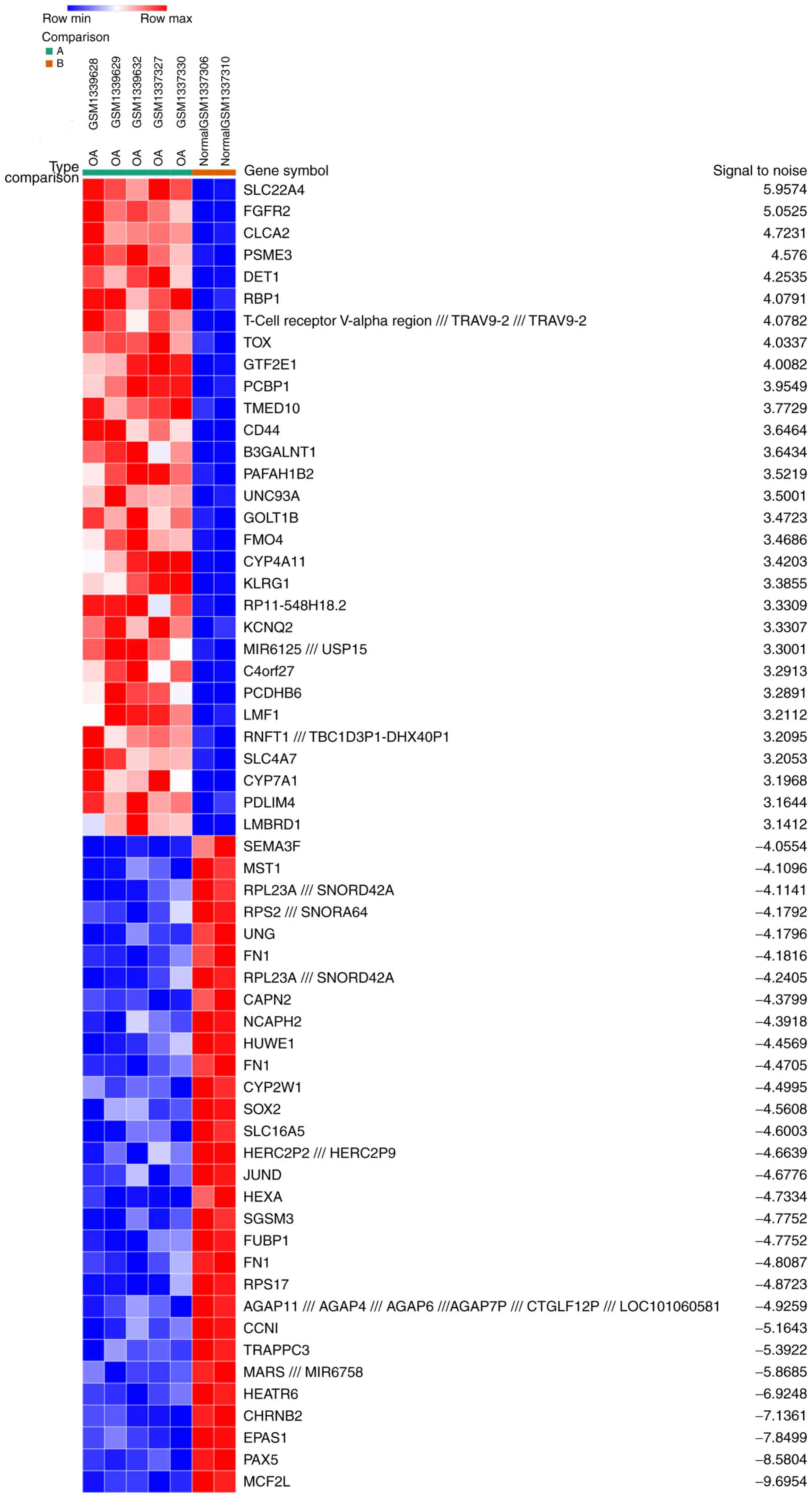
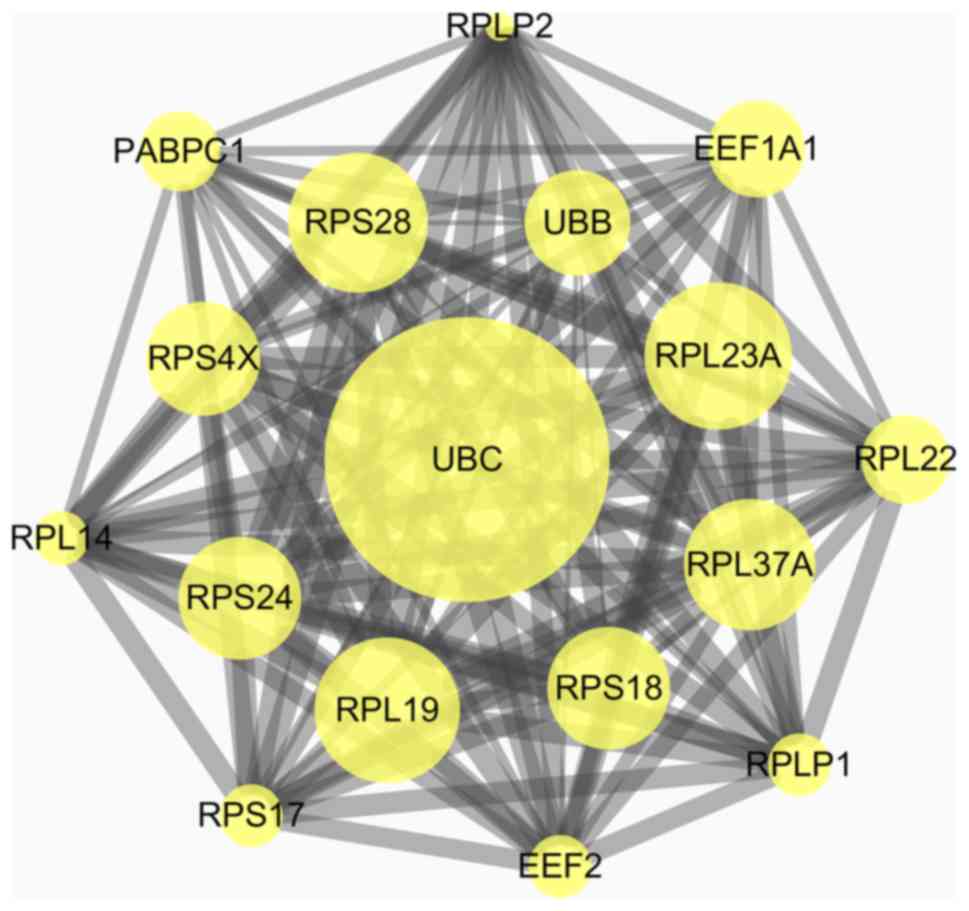
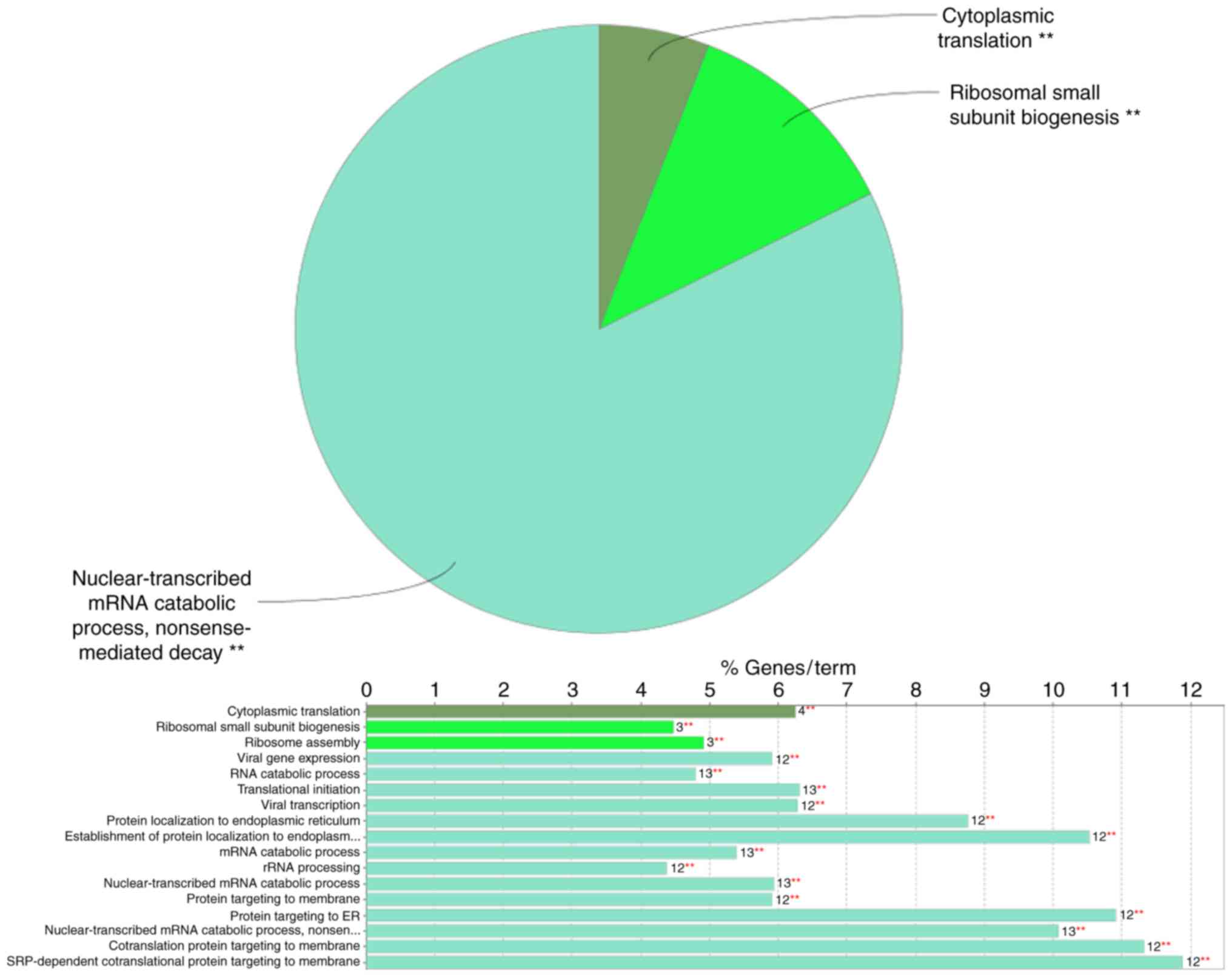
|
1
|
Liu W, He J, Lin R, Liang J and Luo Q:
Differential proteomics of the synovial membrane between bilateral
and unilateral knee osteoarthritis in surgeryinduced rabbit models.
Mol Med Rep. 14:2243–2249. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen S, Fang XQ, Zhang JF, Ma Y, Tang XZ,
Zhou ZJ, Wang JY, Qin A and Fan SW: Lycorine protects cartilage
through suppressing the expression of matrix metalloprotenases in
rat chondrocytes and in a mouse osteoarthritis model. Mol Med Rep.
