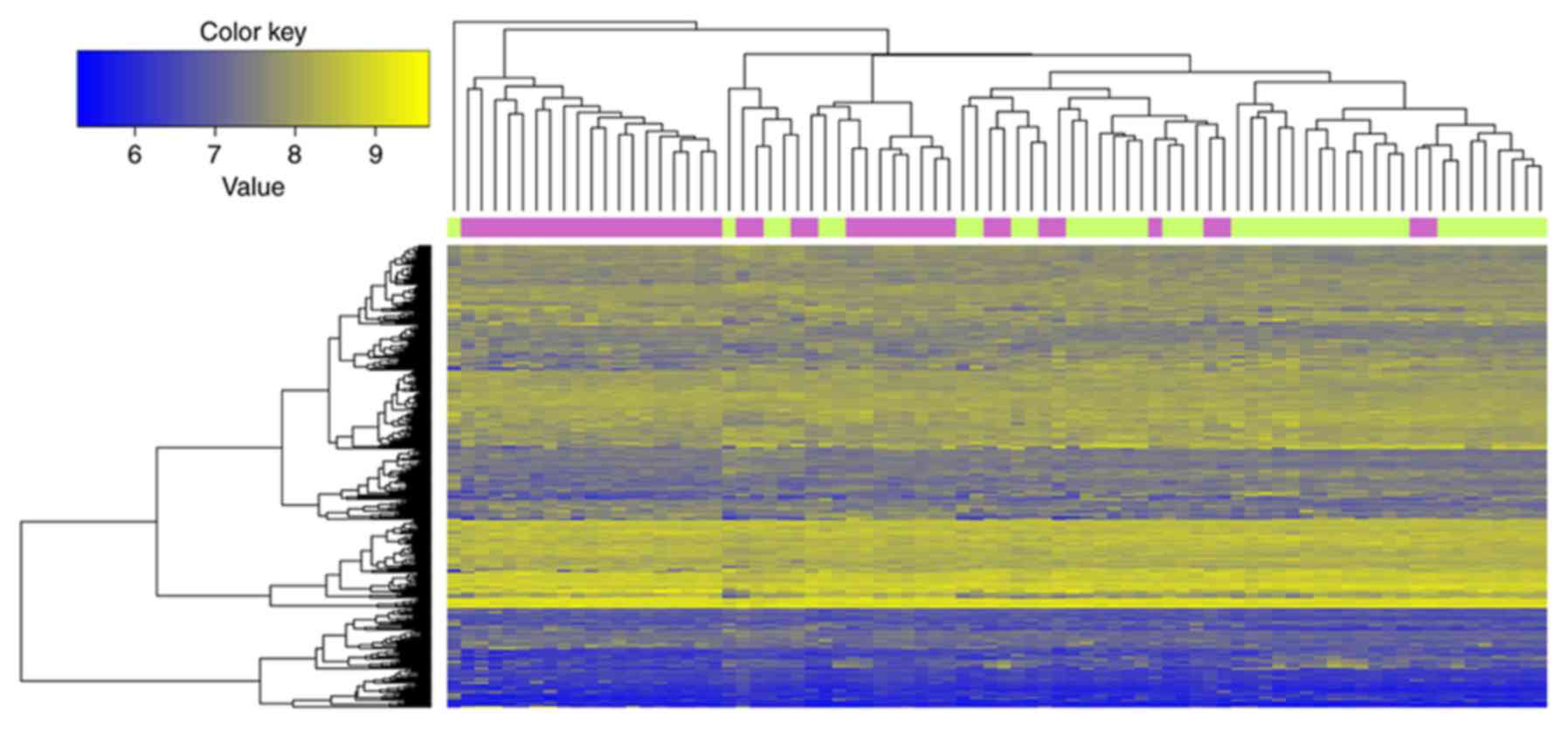
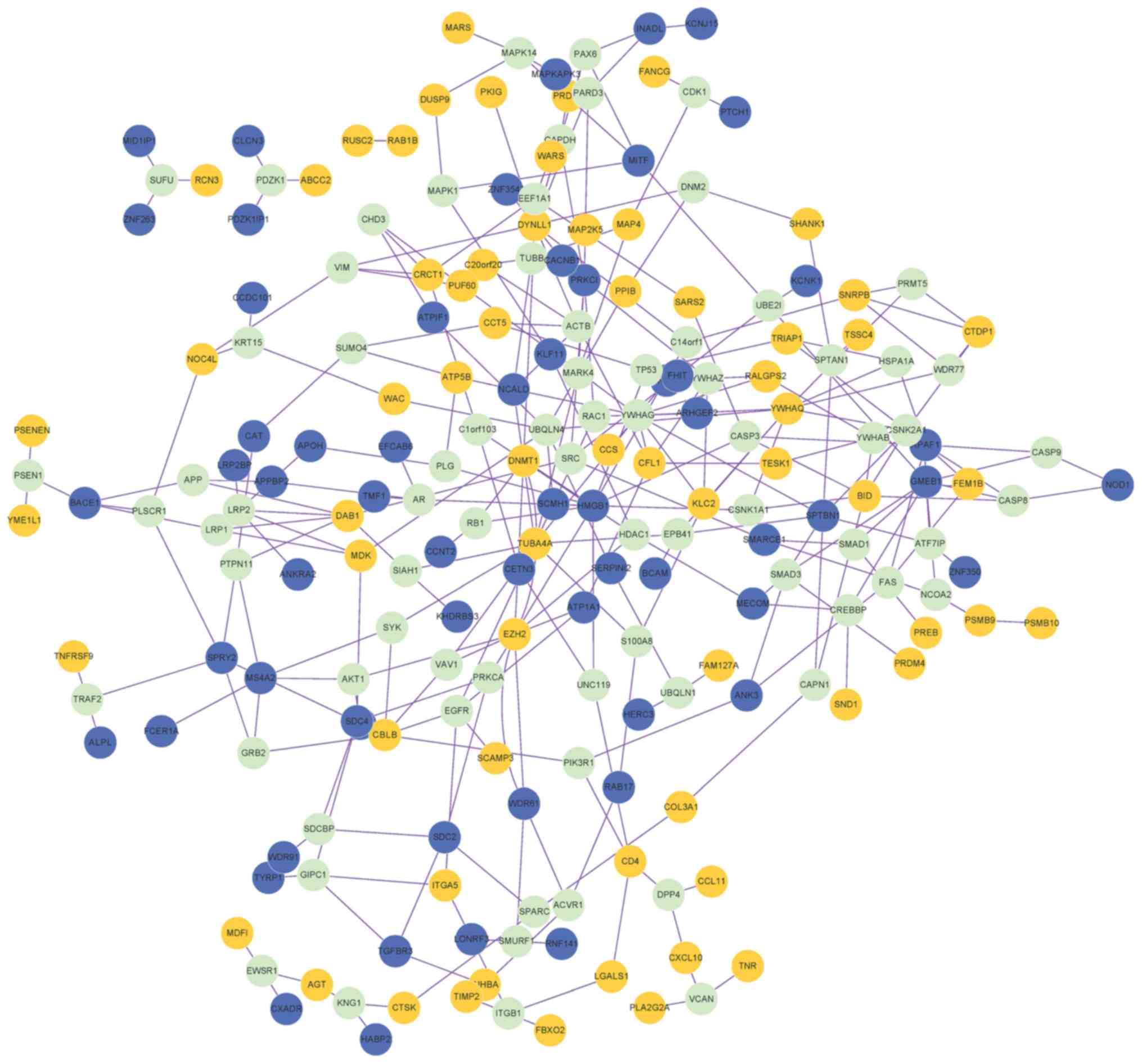
