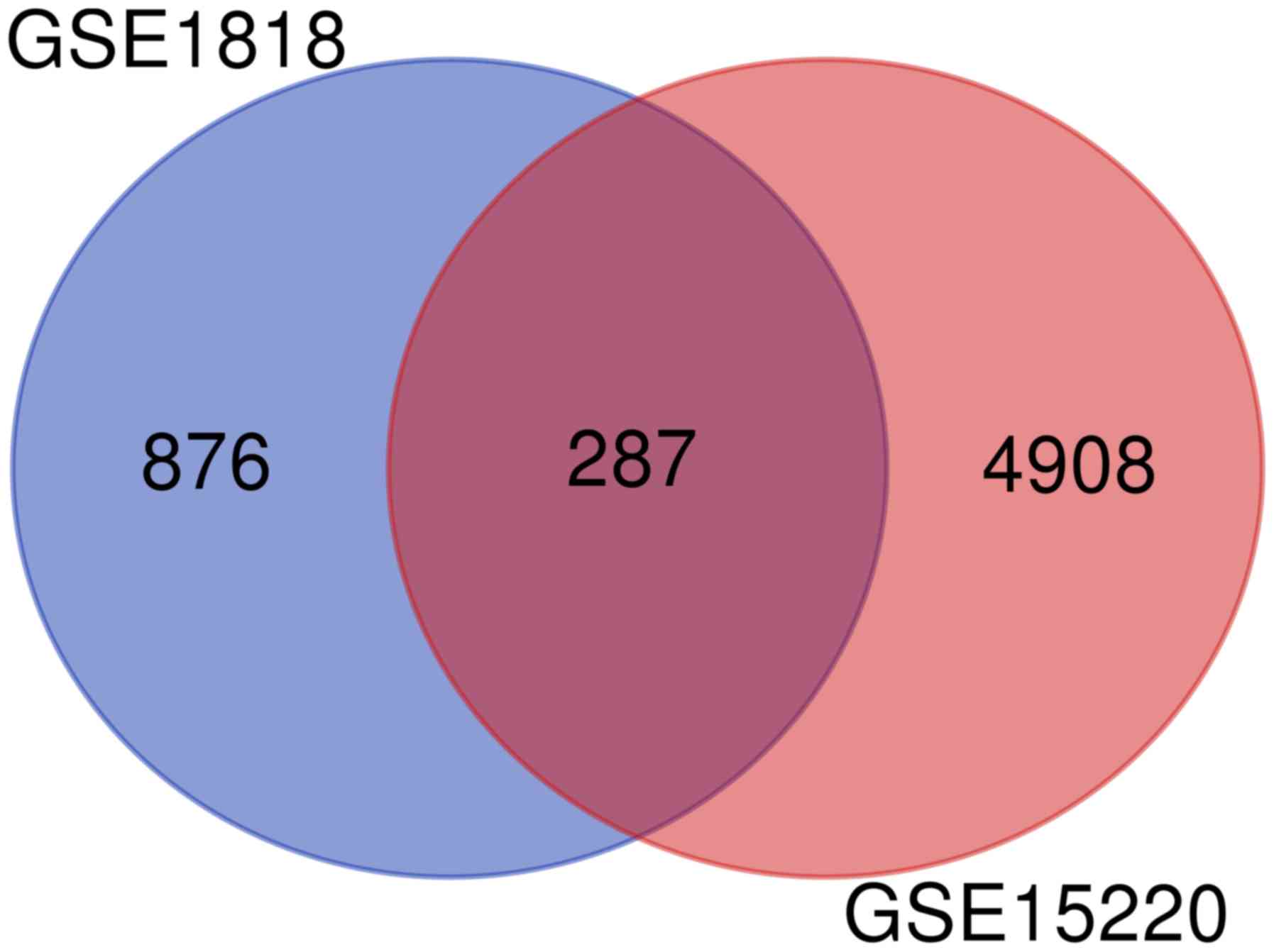
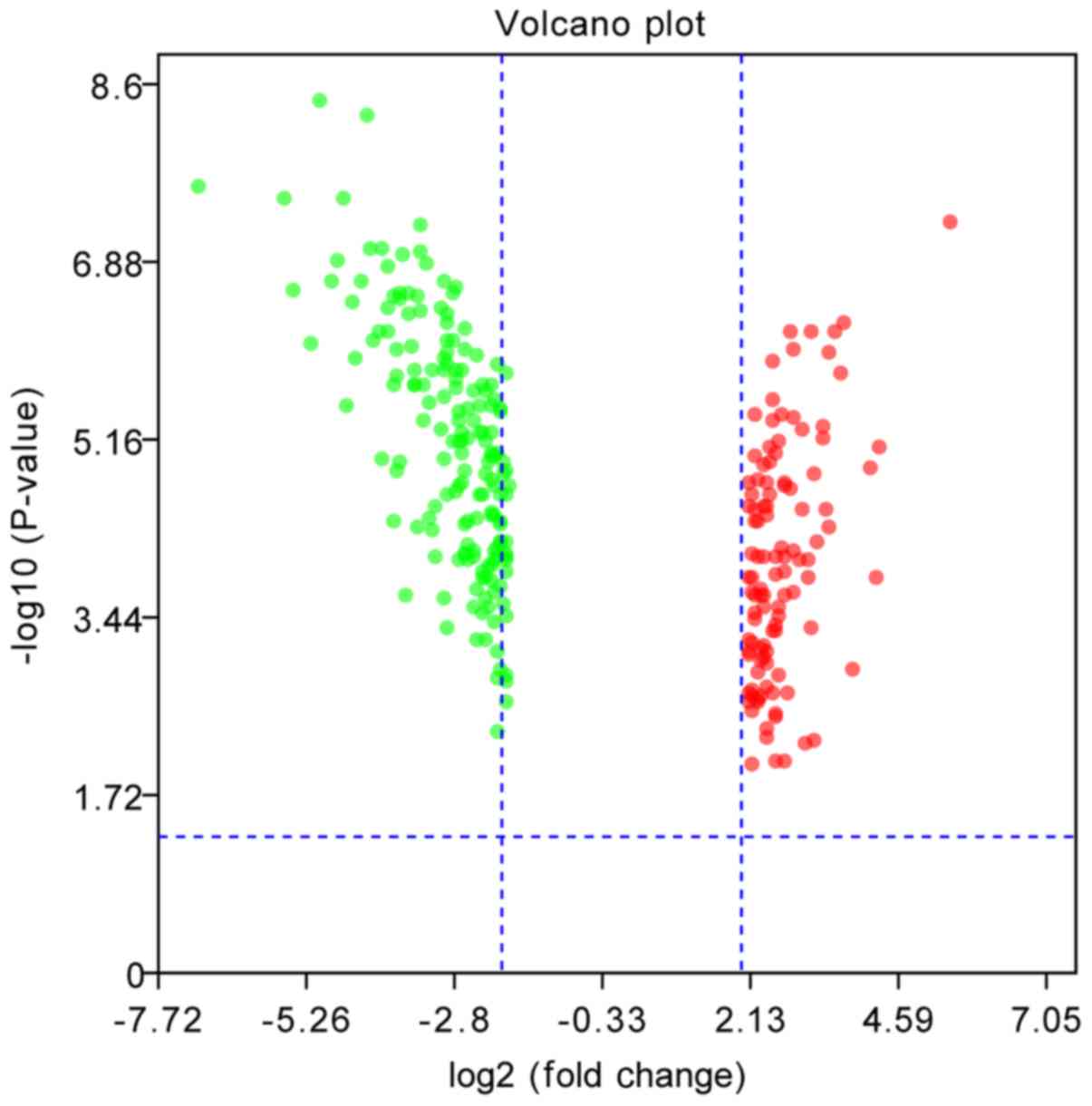
|
1
|
Pedersen MR, Rafaelsen SR, Møller H,
Vedsted P and Osther PJ: Testicular microlithiasis and testicular
cancer: Review of the literature. Int Urol Nephrol. 48:1079–1086.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Smith ZL, Werntz RP and Eggener SE:
Testicular cancer: Epidemiology, diagnosis, and management. Med
Clin North Am. 102:251–264. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ostrowski KA and Walsh TJ: Infertility
with testicular cancer. Urol Clin North Am. 42:409–420. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
McNally RJ, Basta NO, Errington S, James
PW, Norman PD, Hale JP and Pearce MS: Socioeconomic patterning in
the incidence and survival of teenage and young adult men aged
between 15 and 24 years diagnosed with non-seminoma testicular
cancer in northern england. Urol Oncol. 33:506.e9–e14. 2015.
