|
1
|
Moroni RM, Vieira CS, Ferriani RA,
Candido-dos-Reis FJ and Brito LGO: Pharmacological treatment of
uterine fibroids. Ann Med Health Sci Res. 4 Suppl 3:S185–S192.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Razavi MK, Hwang G, Jahed A, Modanlou S
and Chen B: Abdominal myomectomy versus uterine fibroid
embolization in the treatment of symptomatic uterine leiomyomas.
AJR Am J Roentgenol. 180:1571–1575. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
He X: Estrogen receptor and progesterone
receptor expressions at the edge of the targeted region of uterus
fibroids ablated by acoustic power. J Chongqing Med Univ. 36:5–7.
2011.
|
|
4
|
Mäkinen N, Mehine M, Tolvanen J, Kaasinen
E, Li Y, Lehtonen HJ, Gentile M, Yan J, Enge M, Taipale M, et al:
MED12, the mediator complex subunit 12 gene, is mutated at high
frequency in uterine leiomyomas. Science. 334:252–255. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Raimundo N, Vanharanta S, Aaltonen LA,
Hovatta I and Suomalainen A: Downregulation of SRF-FOS-JUNB pathway
in fumarate hydratase deficiency and in uterine leiomyomas.
Oncogene. 28:1261–1273. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shen Y, Wu Y, Lu Q, Zhang P and Ren M:
Transforming growth factor-β signaling pathway cross-talking with
ERα signaling pathway on regulating the growth of uterine leiomyoma
activated by phenolic environmental estrogens in vitro. Tumour
Biol. 37:455–462. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Klotzbücher M, Wasserfall A and Fuhrmann
U: Misexpression of wild-type and truncated isoforms of the
