Introduction
The conidia of Aspergillus fumigatus (A.
fumigatus), a common fungus, are ~2–3 µm in size and easily
spread by wind (1). Humans can
inhale 100s of conidia every day, and conidia can also penetrate
into the alveoli via mucociliary clearance in lung (2). Hosts with healthy immune functions
can phagocytose or kill conidia via alveolar macrophages, thus
infections rarely occur. However, once the body's immune function
is impaired, invasive pulmonary aspergillosis (IPA) occurs and
gradually spreads to the kidneys, brain and other organs; IPA
exhibits a mortality rate as high as 50–90% (3). A. fumigatus is the second
leading cause of invasive fungal diseases worldwide (4).
The lungs are the initial organs that the A.
fumigatus would first come into contact with. Airway epithelial
cells (including lung epithelial cells and tracheal epithelial
cells) are the site of the initial process of A. fumigatus
conidia. Alveolar epithelial cells are composed of alveolar
epithelial type I and type II cells, and inhaled conidia mainly
interact with type II cells, which maintain the alveolar space
(5). An In vitro study has
demonstrated that A. fumigatus conidia first adhere to the
surface of type II lung epithelial cells and are then endocytosed
by lung epithelial cells, which triggers an immune response and
respiratory burst (6). The
surviving conidia remain and germinate into hyphae, causing
infection under suitable conditions (7). The interaction between A.
fumigatus conidia and type II lung epithelial cells serves an
important role in disease progression (6). The present study aimed to explore the
molecular mechanism of the interaction between them.
Isobaric tags for relative and absolute quantitation
(iTRAQ) are an isotopically labeled relative and absolute
quantification technique (8).
Combined with liquid chromatography-mass spectrometry (LC/MS), it
has advantages over traditional protein quantification. iTRAQ is
able to mark most of the proteins in a sample, and the labeling
process is simple, more timesaving and more accurate. At the same
time, this method enables quantitative comparison of proteins in
multiple samples in the same test to obtain a large amount of
protein information.
In the present study, A549 cells were explored as an
in vitro cell model. The proteomes of A549 type II lung
epithelial cells with and without A. fumigatus challenge
were compared using the isobaric tags for relative and absolute
quantification (iTRAQ) technique. Additionally, the functions of
significantly differentially expressed proteins, such as N-myc
downstream-regulated gene 1 (NDRG1) and CD44, were discussed.
Materials and methods
Culture of A. fumigatus strain
IFM40808 and A549 type II lung epithelial cells
A. fumigatus strain IFM40808 was provided by
the Medical Mycology Research Center of Chiba University (Chiba,
Japan). The strain was cultured on potato glucose medium (Difco; BD
Biosciences) for 7 days at 37°C with 5% CO2. The conidia
suspension was collected after being flushed with sterile 0.05%
Tween-20 in 50 mM PBS (pH 7.4) and the conidia concentration was
then adjusted to 2×107 cfu/ml with RPMI-1640 medium
(Gibco; Thermo Fisher Scientific, Inc.).
A549 human type II lung epithelial cells were
purchased from American Type Culture Collection. Cells were
cultured in RPMI-1640 medium containing 10% fetal bovine serum
(Gibco; Thermo Fisher Scientific, Inc.), 50 IU/ml penicillin and 50
µg/ml streptomycin at 37°C with 5% CO2.
Co-incubation of A. fumigatus conidia
and A549 lung epithelial cells
The A549 cell concentration was adjusted to
2×106 cells/ml and the cells were transferred to dishes
in medium without fetal bovine serum, following which culture was
continued for 16–18 h. The A549 cells were infected by A.
fumigatus conidia at an MOI of 10 (i.e., 10 conidia to 1 cell)
and cocultured for 6 h after infection in serum-free medium at 37°C
with 5% CO2.
Protein extraction, digestion and
labelling with iTRAQ reagents
The supernatant following co-incubation was
aspirated and washed with pre-cooled PBS at 4°C. The proteins were
extracted in accordance with the methods of Vuong et al
(9) and She et al (10). Cell total proteins were extracted
by grinding with liquid nitrogen and suspending in lysis buffer [8
M urea, 1% Triton X-100, 65 mM dithiothreitol (DTT) Sigma-Aldrich;
Merck KGaA) and 0.1% protease inhibitor cocktail (Sigma-Aldrich;
Merck KGaA). Proteins were then sonicated on ice five times (5 mins
for one time, and 30 sec on, 30 sec off) for five cycles using
ultrasound sonicator (Ningbo Scientz Biotechnology Co., Ltd.).