View Article : Google Scholar
|
|
5
|
Sharma P, Dhillon J and Sexton WJ:
Intratubular germ cell neoplasia of the testis, bilateral
testicular cancer, and aberrant histologies. Urol Clin North Am.
42:277–285. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Baird DC, Meyers GJ and Hu JS: Testicular
cancer: Diagnosis and treatment. Am Fam Physician. 97:261–268.
2018.PubMed/NCBI
|
|
7
|
Kamel MH, Elfaramawi M, Jadhav S, Saafan
A, Raheem OA and Davis R: Insurance status and differences in
treatment and survival of testicular cancer patients. Urology.
87:140–145. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shen AH, Howell D, Edwards E, Warde P,
Matthew A and Jones JM: The experience of patients with early-stage
testicular cancer during the transition from active treatment to
follow-up surveillance. Urol Oncol. 34:168.e11–e20. 2016.
View Article : Google Scholar
|
|
9
|
Brand S, Williams H and Braybrooke J: How
has early testicular cancer affected your life? A study of sexual
function in men attending active surveillance for stage one
testicular cancer. Eur J Oncol Nurs. 19:278–281. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
McBride D: Surveillance is as effective as
chemotherapy in stage 1 testicular cancer. ONS Connect.
29:102014.
|
|
11
|
Cheung HH, Lee TL, Davis AJ, Taft DH,
Rennert OM and Chan WY: Genome-wide DNA methylation profiling
reveals novel epigenetically regulated genes and non-coding RNAs in
human testicular cancer. Br J Cancer. 102:419–427. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Skotheim RI, Lind GE, Monni O, Nesland JM,
Abeler VM, Fosså SD, Duale N, Brunborg G, Kallioniemi O, Andrews PW
and Lothe RA: Differentiation of human embryonal carcinomas in
vitro and in vivo reveals expression profiles relevant to normal
development. Cancer Res. 65:5588–5598. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT and
Lin CY: CytoHubba: Identifying hub objects and sub-networks from
complex interactome. BMC Syst Biol. 8 (Suppl 4):S112014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45:W98–W102.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bender JL, Wiljer D, To MJ, Bedard PL,
Chung P, Jewett MA, Matthew A, Moore M, Warde P and Gospodarowicz
M: Testicular cancer survivors' supportive care needs and use of
online support: A cross-sectional survey. Support Care Cancer.
20:2737–2746. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Trabert B, Chen J, Devesa SS, Bray F and
McGlynn KA: International patterns and trends in testicular cancer
incidence, overall and by histologic subtype, 1973–2007. Andrology.
3:4–12. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lobo J, Costa AL, Vilela-Salgueiro B,
Rodrigues Â, Guimarães R, Cantante M, Lopes P, Antunes L, Jerónimo
C and Henrique R: Testicular germ cell tumors: Revisiting a series
in light of the new WHO classification and AJCC staging systems,
focusing on challenges for pathologists. Hum Pathol. 82:113–124.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Russell SS: Testicular cancer: Overview
and implications for health care providers. Urol Nurs. 34:172–176,
192. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Delgado JL, Hsieh CM, Chan NL and Hiasa H:
Topoisomerases as anticancer targets. Biochem J. 475:373–398. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Panvichian R, Tantiwetrueangdet A,
Angkathunyakul N and Leelaudomlipi S: TOP2A amplification and
overexpression in hepatocellular carcinoma tissues. Biomed Res Int.