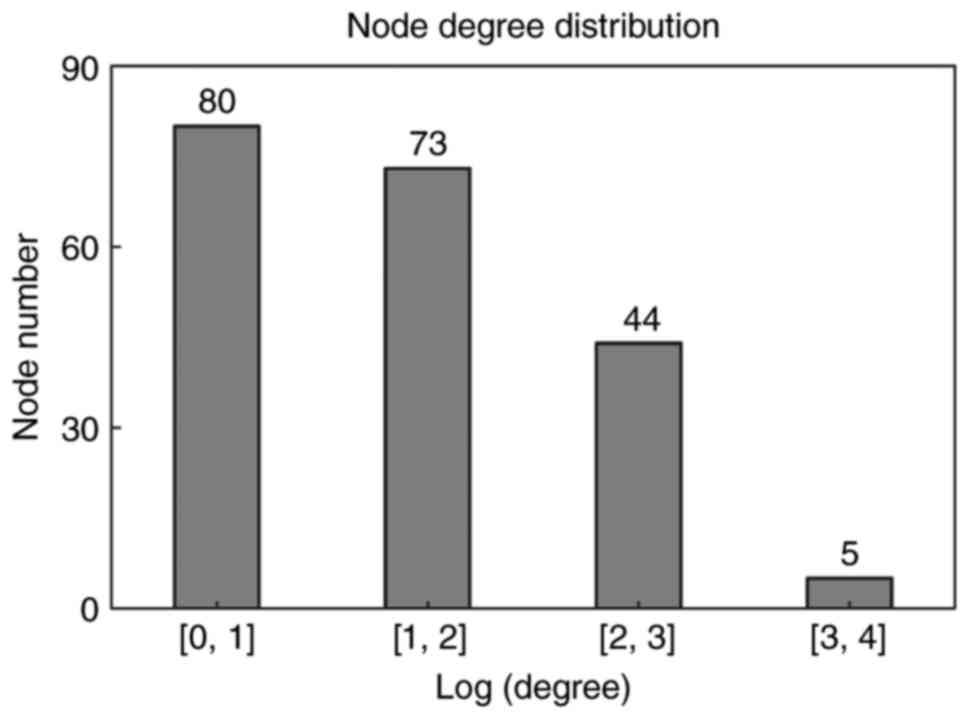
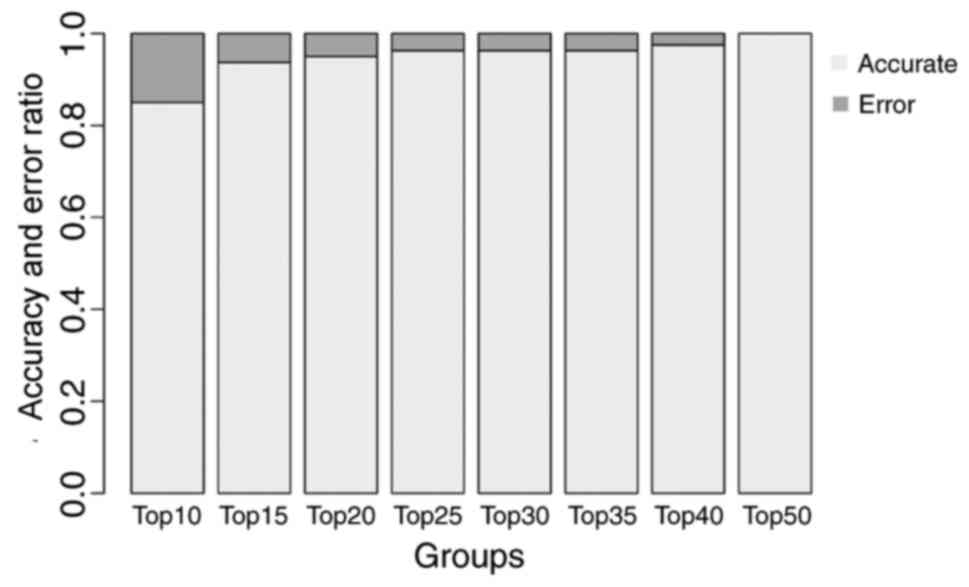