Lysates were centrifuged at 5,000 × g at 4°C for 3 min and the
supernatant was collected. The proteins were precipitated with cold
15% Trichloroacetic acid (Thermo Fisher Scientific, Inc.) for 2 h
at −20°C. The precipitate were collected via centrifugation at
15,000 × g for 10 min at 4°C and rinsed three times with cold
acetone. The air-dried protein pellet was dissolved in lysis buffer
(8 M urea, 100 mM Triethylamine-carbonic acid buffer, pH 8.0) and
the protein concentration was determined using a 2-D Quant kit
(Cytiva), and then adjusted to 2 mg/ml.
Proteolysis and peptide labelling were carried out
in accordance with the methods of She et al (10) and Pizzatti et al (11). For proteolysis, the protein
solution was treated with 10 mM DTT for 1 h at 37°C and 20 mM
iodoacetamide (Sigma-Aldrich; Merck KGaA) for 45 min. The solution
was diluted with TEAB to obtain a final solution containing 1 M
urea. Then, the solution was digested with trypsin (dilution, 1:50)
overnight. Trypsin (Promega Corporation) was added at 1:50 for
digestion overnight and 1:100 for a second 4 h-digestion. Following
enzymatic hydrolysis, the obtained peptides were desalted using a
Strata-X C18 solid phase extraction column (Phenomenex Inc.). After
vacuum freeze-drying, the two samples were labelled according to
the instructions of the iTRAQ Reagent8-Plex kit (AB Sciex Pte.,
Ltd.).
Inspection by LC/MS
The peptides from the two groups were separated
using a Waters 600E reversed-phase high-performance liquid
chromatography system with an Agilent 300 Extend C18 column (5 µm
particle size, 4.6 mm ID, 250 mm length; Agilent Technologies,
Inc.). The column was maintained at room temperature and the flow
rate was 1 ml/min. Briefly, the peptide mixture was first separated
with a gradient of 2–60% acetonitrile in 10 mM ammonium bicarbonate
(pH 10), over 80 min into 80 fractions. The fractions were combined
into eight components and vacuum-dried. The analysis was performed
using an LTQ Orbitrap Elite LC/MS (Thermo Fisher Scientific, Inc.)
equipped with a Dionex UltiMate 3000 nano-UPLC system (Thermo
Fisher Scientific, Inc.). A guard column (C18 PepMap 100, 300 µm ×
1 mm, 5 µm, 100 Å) and an analytical column (Acclaim PepMap C18, 15
cm × 75 µm, 2 µm, 100 Å; Dionex;Thermo Fisher Scientific, Inc.)
were applied. Mobile phase A (ultra-pure water, 0.1% formic acid;
Honeywell Fluka) at a gradient of ~6–44% was used during the run,
followed by mobile phase B [98% acetonitrile (Thermo Fisher
Scientific, Inc.), 0.1% formic acid] at a gradient of 44–98%. After
one cycle had ended, the column was equilibrated with 98% mobile
phase B followed by 6% mobile phase A. The column was maintained by
equilibrating with 6% mobile phase A for 5 min before the next
injection. The technical parameters were in accordance with the
settings of Zhang et al (12). The parameters for MS1 were set as
follows: Cation-positive mode; Orbitraq resolution: 60,000; scan
range, 400 m/z. The parameters for MS2 were set as: Orbitrap
resolution: 15,000; activation type, High Energy Collision
Dissociation; normalized collision energy setting, 35%. The working
conditions were: Nitrogen temperature 350°C at a flow rate of 12
l/min, nebulizer pressure 60 psi and electrospray capillary voltage
+3,000 V.
Protein database alignment and
relative quantification by iTRAQ
The obtained raw protein data were aligned using the
SEQUEST engine built into Proteome discoverer software version 2.0
(Thermo Fisher Scientific, Inc.). The Human International Protein
Index (IPI) protein database (IPI v3.87; ftp://ftp.ebi.ac.uk/pub/databases/IPI; a total of
91,464 records) was referenced for analysis.