2015:3816022015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Coleman LW, Perkins SL, Bronstein IB and
Holden JA: Expression of DNA toposiomerase I and DNA topoisomerase
II-alpha in testicular seminomas. Hum Pathol. 31:728–733. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sano K and Shuhin T: A study of
topoisomerase activity in human testicular cancers. Anticancer Res.
15:2117–2120. 1995.PubMed/NCBI
|
|
23
|
Meng H, Chen R, Li W and Xu L and Xu L:
Correlations of TOP2A gene aberrations and expression of
topoisomerase IIα protein and TOP2A mRNA expression in primary
breast cancer: A retrospective study of 86 cases using fluorescence
in situ hybridization and immunohistochemistry. Pathol Int.
62:391–399. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jensen JD, Knoop A, Ewertz M and Laenkholm
AV: ER, HER2, and TOP2A expression in primary tumor, synchronous
axillary nodes, and asynchronous metastases in breast cancer.
Breast Cancer Res Treat. 132:511–521. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Konecny GE, Pauletti G, Untch M, Wang HJ,
Möbus V, Kuhn W, Thomssen C, Harbeck N, Wang L, Apple S, et al:
Association between HER2, TOP2A, and response to
anthracycline-based preoperative chemotherapy in high-risk primary
breast cancer. Breast Cancer Res Treat. 120:481–489. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Labbé DP, Sweeney CJ, Brown M, Galbo P,
Rosario S, Wadosky KM, Ku SY, Sjöström M, Alshalalfa M, Erho N, et
al: TOP2A and EZH2 provide early detection of an aggressive
prostate cancer subgroup. Clin Cancer Res. 23:7072–7083. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xie C, Powell C, Yao M, Wu J and Dong Q:
Ubiquitin-conjugating enzyme E2C: A potential cancer biomarker. Int
J Biochem Cell Biol. 47:113–117. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mo CH, Gao L, Zhu XF, Wei KL, Zeng JJ,
Chen G and Feng ZB: The clinicopathological significance of UBE2C
in breast cancer: A study based on immunohistochemistry, microarray
and RNA-sequencing data. Cancer Cell Int. 17:832017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang R, Song Y, Liu X, Wang Q, Wang Y, Li
L, Kang C and Zhang Q: UBE2C induces EMT through Wnt/β-catenin and
PI3K/Akt signaling pathways by regulating phosphorylation levels of
Aurora-A. Int J Oncol. 50:1116–1126. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Vang T, Miletic AV, Arimura Y, Tautz L,
Rickert RC and Mustelin T: Protein tyrosine phosphatases in
autoimmunity. Annu Rev Immunol. 26:29–55. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Porcu M, Kleppe M, Gianfelici V, Geerdens
E, De Keersmaecker K, Tartaglia M, Foà R, Soulier J, Cauwelier B,
Uyttebroeck A, et al: Mutation of the receptor tyrosine phosphatase
PTPRC (CD45) in T-cell acute lymphoblastic leukemia. Blood.
119:4476–4479. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Menon SS, Guruvayoorappan C, Sakthivel KM
and Rasmi RR: Ki-67 protein as a tumour proliferation marker. Clin
Chim Acta. 491:39–45. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang CK, Yu TD, Han CY, Qin W, Liao XW, Yu
L, Liu XG, Zhu GZ, Su H, Lu SC, et al: Genome-wide association
study of MKI67 expression and its clinical implications in
HBV-related hepatocellular carcinoma in Southern China. Cell
Physiol Biochem. 42:1342–1357. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bleckmann A, Conradi LC, Menck K, Schmick
NA, Schubert A, Rietkötter E, Arackal J, Middel P, Schambony A,
Liersch T, et al: β-catenin-independent WNT signaling and Ki67 in
contrast to the estrogen receptor status are prognostic and
associated with poor prognosis in breast cancer liver metastases.