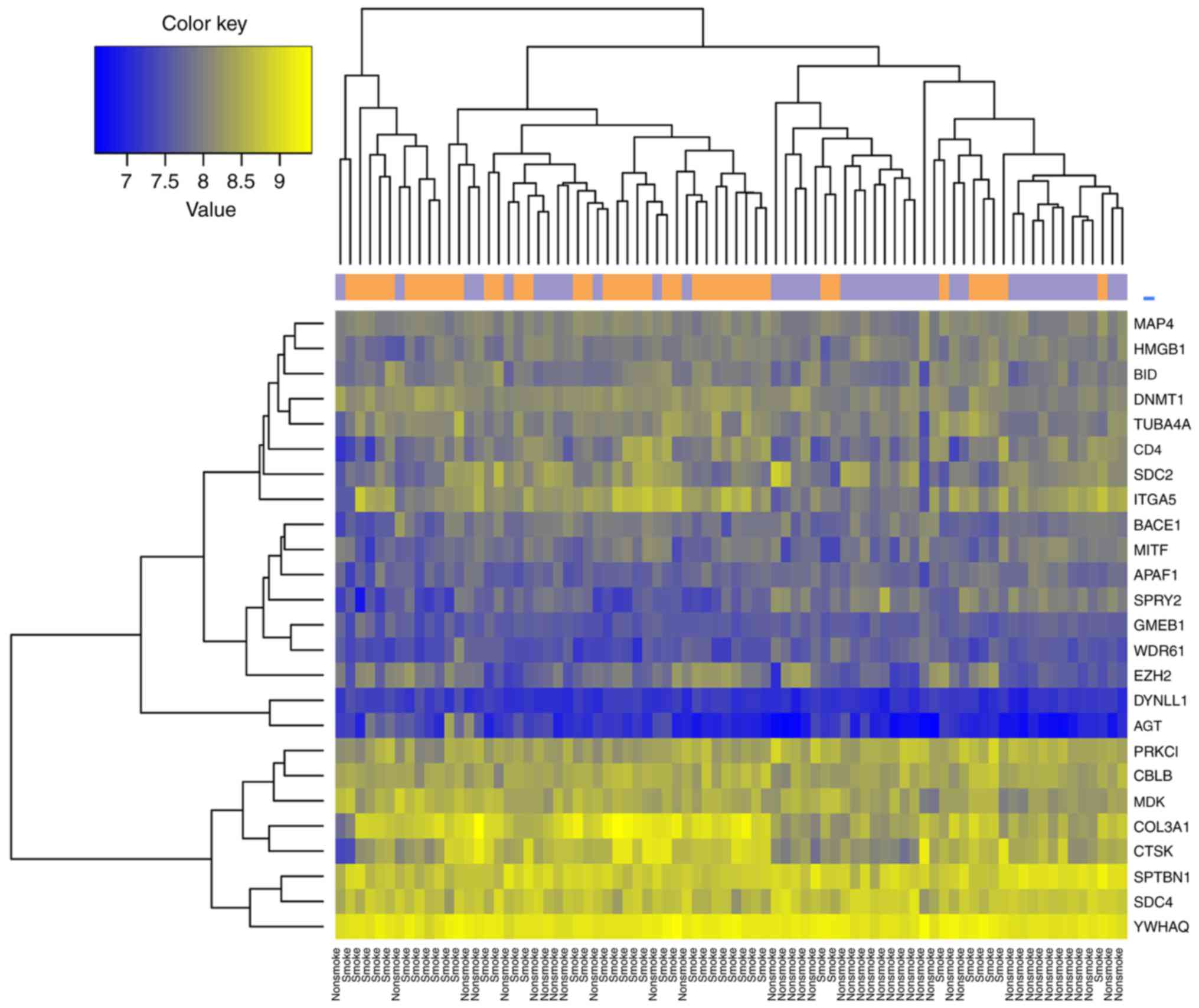
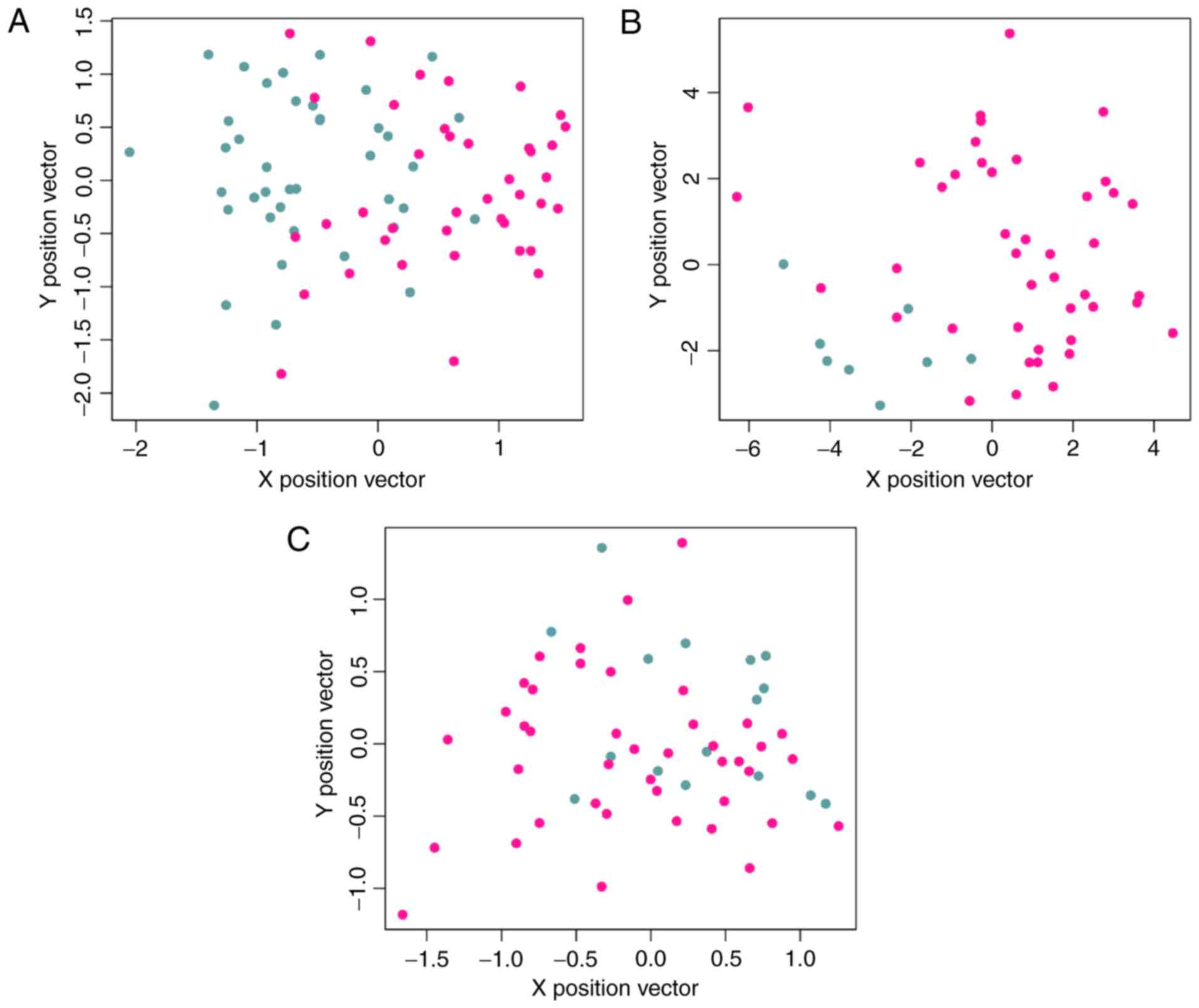
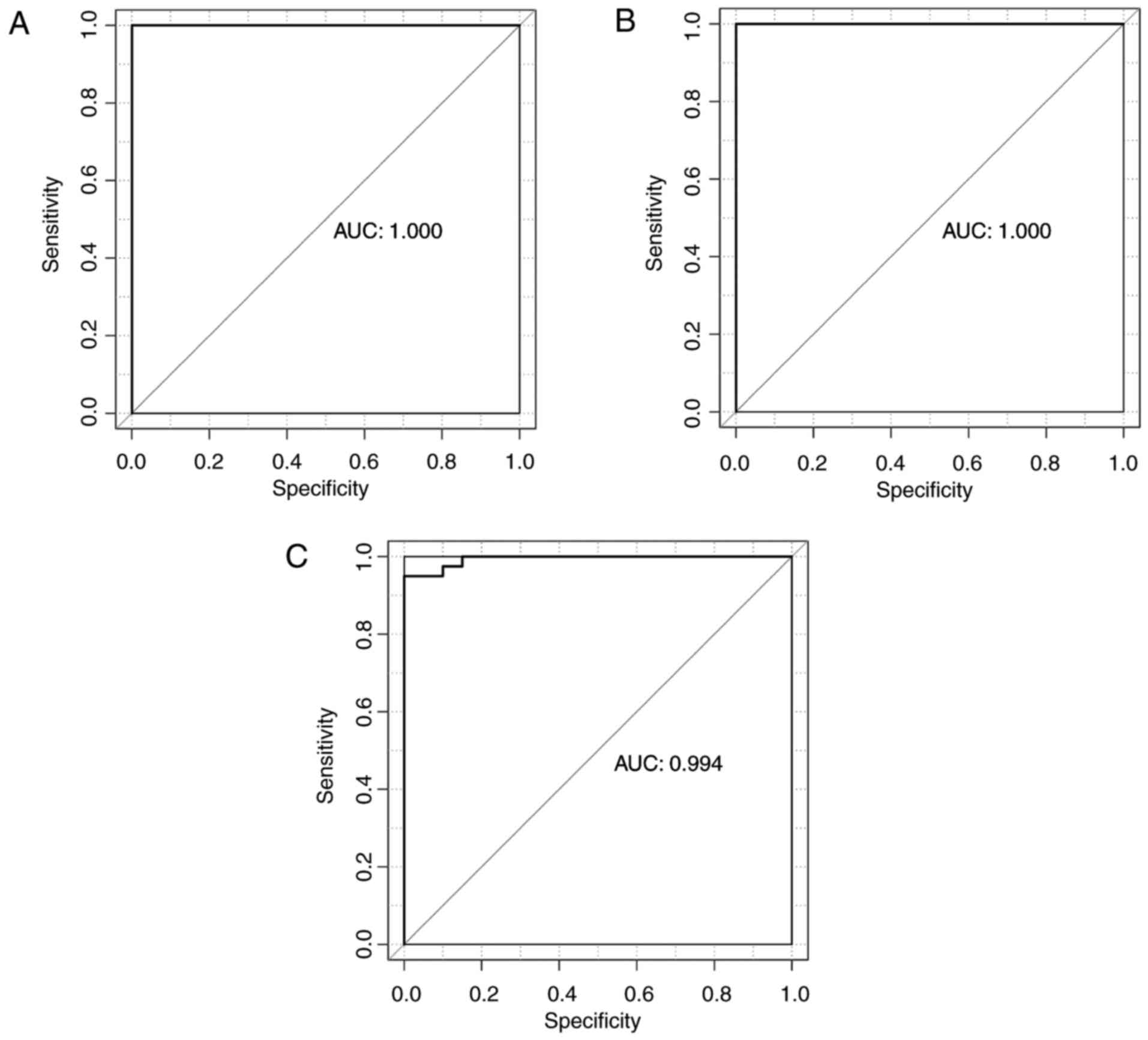
|
1
|
Centers for Disease Control and Prevention
(CDC), . State-specific trends in lung cancer incidence and
smoking-United States, 1999–2008. MMWR Morb Mortal Wkly Rep.