14:3389–3396. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Helmick CG, Felson DT, Lawrence RC,
Gabriel S, Hirsch R, Kwoh CK, Liang MH, Kremers HM, Mayes MD,
Merkel PA, et al: Estimates of the prevalence of arthritis and
other rheumatic conditions in the United States. Part I. Arthritis
Rheum. 58:15–25. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Thomas E, Peat G and Croft P: Defining and
mapping the person with osteoarthritis for population studies and
public health. Rheumatology (Oxford). 53:338–345. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kubosch EJ, Lang GM, Fürst D, Kubosch DC,
Izadpanah K, Rolauffs B, Südkamp NP and Schmal H: The potential for
synovium-derived stem cells in cartilage repair. Curr Stem Cell Res
Ther. Oct 2–2017.(Epub ahead of print). PubMed/NCBI
|
|
6
|
Mathiessen A and Conaghan PG: Synovitis in
osteoarthritis: Current understanding with therapeutic
implications. Arthritis Res Ther. 19:182017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Prieto-Potin I, Largo R, Roman-Blas JA,
Herrero-Beaumont G and Walsh DA: Characterization of multinucleated
giant cells in synovium and subchondral bone in knee osteoarthritis
and rheumatoid arthritis. BMC Musculoskelet Disord. 16:2262015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Deligne C, Casulli S, Pigenet A, Bougault
C, Campillo-Gimenez L, Nourissat G, Berenbaum F, Elbim C and Houard
X: Differential expression of interleukin-17 and interleukin-22 in
inflamed and non-inflamed synovium from osteoarthritis patients.
Osteoarthritis Cartilage. 23:1843–1852. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kuca-Warnawin EH, Kurowska WJ, Radzikowska
A, Massalska MA, Burakowski T, Kontny E, Słowińska I, Gasik R and
Maśliński W: Different expression of chemokines in rheumatoid
arthritis and osteoarthritis bone marrow. Reumatologia. 54:51–53.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Takano S, Uchida K, Miyagi M, Inoue G,
Fujimaki H, Aikawa J, Iwase D, Minatani A, Iwabuchi K and Takaso M:
Nerve growth factor regulation by TNF-α and IL-1β in synovial
macrophages and fibroblasts in osteoarthritic mice. J Immunol Res.
2016:57063592016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Salaffi F, Ciapetti A and Carotti M: The
sources of pain in osteoarthritis: A pathophysiological review.
Reumatismo. 66:57–71. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Das DK, Ali T, Krampis K and Ogunwobi OO:
Fibronectin and androgen receptor expression data in prostate
cancer obtained from a RNA-sequencing bioinformatics analysis. Data
Brief. 11:131–135. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gong C, Sun S, Liu B, Wang J and Chen X:
Identification of potential therapeutic target genes, key miRNAs
and mechanisms in oral lichen planus by bioinformatics analysis.
Arch Oral Biol. 78:122–128. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ramos YF, Bos SD, Lakenberg N, Böhringer
S, den Hollander WJ, Kloppenburg M, Slagboom PE and Meulenbelt I:
Genes expressed in blood link osteoarthritis with apoptotic
pathways. Ann Rheum Dis. 73:1844–1853. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ungethuem U, Haeupl T, Witt H, Koczan D,
Krenn V, Huber H, von Helversen TM, Drungowski M, Seyfert C, Zacher
J, et al: Molecular signatures and new candidates to target the
pathogenesis of rheumatoid arthritis. Physiol Genomics. 42A:1–282.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Blagojevic M, Jinks C, Jeffery A and
Jordan KP: Risk factors for onset of osteoarthritis of the knee in
older adults: A systematic review and meta-analysis. Osteoarthritis
Cartilage. 18:24–33. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kee CC: Osteoarthritis: Manageable scourge
of aging. Nurs Clin North Am. 35:199–208. 2000.PubMed/NCBI
|
|
18
|
Venet D, Detours V and Bersini H: A
measure of the signal-to-noise ratio of microarray samples and
studies using gene correlations. PLoS One. 7:e510132012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bindea G, Mlecnik B, Hackl H, Charoentong
P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z and
Galon J: ClueGO: A Cytoscape plug-in to decipher functionally
grouped gene ontology and pathway annotation networks.
Bioinformatics. 25:1091–1032. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Oh C, Park S, Lee EK and Yoo YJ:
Downregulation of ubiquitin level via knockdown of polyubiquitin
gene Ubb as potential cancer therapeutic intervention. Sci Rep.