Clin Exp Metastasis. 33:309–323. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li LT, Jiang G, Chen Q and Zheng JN: Ki67
is a promising molecular target in the diagnosis of cancer
(review). Mol Med Rep. 11:1566–1572. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gallegos I, Valdevenito JP, Miranda R and
Fernandez C: Immunohistochemistry expression of P53, Ki67, CD30,
and CD117 and presence of clinical metastasis at diagnosis of
testicular seminoma. Appl Immunohistochem Mol Morphol. 19:147–152.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhou J, Dai W and Song J: miR-1182
inhibits growth and mediates the chemosensitivity of bladder cancer
by targeting hTERT. Biochem Biophys Res Commun. 470:445–452. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang T, Zhao D, Wang Q, Yu X, Cui Y, Guo
L and Lu SH: MicroRNA-1322 regulates ECRG2 allele specifically and
acts as a potential biomarker in patients with esophageal squamous
cell carcinoma. Mol Carcinog. 52:581–590. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhou C, Cui F, Li J, Wang D, Wei Y, Wu Y,
Wang J, Zhu H and Wang S: MiR-650 represses high-risk
non-metastatic colorectal cancer progression via inhibition of
AKT2/GSK3β/E-cadherin pathway. Oncotarget. 8:49534–49547.
2017.PubMed/NCBI
|
|
40
|
Ningning S, Libo S, Chuanbin W, Haijiang S
and Qing Z: MiR-650 regulates the proliferation, migration and
invasion of human oral cancer by targeting growth factor
independent 1 (Gfi1). Biochimie. 156:69–78. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xu X, Chen H, Zhang Q, Xu J, Shi Q and
Wang M: MiR-650 inhibits proliferation, migration and invasion of
rheumatoid arthritis synovial fibroblasts by targeting AKT2. Biomed
Pharmacother. 88:535–541. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yang Z, Liu Y, Shi C, Zhang Y, Lv R, Zhang
R, Wang Q and Wang Y: Suppression of PTEN/AKT signaling decreases
the expression of TUBB3 and TOP2A with subsequent inhibition of
cell growth and induction of apoptosis in human breast cancer MCF-7
cells via ATP and caspase-3 signaling pathways. Oncol Rep.
37:1011–1019. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Xu J, Gu X, Yang X and Meng Y: MiR-1204
promotes ovarian squamous cell carcinoma growth by increasing
glucose uptake. Biosci Biotechnol Biochem. 1–6. 2018.(Epub ahead of
print).
|
|
44
|
Biermann K, Heukamp LC, Steger K, Zhou H,
Franke FE, Guetgemann I, Sonnack V, Brehm R, Berg J, Bastian PJ, et
al: Gene expression profiling identifies new biological markers of
neoplastic germ cells. Anticancer Res. 27:3091–3100.
2007.PubMed/NCBI
|
|
45
|
Liu R, Zhang W, Liu ZQ and Zhou HH:
Associating transcriptional modules with colon cancer survival
through weighted gene co-expression network analysis. Bmc Genomics.
18:3612017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Collins CM, Malacrida B, Burke C, Kiely PA
and Dunleavy EM: ATP synthase F1 subunits recruited to
centromeres by CENP-A are required for male meiosis. Nat Commun.
9:27022018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Liu F, Cai Y, Rong X, Chen J, Zheng D,
Chen L, Zhang J, Luo R, Zhao P and Ruan J: MiR-661 promotes tumor
invasion and metastasis by directly inhibiting RB1 in non small
cell lung cancer. Mol Cancer. 16:1222017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhu T, Yuan J, Wang Y, Gong C, Xie Y and
Li H: MiR-661 contributed to cell proliferation of human ovarian
cancer cells by repressing INPP5J expression. Biomed Pharmacother.