60:1243–1247. 2011.PubMed/NCBI
|
|
2
|
Nanavaty P, Alvarez MS and Alberts WM:
Lung cancer screening: Advantages, controversies, and applications.
Cancer Control. 21:9–14. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bryant A and Cerfolio RJ: Differences in
epidemiology, histology, and survival between cigarette smokers and
never-smokers who develop non-small cell lung cancer. Chest.
132:185–192. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nakamura H and Saji H: Worldwide trend of
increasing primary adenocarcinoma of the lung. Surg Today.
44:1004–1012. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kong J, Xu F, He M, Chen K and Qian B: The
incidence of lung cancer by histological type: A population-based
study in Tianjin, China during 1981–2005. Respirology.
19:1222–1228. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cancer Genome Atlas Research Network, .
Comprehensive molecular profiling of lung adenocarcinoma. Nature.
511:543–550. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Devarakonda S, Morgensztern D and Govindan
R: Genomic alterations in lung adenocarcinoma. Lancet Oncol.
16:e342–e351. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gautschi O, Hugli B, Ziegler A, Bigosch C,
Bowers NL, Ratschiller D, Jermann M, Stahel RA, Heighway J and
Betticher DC: Cyclin D1 (CCND1) A870G gene polymorphism modulates
smoking-induced lung cancer risk and response to platinum-based
chemotherapy in non-small cell lung cancer (NSCLC) patients. Lung
Cancer. 51:303–311. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li MY, Liu Y, Liu LZ, Kong AW, Zhao Z, Wu
B, Long X, Wu J, Ng CS, Wan IY, et al: Estrogen receptor alpha
promotes smoking-carcinogen-induced lung carcinogenesis via
cytochrome P450 1B1. J Mol Med. 93:1221–1233. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cheng Z, Wang W, Song YN, Kang Y and Xia
J: hOGG1, p53 genes, and smoking interactions are associated with
the development of lung cancer. Asian Pac J Cancer Prev.
13:1803–1808. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang J, Chen D, Shen QM, Tian DL, Jiang
YH, Yin HN and Li HW: Association between cigarette smoking and
FHIT gene alterations in Chinese lung cancer. Lung Cancer.
29:2352000. View Article : Google Scholar
|
|
12
|
Zhou W, Liu G, Park S, Wang Z, Wain JC,
Lynch TJ, Su L and Christiani DC: Gene-smoking interaction
associations for the ERCC1 polymorphisms in the risk of lung
cancer. Cancer Epidemiol Biomarkers Prev. 14:491–496. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhou C, Chen H, Han L, Xue F, Wang A and
Liang YJ: Screening of genes related to lung cancer caused by
smoking with RNA-Seq. Eur Rev Med Pharmacol Sci. 18:117–125.
2014.PubMed/NCBI
|
|
14
|
Kabbout M, Garcia MM, Fujimoto J, Liu DD,
Woods D, Chow CW, Mendoza G, Momin AA, James BP, Solis L, et al:
ETS2 mediated tumor suppressive function and MET oncogene
inhibition in human non-small cell lung cancer. Clin Cancer Res.
19:3383–3395. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Landi MT, Dracheva T, Rotunno M, Figueroa
JD, Liu H, Dasgupta A, Mann FE, Fukuoka J, Hames M, Bergen AW, et
al: Gene expression signature of cigarette smoking and its role in
lung adenocarcinoma development and survival. PLoS One.