Function analysis of differentially
expressed proteins
Data on differentially expressed proteins were
entered into the Database for Annotation, Visualization and
Integrated Discovery (version 6.7; http://david.abcc.ncifcrf.gov/). Gene Ontology (GO;
http://wego.genomics.org.cn/cgi-bin/wego/index.pl) and
Kyoto Encyclopaedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/) analyses were applied for
functional prediction of the obtained differentially expressed
proteins. The obtained GO terms and KEGG pathways were considered
enriched in the differentially expressed proteins when P<0.05 as
determined by tests of statistical significance. Protein
interaction network analysis of differentially expressed proteins
was performed using the protein interaction network online database
Search Tool for the Retrieval of Interacting Genes/Proteins
(STRING; version 11.0; http://string.embl.de). The graphs were visualized
using Cytoscape 2.8 (http://www.cytoscape.org/) (13).
Western blot analysis
The iTRAQ results obtained from the infected group
and the control group were verified by western blotting. Following
infection with A. fumigatus conidia at an MOI of 10 for 6 h,
the infected A549 cells were collected. The protein in cells was
extracted using RIPA buffer (Beyotime Institute of Biotechnology)
and evaluated using the BCA Protein Assay kit (Beyotime Institute
of Biotechnology). Equal amounts (40 ng) of protein were separated
via 12% SDS-PAGE and transferred to PVDF membrane (EMD Millipore).
The membrane was then blocked with 5% nonfat milk powder in PBS
with 0.1% (v/v) Tween (PBST) for 1 h at room temperature, followed
by rabbit anti-human primary antibodies (dilution, 1:500) against
N-myc downstream-regulated gene 1 (NDRG1; cat. no. 9485S, Cell
Signaling Technology, Inc.), isoform 1 of STE20-like
serine/threonine-protein kinase (SLK; cat. no. 41255S; Cell
Signaling Technology, Inc.) and CD44 (cat. no. 37259S; Cell
Signaling Technology, Inc.), and the internal reference protein
GAPDH (cat. no. AF7021; dilution, 1:1,000; Affinity Biosciences) at
4°C overnight. The membrane was incubated with goat anti-rabbit IgG
(H+L)-HRP (cat. no. S0001; dilution, 1:5,000; Affinity Biosciences)
secondary antibody at room temperature for 1 h. The expression
levels of target proteins were detected by ECL (PerkinElmer, Inc.).
Proteins were observed and images were captured with a
chemiluminescence gel imager. Grayscale analysis was performed
using ImageJ software (Version 1.52a; National Institutes of
Health).
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
A549 cells (1×106) with or without A.
fumigatus infection were harvested and the total RNA extraction
performed with TRIzol® (Invitrogen; Thermo Fisher
Scientific, Inc.). The total RNA (1 µg) was then reverse
transcribed into cDNA using the TransCript One-step gDNA Removal
and cDNA Synthesis SuperMix kit (Beijing Transgen Biotech Co.,
Ltd.) according to the manufacturer's instructions. qPCR was
conducted using the SYBR Green PCR Master Mix (Roche Diagnostics,
Inc.) in a final reaction volume of 20 µl. Cyclic parameters of
qPCR were as follows: Initial denaturation temperature at 95°C for
5 min, followed by 40 cycles of 15 sec denaturation at 95°C, 30 sec
annealing at 60°C and 45 sec extension at 72°C. Specific primer
sequences are listed in Table I.
The ΔΔCq method was used to calculate relative
transcript levels (14). Relative
gene expression was normalized to the expression levels of GAPDH
and the experiment was repeated three times.
 | Table I.Primers for reverse
transcription-quantitative PCR. |
Table I.
Primers for reverse
transcription-quantitative PCR.
| Gene | Forward primer
sequence (5′-3′) | Reverse primer
sequence (5′-3′) | Amplicon
size(bp) |
|---|
| NDRG1 |
GTCCTTATCAACGTGAACCCTT |
GCATTGGTCGCTCAATCTCCA | 250 |
| SLK |
CAATGCGAGTGCTGCTAAAAAT |
CGGATGGGTTTGTTGGAATCA | 184 |
| CD44 |
CTGCCGCTTTGCAGGTGTA |
CATTGTGGGCAAGGTGCTATT | 109 |
| GAPDH |
TGTGGGCATCAATGGATTTGG |
ACACCATGTATTCCGGGTCAAT | 116 |
RNA interference
A549 cells were transfected with small interfering
RNAs (siRNAs) using siTRAN 1.0 transfection reagent (cat. no.