high-mobility group I proteins HMGI-C and HMGI(Y) in uterine
leiomyomas. Am J Pathol. 155:1535–1542. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chuang TY, Min J, Wu HL, McCrary C, Layman
LC, Diamond MP, Azziz R, Al-Hendy A and Chen YH: Berberine inhibits
uterine leiomyoma cell proliferation via downregulation of
cyclooxygenase 2 and pituitary tumor-transforming gene 1. Reprod
Sci. 24:1005–1013. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huan JL, Gao X, Xing L, Qin XJ, Qian HX,
Zhou Q and Zhu L: Screening for key genes associated with invasive
ductal carcinoma of the breast via microarray data analysis. Genet
Mol Res. 13:7919–7925. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen L, Chu C, Lu J, Kong X, Huang T and
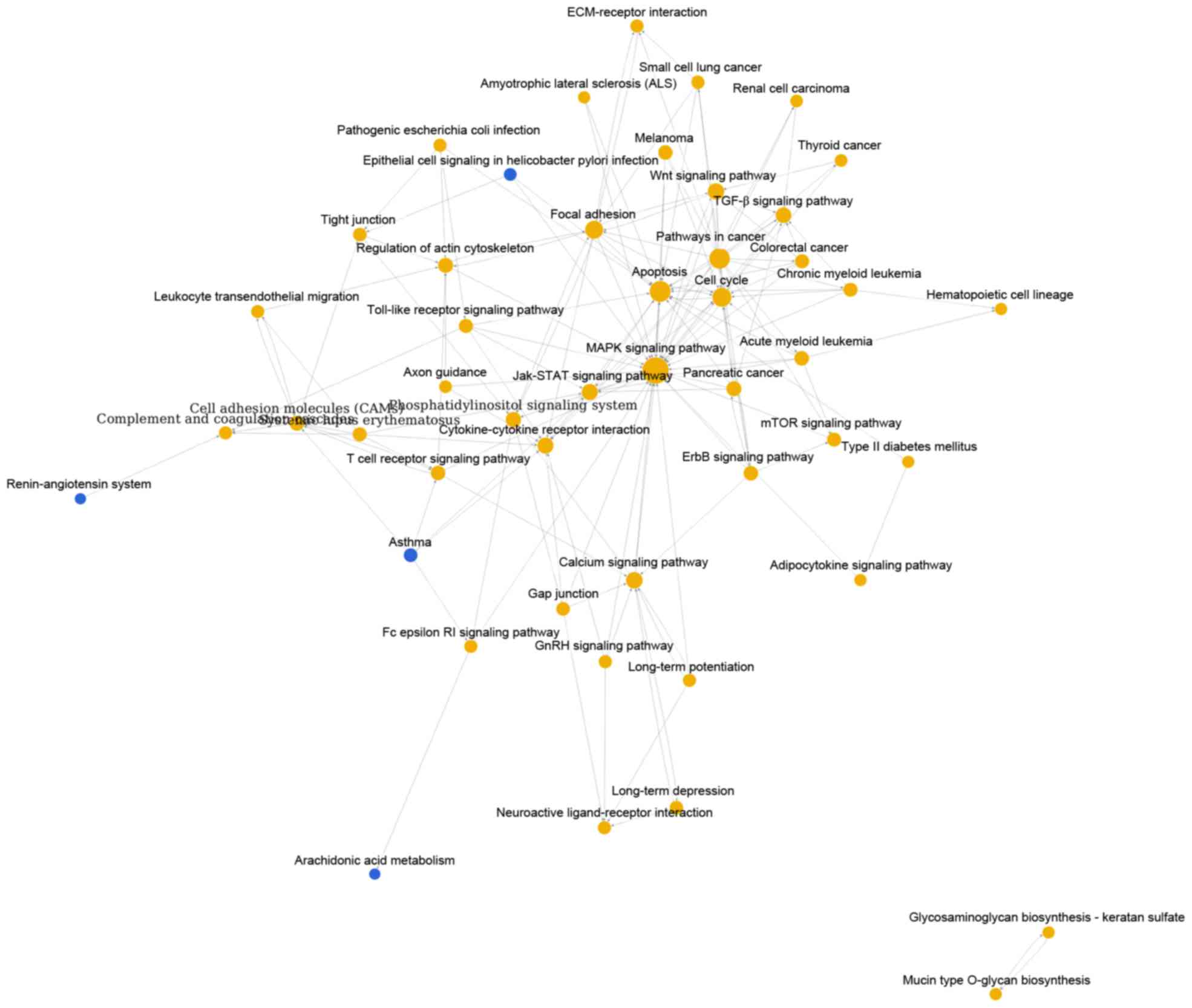