3:e16512008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ding L, Getz G, Wheeler DA, Mardis ER,
McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan
MB, et al: Somatic mutations affect key pathways in lung
adenocarcinoma. Nature. 455:1069–1075. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: Affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Smyth GK: Limma: Linear models for
microarray dataBioinformatics and Computational Biology Solutions
Using R and Bioconductor. Gentleman R, Carey V, Huber W, Irizarry R
and Dudoit S: Springer; New York: pp. 397–420. 2004
|
|
19
|
Wang X, Kang DD, Shen K, Song C, Lu S,
Chang LC, Liao SG, Huo Z, Tang S, Ding Y, et al: An R package suite
for microarray meta-analysis in quality control, differentially
expressed gene analysis and pathway enrichment detection.
Bioinformatics. 28:2534–2536. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang L, Cao C, Ma Q, Zeng Q, Wang H, Cheng
Z, Zhu G, Qi J, Ma H, Nian H and Wang Y: RNA-seq analyses of
multiple meristems of soybean: Novel and alternative transcripts,
evolutionary and functional implications. BMC Plant Biol.
14:1692014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Keshava Prasad TS, Goel R, Kandasamy K,
Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R,
Shafreen B, Venugopal A, et al: Human protein reference
database-2009 update. Nucleic Acids Res. 37(Database issue):
D767–D772. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Barthélemy M: Betweenness centrality in
large complex networks. Eur Phy J Conden Matter Com Sys.
38:163–168. 2004.
|
|
24
|
Zhang HH, Ahn J, Lin X and Park C: Gene
selection using support vector machines with non-convex penalty.
Bioinformatics. 22:88–95. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Brown MP, Grundy WN, Lin D, Cristianini N,
Sugnet CW, Furey TS, Ares M Jr and Haussler D: Knowledge-based
analysis of microarray gene expression data using support vector
machines. Proc Natl Acad Sci USA. 97:pp. 262–267. 2000; View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Stojanović M, Andjelković Apostolović M,
Stojanović D, Milosević Z, Ignjatović A, Lakusić VM and Golubović
M: Understanding sensitivity, specificity and predictive values.
Vojnosanit Pregl. 71:1062–1065. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Parikh R, Mathai A, Parikh S, Chandra
Sekhar G and Thomas R: Understanding and using sensitivity,
specificity and predictive values. Indian J Ophthalmol. 56:45–50.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Orru G, Pettersson-Yeo W, Marquand AF,
Sartori G and Mechelli A: Using support vector machine to identify
imaging biomarkers of neurological and psychiatric disease: A
critical review. Neurosci Biobehav Rev. 36:1140–1152. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fan XJ, Wan XB, Huang Y, Cai HM, Fu XH,
Yang ZL, Chen DK, Song SX, Wu PH, Liu Q, et al:
Epithelial-mesenchymal transition biomarkers and support vector
machine guided model in preoperatively predicting regional lymph
node metastasis for rectal cancer. Br J Cancer. 106:1735–1741.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Han M, Dai J, Zhang Y, Lin Q, Jiang M, Xu
X, Liu Q and Jia J: Support vector machines coupled with proteomics
approaches for detecting biomarkers predicting chemotherapy
resistance in small cell lung cancer. Oncol Rep. 28:2233–2238.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Guyon I, Weston J and Barnhill S: Gene
selection for cancer classification using support vector machines.
Machine Learning. 46:389–422. 2002. View Article : Google Scholar
|
|
32
|
Yousef M, Ketany M, Manevitz L, Showe LC
and Showe MK: Classification and biomarker identification using
gene network modules and support vector machines. BMC
Bioinformatics. 10:3372009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Guan P, Huang D, He M and Zhou B: Lung
cancer gene expression database analysis incorporating prior
knowledge with support vector machine-based classification method.
J Exp Clin Cancer Res. 28:1032009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li P, Wang X, Liu Z, Liu H, Xu T, Wang H,
Gomez DR, Nguyen QN, Wang LE, Teng Y, et al: Single nucleotide
polymorphisms in CBLB, a regulator of T-cell response, predict
radiation pneumonitis and outcomes after definitive radiotherapy
for non-small-cell lung cancer. Clin Lung Cancer. 17:253–262. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mu X, Zhang Y, Qu X, Hou K, Kang J, Hu X
and Liu Y: Ubiquitin ligase Cbl-b is involved in icotinib
(BPI-2009H)-induced apoptosis and G1 phase arrest of EGFR
mutation-positive non-small-cell lung cancer. Biomed Res Int.