TT300001; OriGene Technologies, Inc.) (15). The siRNAs targeted to the
CD44 (cat. no. SR319622) and NDRG1 (cat. no.
SR307046) gene, and scrambled negative control siRNA (cat. no.
SR30004) were purchased from OriGene Technologies, Inc. The sense
sequence of siCD44 5′-GAACGAAUCCUGAAGACAUCU-3′ and siNDRG1
5′-GCUGAAGCUCGUCAGUUCACCAUCC-3′ were used in studies. The procedure
was performed according to OriGeneproduct manuals. Briefly, 5 µl
siRNA and 20 µl transfection reagent were mixed with 160 µl
Opti-MEM I Reduced Serum Medium (Invitrogen; Thermo Fisher
Scientific, Inc.). Then, the mixture was incubated at room
temperature for 10 min before being added to 6-well plates
containing cells at confluence 60–70% for 48 h. Cells were then
subjected to further assays. Western blotting was performed to
confirm CD44 and NDRG1 siRNA-mediated knockdown.
Internalization
The pathway by which A. fumigatus is
internalized before infecting A549 cells was studied by a nystatin
protection assay according to the method of Bao et al
(16). A549 cells transfected with
CD44 or NDRG1 siRNA were seeded in 24-well plates at a density of
2×104 cells/well 24 h prior to the experiment (Nest
Biotech Co., Ltd.). The cells were incubated with A.
fumigatus conidia at a MOI of 10 for 1 h at 37°C with 5%
CO2. Then, the cells were washed three times with PBST
and incubated with nystatin in DMEM at 37°C for 4 h. Subsequently,
monolayer cells were washed three times with PBST and lysed by
incubation in 0.25% Triton X-100 (BBI Life Sciences Corp.) diluted
in PBS for 15 min. The lysate was then serially diluted and plated
on PDA plates (three replicate plates/well) and incubated at 37°C
for 4 days before individual colony forming units were counted. The
colonies were counted to determine the total intracellular conidia.
The conidia internalization rate was calculated based on the
formula: Conidia internalization rate = the total intracellular
conidia/the initial inoculum ×100%.
Determination of cytokine levels
A549 cells transfected with CD44 or NDRG1 siRNAwere
seeded in 6-well plates at a density of 2×106 cells/well
24 h prior to the experiment (Nest Biotech Co., Ltd.). The cells
were incubated with A. fumigatus conidia at a MOI of 10 for
12 h at 37°C. The supernatants were collected and centrifuged at
1,500 × g at 4°C for 15 min twice. IL-6 (cat. no. 88-7066-22;
Thermo Fisher Scientific, Inc.), IL-8 (cat. no. 88-8086-22; Thermo
Fisher Scientific, Inc.), TNF-α (cat. no. 88-7346-22; Thermo Fisher
Scientific, Inc.) and IL-1β (cat. no. 88-7261-22; Thermo Fisher
Scientific, Inc.) were assayed with ELISA kits according to the
manufacturer's protocols.
Statistical analysis
All data are presented as the mean ± SD of at least
three replicate measurements. Mean values were compared using
unpaired t-tests (two groups), followed by Bonferroni correction
for multiple comparisons using one-way ANOVA and Tukey's post hoc.
All statistical tests were performed using Prism software (v5.0;
GraphPad Software, Inc.). P<0.05 was considered to indicate a
statistically significant difference.
Results
Data analysis of the proteome of A549
cells infected with A. fumigatus
A total of 1,582 proteins were obtained after
comparing the proteomes of A549 cells with or without A.
fumigatus infection determined by iTRAQ labelling with the
human protein database. A total of 111 differentially expressed
proteins were identified, among which 18 were upregulated (>1.5)
and 93 were downregulated (<0.75; Table SI), including: NDRG1 (fold
change,1.88; function, Rab GTPase binding); proteasome subunit α
type-1 (1.74; threonine-type endopeptidase activity); SLK (1.69;
protein serine/threonine kinase activity and adenyl
ribonucleotide-binding ability); CD44 (0.47; primarily cytokine
receptor activity and hyaluronic glucosaminidase activity),
RNA-binding protein EWS isoform 1 (0.53; zinc ion binding). NDRG1
and CD44 were the most significantly upregulated and downregulated
genes, respectively.