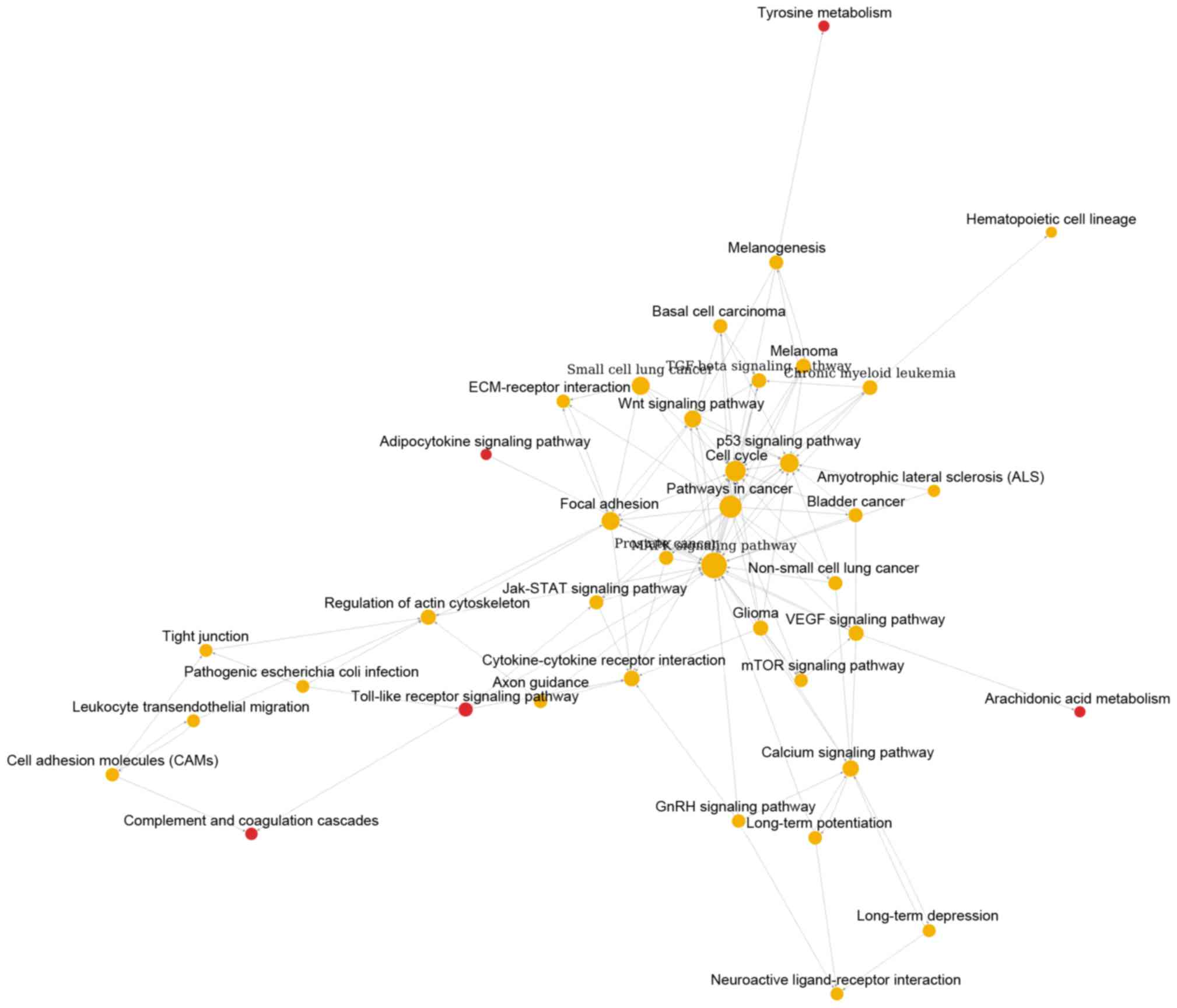
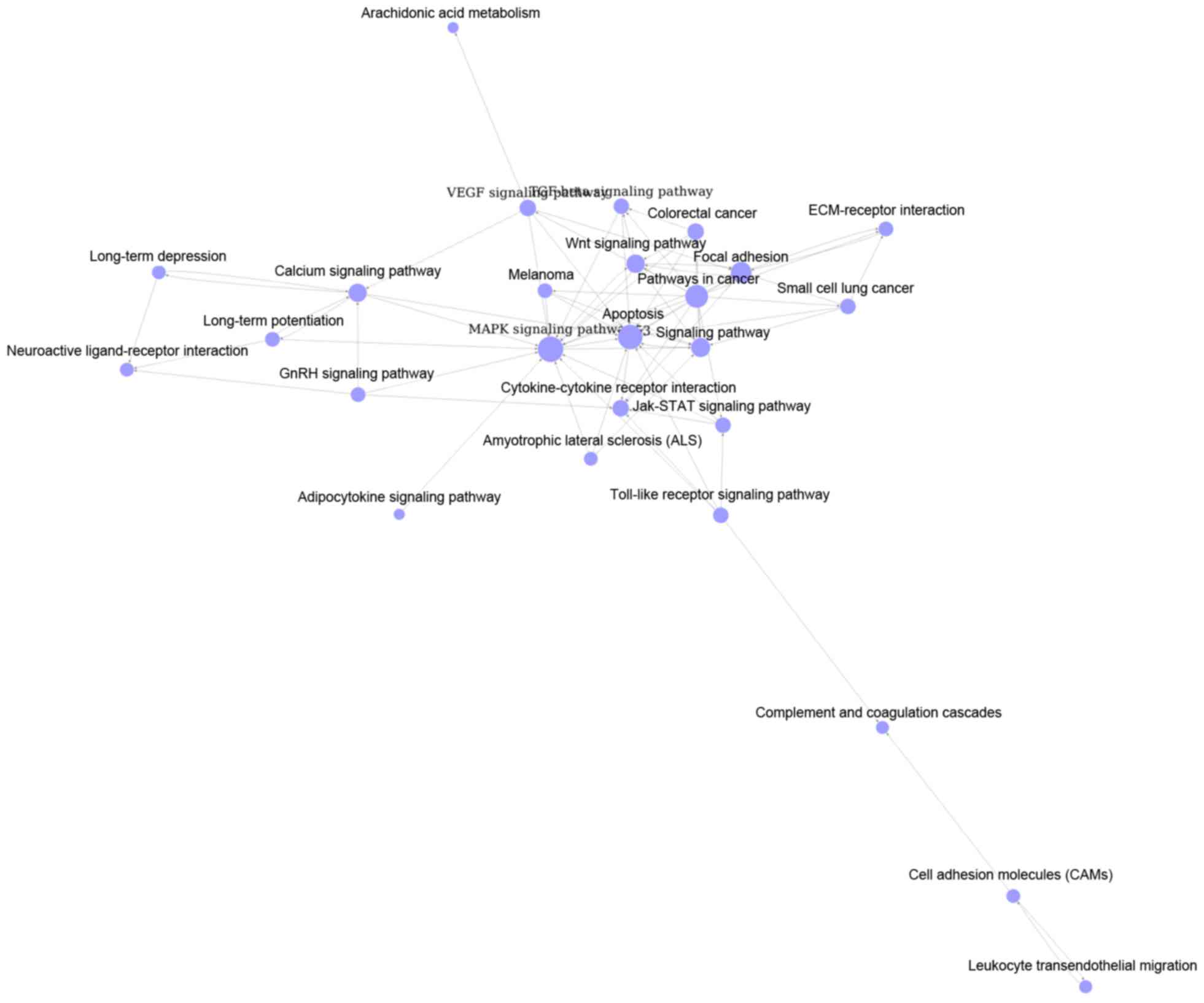
Cai YD: Gene ontology and KEGG pathway enrichment analysis of a
drug target-based classification system. PLoS One. 10:e01264922015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
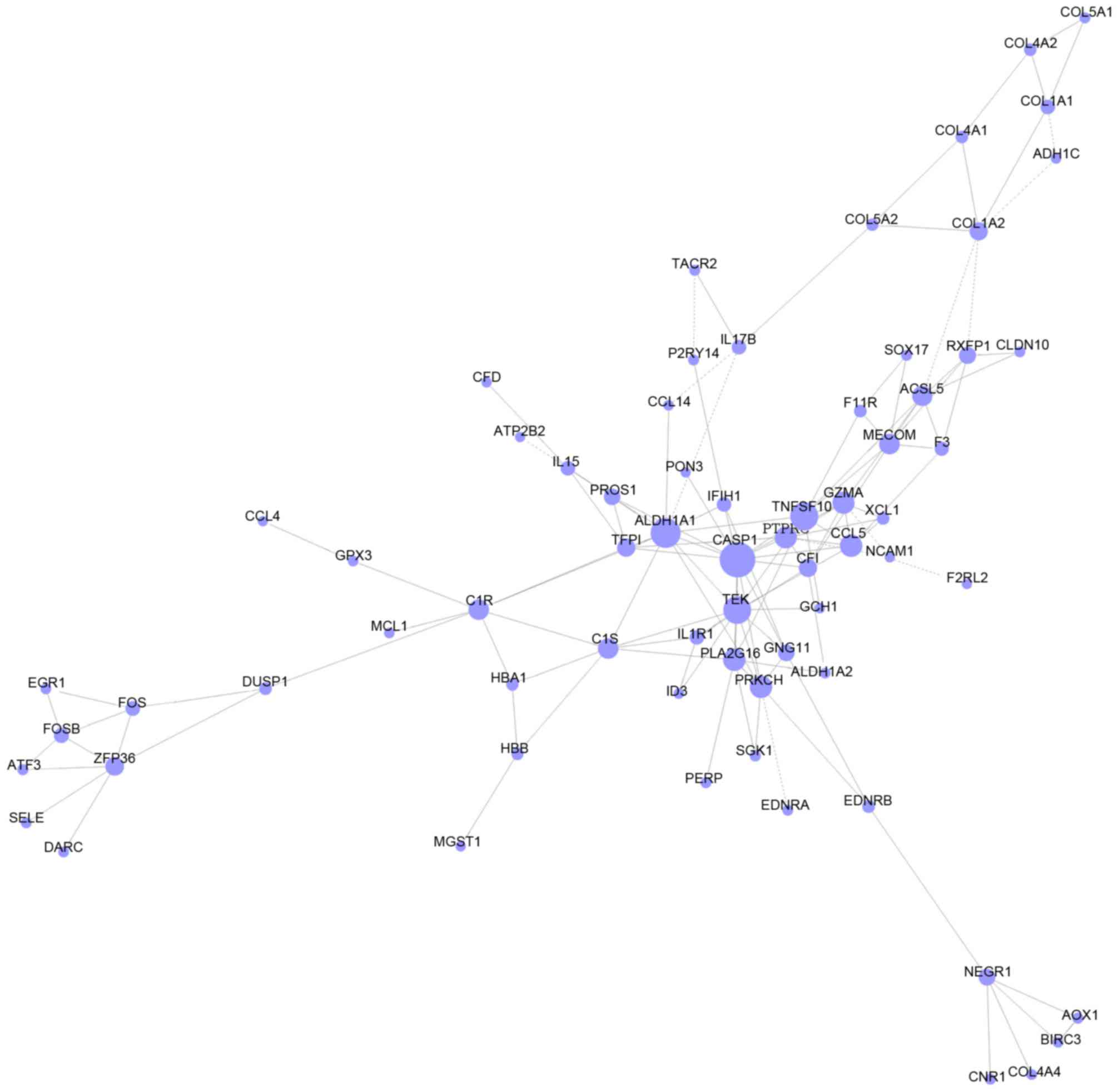
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Southby J, Murphy LM, Martin TJ and
Gillespie MT: Cell-specific and regulator-induced promoter usage
and messenger ribonucleic acid splicing for parathyroid
hormone-related protein. Endocrinology. 137:1349–1357. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Commandeur AE, Styer AK and Teixeira JM:
Epidemiological and genetic clues for molecular mechanisms involved
in uterine leiomyoma development and growth. Hum Reprod Update.