75:123–128. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Hoffman Y, Bublik DR, Pilpel Y and Oren M:
miR-661 downregulates both Mdm2 and Mdm4 to activate p53. Cell
Death Differ. 21:302–309. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Haldrup C, Kosaka N, Ochiya T, Borre M,
Høyer S, Orntoft TF and Sorensen KD: Profiling of circulating
microRNAs for prostate cancer biomarker discovery. Drug Deliv
Transl Res. 4:19–30. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Okumura T, Shimada Y, Omura T, Hirano K,
Nagata T and Tsukada K: MicroRNA profiles to predict postoperative
prognosis in patients with small cell carcinoma of the esophagus.
Anticancer Res. 35:719–727. 2015.PubMed/NCBI
|
|
52
|
Okumura T, Kojima H, Miwa T, Sekine S,
Hashimoto I, Hojo S, Nagata T and Shimada Y: The expression of
microRNA 574-3p as a predictor of postoperative outcome in patients
with esophageal squamous cell carcinoma. World J Surg Oncol.
14:2282016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Prashad N: miR-665 targets c-MYC and HDAC8
to inhibit murine neuroblastoma cell growth. Oncotarget.
9:33186–33201. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Dong C, Du Q, Wang Z, Wang Y, Wu S and
Wang A: MicroRNA-665 suppressed the invasion and metastasis of
osteosarcoma by directly inhibiting RAB23. Am J Transl Res.
8:4975–4981. 2016.PubMed/NCBI
|
|
55
|
Li K, Pan J, Wang J, Liu F and Wang L:
MiR-665 regulates VSMCs proliferation via targeting FGF9 and MEF2D
and modulating activities of Wnt/β-catenin signaling. Am J Transl
Res. 9:4402–4414. 2017.PubMed/NCBI
|
|
56
|
Li X, Lu Y, Chen Y, Lu W and Xie X:
MicroRNA profile of paclitaxel-resistant serous ovarian carcinoma
based on formalin-fixed paraffin-embedded samples. Bmc Cancer.
13:2162013. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zhou Y, Li XH, Zhang CC, Wang MJ, Xue WL,
Wu DD, Ma FF, Li WW, Tao BB and Zhu YC: Hydrogen sulfide promotes
angiogenesis by downregulating miR-640 via the VEGFR2/mTOR pathway.
Am J Physiol Cell Physiol. 310:C305–C317. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Yanai A, Inoue N, Yagi T, Nishimukai A,
Miyagawa Y, Murase K, Imamura M, Enomoto Y, Takatsuka Y, Watanabe
T, et al: Activation of mTOR/S6K But Not MAPK pathways might be
associated with high Ki-67, ER(+), and HER2(−) breast cancer. Clin
Breast Cancer. 15:197–203. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Dimov ND, Zynger DL, Luan C, Kozlowski JM
and Yang XJ: Topoisomerase II alpha expression in testicular germ
cell tumors. Urology. 69:955–961. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Bo H, Cao K, Tang R, Zhang H, Gong Z, Liu
Z, Liu J, Li J and Fan L: A network-based approach to identify DNA
methylation and its involved molecular pathways in testicular germ
cell tumors. J Cancer. 10:893–902. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Yang Q, Jiang W and Hou P: Emerging role
of PI3K/AKT in tumor-related epigenetic regulation. Semin Cancer
Biol. Apr 2–2019.(Epub ahead of print). View Article : Google Scholar
|
|
62
|
Nusse R and Clevers H: Wnt/β-catenin
signaling, disease, and emerging therapeutic modalities. Cell.
169:985–999. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Fruman DA and Rommel C: PI3K and cancer:
Lessons, challenges and opportunities. Nat Rev Drug Discov.
13:140–156. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Rajpert-De Meyts E, McGlynn KA, Okamoto K,
Jewett MA and Bokemeyer C: Testicular germ cell tumours. Lancet.
387:1762–1774. 2016. View Article : Google Scholar : PubMed/NCBI
|