GO annotation and enrichment analyses of the
differentially expressed proteins were performed from three
perspectives: Biological process (BP), cellular component (CC) and
molecular function (MF). Fig. 1
shows the top 20 most significantly enriched GO terms in the three
categories (BP, CC and MF) determined by enrichment analysis. The
aforementioned differentially expressed proteins were found to be
involved in 1,169 biological processes (P<0.05; Table SII). The BPs enriched in the
largest number (n) of proteins were ‘single-organism process’
(n=96), ‘single-organism cellular process’ (n=91), ‘cellular
metabolic process’ (n=86), ‘single-organism metabolic process’
(n=66), ‘cellular component organization or biogenesis’ (n=51) and
‘response to stress’ (n=40). These differentially expressed
proteins in A549 cells in the early stage of A. fumigatus
infection compared with uninfected A549 cells are primarily related
to biological processes, such as cell metabolism, synthesis and the
cellular stress response. The differentially expressed proteins
were primarily associated with binding-related functions, including
‘protein binding’ (n=75), ‘organic cyclic compound binding’ (n=55)
and ‘heterocyclic compound binding’ (n=54), and catalytic
functions, including ‘hydrolase activity’ (n=29), ‘oxidoreductase
activity’ (n=17) and ‘transferase activity’ (n=13; Table SIII). The majority of these
proteins were located in ‘membrane-bounded organelle’ (n=105),
‘intracellular organelle’ (n=100), ‘intracellular membrane-bounded
organelle’ (n=98) and ‘cytoplasm’ (n=96), or ‘intracellular
organelle lumen’ (n=60; Table
SIV).
NDRG1, SLK and CD44 were the three proteins with the
most significant differences in expression and primary function.
These proteins are distributed in multiple parts of the cell and
are involved in mainly biological processes, such as ‘biological
regulation’ and ‘response to stimulus’.
Results of KEGG pathway analysis
KEGG pathway enrichment analysis was performed for
the 111 differentially expressed proteins using the KEGG pathway
database. The results revealed 126 pathways primarily enriched in
the differentially expressed proteins of A549 cells infected with
A. fumigatus (Table SV). A
total of 19 pathways (P<0.05) were obtained and the top 10
pathways were presented in Fig. 2.