3:26232013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tian Y, Ding W, Wang Y, Ji T, Sun S, Mo Q,
Chen P, Fang Y, Liu J, Wang B, et al: Correction: Ubiquitin B in
cervical cancer: Critical for the maintenance of cancer stem-like
cell characters. PLoS One. 11:e01528132016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang Y, Vasheghani F, Li YH, Blati M,
Simeone K, Fahmi H, Lussier B, Roughley P, Lagares D, Pelletier JP,
et al: Cartilage-specific deletion of mTOR upregulates autophagy
and protects mice from osteoarthritis. Ann Rheum Dis. 74:1432–1440.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ribeiro M, López de Figueroa P,
Nogueira-Recalde U, Centeno A, Mendes AF, Blanco FJ and Caramés B:
Diabetes-accelerated experimental osteoarthritis is prevented by
autophagy activation. Osteoarthritis Cartilage. 24:2116–2125. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ito Y, Hashimoto M, Hirota K, Ohkura N,
Morikawa H, Nishikawa H, Tanaka A, Furu M, Ito H, Fujii T, et al:
Detection of T cell responses to a ubiquitous cellular protein in
autoimmune disease. Science. 346:363–368. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tang YP and Wade J: Sex- and age-related
differences in ribosomal proteins L17 and L37, as well as androgen
receptor protein, in the song control system of zebra finches.
Neuroscience. 171:1131–1140. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang Y, Sui J, Li X, Cao F, He J, Yang B,
Zhu X, Sun Y and Pu YD: RPS24 knockdown inhibits colorectal cancer
cell migration and proliferation in vitro. Gene. 571:286–291. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Badhai J, Fröjmark ASJ, Davey E, Schuster
J and Dahl N: Ribosomal protein S19 and S24 insufficiency cause
distinct cell cycle defects in Diamond-Blackfan anemia. Biochim
Biophys Acta. 1792:1036–1042. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Anthony RA and Liebman SW: Alterations in
ribosomal protein RPS28 can diversely affect translational accuracy
in Saccharomyces cerevisiae. Genetics. 140:1247–1258.
1995.PubMed/NCBI
|
|
29
|
Doherty L, Sheen MR, Vlachos A, Choesmel
V, O'Donohue MF, Clinton C, Schneider HE, Sieff CA, Newburger PE,
Ball SE, et al: Ribosomal protein genes RPS10 and RPS26 are
commonly mutated in Diamond-Blackfan anemia. Am J Hum Genet.
86:222–228. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gazda HT, Preti M, Sheen MR, O'Donohue MF,
Vlachos A, Davies SM, Kattamis A, Doherty L, Landowski M, Buros C,
et al: Frameshift mutation in p53 regulator RPL26 is associated
with multiple physical abnormalities and a specific pre-ribosomal
RNA processing defect in diamond-blackfan anemia. Hum Mutat.
33:1037–1044. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gripp KW, Curry C, Olney AH, Sandoval C,
Fisher J, Chong JX; UW Center for Mendelian Genomics, ; Pilchman L,
Sahraoui R, Stabley DL and Sol-Church K: Diamond-Blackfan anemia
with mandibulofacial dystostosis is heterogeneous, including the
novel DBA genes TSR2 and RPS28. Am J Med Genet A. 164A:1–2249.
2014.PubMed/NCBI
|
|
32
|
Shen H, Zhu H, Song M, Tian Y, Huang Y,
Zheng H, Cao R, Lin J, Bi Z and Zhong W: A selenosemicarbazone
complex with copper efficiently down-regulates the 90-kDa heat
shock protein HSP90AA1 and its client proteins in cancer cells. BMC
Cancer. 14:6292014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hentze MW and Kulozik AE: A perfect
message: RNA surveillance and nonsense-mediated decay. Cell.
96:307–310. 1999. View Article : Google Scholar : PubMed/NCBI
|