2013:7263752013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Rountree MR, Bachman KE, Herman JG and
Baylin SB: DNA methylation, chromatin inheritance, and cancer.
Oncogene. 20:3156–3165. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kerr KM, Galler JS, Hagen JA, Laird PW and
Laird-Offringa IA: The role of DNA methylation in the development
and progression of lung adenocarcinoma. Dis Markers. 23:5–30. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lin RK, Hsieh YS, Lin P, Hsu HS, Chen CY,
Tang YA, Lee CF and Wang YC: The tobacco-specific carcinogen NNK
induces DNA methyltransferase 1 accumulation and tumor suppressor
gene hypermethylation in mice and lung cancer patients. J Clin
Invest. 120:521–532. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tennis MA, Vanscoyk MM, Wilson LA, Kelley
N and Winn RA: Methylation of Wnt7a is modulated by DNMT1 and
cigarette smoke condensate in non-small cell lung cancer. PLoS One.
7:e329212012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
McCabe MT and Creasy CL: EZH2 as a
potential target in cancer therapy. Epigenomics. 6:341–351. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yoon KA, Gil HJ, Han J, Park J and Lee JS:
Genetic polymorphisms in the polycomb group gene EZH2 and the risk
of lung cancer. J Thorac Oncol. 5:10–16. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhang H, Zhang H, Zhao M, Lv Z, Zhang X,
Qin X, Wang H, Wang S, Su J, Lv X, et al: miR-138 inhibits tumor
growth through repression of EZH2 in non-small cell lung cancer.
Cell Physiol Biochem. 31:56–65. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Xia H, Zhang W, Li Y, Guo N and Yu C: EZH2
silencing with RNA interference induces G2/M arrest in human lung
cancer cells in vitro. Biomed Res Int. 2014:3487282014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang X, Zhao H, Lv L, Bao L, Wang X and
Han S: Prognostic significance of EZH2 expression in non-small cell
lung cancer: A meta-analysis. Sci Rep. 6:192392016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang C, Ge S, Hu C, Yang N and Zhang J:
miRNA-218, a new regulator of HMGB1, suppresses cell migration and
invasion in non-small cell lung cancer. Acta Biochim Biophys Sin
(Shanghai). 45:1055–1061. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Shen X, Hong L, Sun H, Shi M and Song Y:
The expression of high-mobility group protein box 1 correlates with
the progression of non-small cell lung cancer. Oncol Rep.
22:535–539. 2009.PubMed/NCBI
|
|
47
|
Zheng W, Jiang C and Li R: Integrin and
gene network analysis reveals that ITGA5 and ITGB1 are prognostic
in non-small-cell lung cancer. Onco Targets Ther. 9:2317–2327.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Jono H and Ando Y: Midkine: A novel
prognostic biomarker for cancer. Cancers. 2:624–641. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yuan K, Chen Z, Li W, Gao CE, Li G, Guo G,
Yang Y, Ai Y, Wu L and Zhang M: MDK protein overexpression
correlates with the malignant status and prognosis of non-small
cell lung cancer. Arch Med Res. 46:635–641. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Hao H, Maeda Y, Fukazawa T, Yamatsuji T,
Takaoka M, Bao XH, Matsuoka J, Okui T, Shimo T, Takigawa N, et al:
Inhibition of the growth factor MDK/midkine by a novel small
molecule compound to treat non-small cell lung cancer. PLoS One.
8:e710932013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Regala RP, Davis RK, Kunz A, Khoor A,
Leitges M and Fields AP: Atypical protein kinase C{iota} is
required for bronchioalveolar stem cell expansion and lung
tumorigenesis. Cancer Res. 69:7603–7611. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Sutterlüty H, Mayer CE, Attems J, Setinek
U, Mikula M, Mikulits W, Micksche M and Berger W: Inhibition of
cell migration and proliferation in non-small cell lung cancer
(NSCLC) by Sprouty 2 (Spry2) via K-Ras dependent and independent
pathways. Cancer Res. 66 Suppl 8:S349–S350. 2006.
|
|
53
|
Sutterlüty H, Mayer CE, Setinek U, Attems
J, Ovtcharov S, Mikula M, Mikulits W, Micksche M and Berger W:
Down-regulation of Sprouty2 in non-small cell lung cancer
contributes to tumor malignancy via extracellular signal-regulated
kinase pathway-dependent and -independent mechanisms. Mol Cancer
Res. 5:509–520. 2007. View Article : Google Scholar : PubMed/NCBI
|