These pathways mainly concerned the ‘TCA cycle’, ‘glyoxylate and
dicarboxylate metabolism’, ‘pyruvate metabolism’, ‘carbon
metabolism’ and ‘protein processing in endoplasmic reticulum’. The
involved differential proteins had lower expression levels.
Interaction analysis of differentially
expressed proteins
To further understand the protein-protein
interactions between differentially expressed proteins, they were
submitted to the STRING database (http://string.embl.de/) and subjected to analysis by
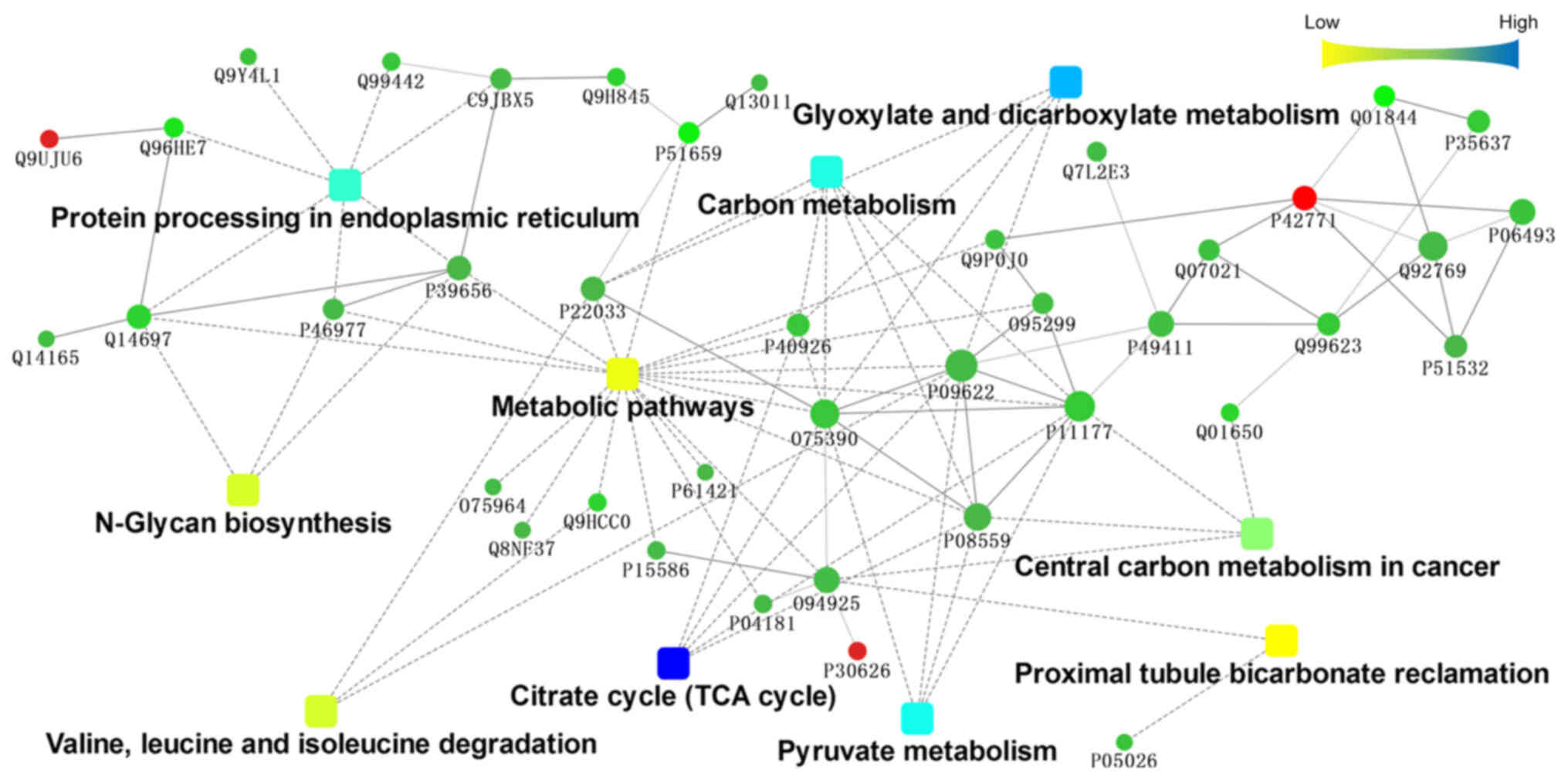
Cytoscape software (http://www.cytoscape.org/). Fig. 3 illustrated the interaction network
among the top 10 KEGG pathways significantly enriched in the
involved proteins. As indicated in the figure, pathways related to
metabolism, such as ‘metabolic pathways’, ‘carbon metabolism’ and
‘central carbon metabolism in cancer’, were at the centre of the
interaction network. Citrate synthase, mitochondrial, isoform 1 of
pyruvate dehydrogenase E1 component subunit β, mitochondrial,
pyruvate dehydrogenase E1 component subunit α, somatic form,
mitochondrial isoform 2 precursor, dihydrolipoyl dehydrogenase,
mitochondrial and malate dehydrogenase, mitochondrial were found to
be involved in multiple metabolic pathways. These molecules are
located mainly in the mitochondria.
Validation of the identified proteins
and their quantification by western blotting and RT-qPCR
Western blotting and qPCR were employed to verify
the accuracy of iTRAQ-mediated detection. NDRG1, SLK and CD44
levels in A549 cells infected with A. fumigatus determined
by western blot analysis were consistent with the results of iTRAQ
detection (Fig. 4A). The mRNA
levels were consistent with their protein levels (Fig. 4B for protein and 4C for mRNA).
Knockdown of NDRG1 affects the
internalization efficiency of A549 cells for A. fumigatus
To evaluate the role of NDRG1 and CD44
in the internalization of A. fumigatus by A549 cells, siRNAs
were employed to knock down NDRG1 and CD44
expression. The results demonstrated that the internalization
efficiency was significantly lower following NDRG1-knockdown
compared with the control group (P=0.0057; Fig. 5A and B). The internalization
efficiency tended to increase following CD44-knockdown, but
this difference was not statistically significant (P=0.2655;
Fig. 5C and D).