21:593–615. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kessel PdAHMGv, Pauwels PAA and
Schoenmakers EFPM: Molecular genetic and functional
characterization of signalling networks that govern the development
of human uterine leiomyomas. IEEE International Symposium on
Multiple-valued Logic. pp. 2331998;
|
|
16
|
Commandeur AE, Styer AK and Teixeira JM:
Epidemiological and genetic clues for molecular mechanisms involved
in uterine leiomyoma development and growth. Hum Reprod Update.
21:593–615. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rice KE, Secrist JR, Woodrow EL, Hallock
LM and Neal JL: Etiology, diagnosis, and management of uterine
leiomyomas. J Midwifery Womens Health. 57:241–247. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Thirupathi A, Elango T, Subramanian S and
Gnanaraj P: Methotrexate regulates Th-1 response by suppressing
caspase-1 and cytokines in psoriasis patients. Clin Chim Acta.
453:164–169. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gloria-Bottini F, Pietropolli A, Ammendola
M, Saccucci P and Bottini E: PTPN22 and uterine leiomyomas. Eur J
Obstet Gynecol Reprod Biol. 185:96–98. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Christman GM, Tang H, Ahmad I and Stribley
JM: Differential expression of the Notch signal transduction
pathway: Ligands, receptors and Numb in uterine leiomyomas vs.
myometrium. Fertil Steril. 88 Suppl 1:S722007. View Article : Google Scholar
|
|
21
|
Zaitseva M, Vollenhoven BJ and Rogers PAW:
443. The fibroblast-smooth muscle cell relationship is altered in
uterine leiomyoma. Reprod Fertil Dev. 20:1232008. View Article : Google Scholar
|
|
22
|
Shveiky D and Rojansky N: 255: Alcohol has
a permissive effect on the growth of uterine leiomyomata cells in
tissue cultures. J Minim Invasive Gynecol. 14 Suppl:S92–S93. 2007.
View Article : Google Scholar
|
|
23
|
Luo X and Chegini N: The expression and
potential regulatory function of microRNAs in the pathogenesis of
leiomyoma. Semin Reprod Med. 26:500–514. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Moreb JS, Baker HV, Chang LJ, Amaya M,
Lopez MC, Ostmark B and Chou W: ALDH isozymes downregulation
affects cell growth, cell motility and gene expression in lung
cancer cells. Mol Cancer. 7:872008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Takeda T, Sakata M, Isobe A, Miyake A,
Nishimoto F, Ota Y, Kamiura S and Kimura T: Relationship between
metabolic syndrome and uterine leiomyomas: A case-control study.
Gynecol Obstet Invest. 66:14–17. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Castoldi E and Hackeng TM: Regulation of
coagulation by protein S. Curr Opin Hematol. 15:529–536. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fang BQ and Zhang JW: Molecular mechanisms
of protein S deficiency caused by a novel nonsense mutation. Chin J
Vasc Surg. 2:32–35. 2015.
|
|
28
|
Cunin P, Beauvillain C, Miot C, Augusto
JF, Preisser L, Blanchard S, Pignon P, Scotet M, Garo E, Fremaux I,
et al: Clusterin facilitates apoptotic cell clearance and prevents
apoptotic cell-induced autoimmune responses. Cell Death Dis.
7:e22152016. View Article : Google Scholar : PubMed/NCBI
|