernstein EA, Fuchs S, Liu GY and Bernstein KE:
Angiotensin-converting enzyme overexpression in mouse
myelomonocytic cells augments resistance to Listeria and
methicillin-resistant Staphylococcus aureus. J Biol Chem.
285:39051–39060. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Song GG, Kim JH and Lee YH: Associations
between the insertion/deletion polymorphism of the
angiotensin-converting enzyme and susceptibility to aortic
aneurysms: A meta-analysis. J Renin Angiotensin Aldosterone Syst.
16:211–218. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lin H, Lin D and Zheng CQ:
Angiotensin-converting enzyme insertion/deletion polymorphism
associated with allergic rhinitis susceptibility: Evidence from
1410 subjects. J Renin Angiotensin Aldosterone Syst. 15:593–600.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Evans RM: The steroid and thyroid hormone
receptor superfamily. Science. 240:889–895. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
LaForce C: Use of nasal steroids in
managing allergic rhinitis. J Allergy Clin Immunol. 103:S388–S394.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mygind N: Topical steroid treatment for
allergic rhinitis and allied conditions. Clin Otolaryngol Allied
Sci. 7:343–352. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Benson M, Strannegård IL, Strannegård O
and Wennergren G: Topical steroid treatment of allergic rhinitis
decreases nasal fluid TH2 cytokines, eosinophils, eosinophil
cationic protein and IgE but has no significant effect on
IFN-gamma, IL-1beta, TNF-alpha, or neutrophils. J Allergy Clin
Immunol. 106:307–312. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Proud D, Baumgarten CR, Naclerio RM and
Ward PE: Kinin metabolism in human nasal secretions during
experimentally induced allergic rhinitis. J Immunol. 138:428–434.
1987.PubMed/NCBI
|
|
30
|
Ciprandi G, De Amici M, Tosca M and Fuchs
D: Tryptophan metabolism in allergic rhinitis: The effect of pollen
allergen exposure. Hum Immunol. 71:911–915. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wjst M and Hyppönen E: Vitamin D serum
levels and allergic rhinitis. Allergy. 62:1085–1086. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shi J, Zhang Y, Qi S, Liu G, Dong X, Huang
N, Li W, Chen H and Zhu B: Identification of potential crucial gene
network related to seasonal allergic rhinitis using microarray
data. Eur Arch Otorhinolaryngol. 274:231–237. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hauswald B and Yarin YM: Acupuncture in
allergic rhinitis: A mini-review. Allergo J Int. 23:115–119. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yu Y, Wu A, Zhang Z, Yan G, Zhang F, Zhang
L, Shen X, Hu R, Zhang Y, Zhang K and Wang F: Characterization of
the GufA subfamily member SLC39A11/Zip11 as a zinc transporter. J
Nutr Biochem. 24:1697–1708. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bao S and Knoell DL: Zinc modulates airway
epithelium susceptibility to death receptor-mediated apoptosis. Am
J Physiol Lung Cell Mol Physiol. 290:L433–L441. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Besecker B, Bao S, Bohacova B, Papp A,
Sadee W and Knoell DL: The human zinc transporter SLC39A8 (Zip8) is
critical in zinc-mediated cytoprotection in lung epithelia. Am J
Physiol Lung Cell Mol Physiol. 294:L1127–L1136. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Prasad AS: Clinical, immunological,
anti-inflammatory and antioxidant roles of zinc. Exp Gerontol.
43:370–377. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
AlKhyat THAT, Alturaihy SH and Ali ZA: The
relationship between tumour necrosis factor -alpha and zinc/copper
ratio in iraqi patients with allergic rhinitis. Glob J Sci Front
Res. 13:2013.
|
|
39
|
Hirsch DS, Pirone DM and Burbelo PD: A new
family of Cdc42 effector proteins, CEPs, function in fibroblast and
epithelial cell shape changes. J Biol Chem. 276:875–883. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hall A: Rho GTPases and the actin
cytoskeleton. Science. 279:509–514. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wesselborg S, Bauer MK, Vogt M, Schmitz ML
and Schulze-Osthoff K: Activation of transcription factor NF-kappaB
and p38 mitogen-activated protein kinase is mediated by distinct
and separate stress effector pathways. J Biol Chem.
272:12422–12429. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Puls A, Eliopoulos AG, Nobes CD, Bridges
T, Young LS and Hall A: Activation of the small GTPase Cdc42 by the
inflammatory cytokines TNF(alpha) and IL-1, and by the Epstein-Barr
virus transforming protein LMP1. J Cell Sci. 112:2983–2992.
1999.PubMed/NCBI
|
|
43
|
Ito TK, Yokoyama M, Yoshida Y, Nojima A,
Kassai H, Oishi K, Okada S, Kinoshita D, Kobayashi Y, Fruttiger M,
et al: A crucial role for CDC42 in senescence-associated
inflammation and atherosclerosis. PLoS One. 9:e1021862014.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kim KC and Huang S: Histone
methyltransferases in tumor suppression. Cancer Biol Ther.
2:491–499. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Huang S: Histone methyltransferases, diet
nutrients and tumour suppressors. Nat Rev Cancer. 2:469–476. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Park JA, Kim TH, Lee B, Kwon E and Kim KC:
Expression of PRDM10 in arthritic synovial derived tissues. Genes
Genomics. 35:685–691. 2013. View Article : Google Scholar
|
|
47
|
Center CM: Methods of using prdm1 genetic
variants to prognose, diagnose and treat inflammatory bowel
disease. 2011.
|
|
48
|
Horsch M, Aguilar-Pimentel JA, Bönisch C,
Côme C, Kolster-Fog C, Jensen KT, Lund AH, Lee I, Grossman LI,
Sinkler C, et al: Cox4i2, Ifit2, and Prdm11 Mutant Mice: Effective
selection of genes predisposing to an altered airway inflammatory
response from a large compendium of mutant mouse lines. PLoS One.
10:e01345032015. View Article : Google Scholar : PubMed/NCBI
|