Knockdown of CD44 reduces the
secretion of IL-8 and IL-6 from A549 cells
To further investigate the effect of NDRG1
and CD44 in A549 cells on the expression of cytokines in
response to A. fumigatus infection, ELISA was performed. The
results demonstrated that the presence of A. fumigatus led
to the increased secretion of IL-8, IL-6, TNF-α and IL-1β from A549
cells. IL-6 (P=0.0461) and IL-8 (P=0.0014) were decreased to
different extents after CD44 knockdown, but there was no
significant change in IL-1β and TNF-α levels. However, after
NDRG1 was knocked down, only IL-8 secretion was reduced
(P=0.0096) and the expression of the other three cytokines examined
remained constant (Fig. 6).
 | Figure 6.NDRG1 and CD44
knockdown affects the expression levels of cytokines. A549 cells
were incubated with A. fumigatus conidia at a multiplicity
of infection of 10 for a total of 12 h. Differences in the
expression levels of (A) IL-8, (B) IL-6, (C) TNF-α and (D) IL-1β in
the cell supernatants of the uninfected group (Con), the A.
fumigatus conidia-infected group (AF), the NC siRNA-transfected
group (AF and NC siRNA), the NDRG1-knockdown and A.
fumigatus conidia-infected group (AF and NDRG1 siRNA) and the
CD44-knockdown and A. fumigatus conidia-infected group (AF
and CD44 siRNA) were determined. Data are presented as the mean ±
standard error of the mean (n=6). Data analysis was performed using
one-way ANOVA and Tukey's post hoc multiple comparison tests.
*P<0.05, **P<0.01 and ***P<0.001 vs. AF group;
#P<0.05, ##P<0.01 and
###P<0.001 vs. Control group. A. fumigatus,
Aspergillus fumigatus; IL, interleukin; TNF-α, tumor necrosis
factor α; NC, negative control; siRNA, small interfering RNA. |
Discussion
The successful clearance of A. fumigatus
conidia from the lungs depends on immune cells, such as
monocytes/macrophages, neutrophils and airway epithelial cells,
including bronchial epithelial cells and alveolar epithelial cells
(17,18). Additionally, epithelial cells serve
an important role in clearing A. fumigatus (19). Studies have demonstrated that
conidia adhere to the surface of A549 cells incubated with A.
fumigatus conidia for 2 h with the assistance of RodA and CspA,
surface components of the cell wall (3,20).
The internalization of the conidia by the cells at 4 h after
infection is mediated by the activation of pattern recognition
receptors, such as dectin-1, mannose receptors, or Toll-like
receptors on endothelial cells (3). When co-incubated for 4 h, the conidia
begin to swell after being taken up by the endothelial cells to
avoid being killed by immune cells and to ensure their survival
(21). After 6–8 h post-infection,
the conidia on the epithelial cells are used as ‘kindling’ that
eventually stimulates germination and the formation of germ tubes
(20), although the survival rate
of conidia on the epithelial cells decreases sharply. In the
present study, the iTRAQ method was used to observe changes in the
proteome of A549 cells following infection with A. fumigatus
conidia for 6 h and the molecular mechanism of A. fumigatus
conidia infection, and excluded the effects of conidia swelling and
the germination of germ tubes.
GO enrichment and KEGG pathway analyses revealed
that most differentially expressed proteins were involved in basic
biological processes, including cell energy metabolism, substance
metabolism and cell synthesis. The expression levels of some
proteins related to cytoskeletal rearrangement, such as stathmin
(fold change of 1.67), CAP-Gly domain containing linker protein 1
(1.66), protein phosphatase 1 regulatory subunit 12A (1.54),
kinesin family member 2A (0.71) and cyclin-dependent kinase 1
(0.69), changed with A. fumigatus infection. These molecules
are mainly involved in the upstream regulatory process. Chen et
al (21) analyzed changes in
the transcriptome of A549 cells following infection with A.
fumigatus conidia for 8 h and reported that expression of the
molecules leukocytespecific transcript 1, activity-regulated
cytoskeleton-associated protein, early growth response protein
(EGR)1 and EGR4, which are involved in the rearrangement of the
cytoskeleton, change when A549 cells are infected with A.
fumigatus conidia for 8 h. The results of the present study
indicated changes in molecular regulation at 6 h following
infection in preparation for endocytosis.
The molecules cytochrome b-245 light chain (0.7),
cathepsin S (0.7) and complement component 1 Q subcomponent-binding
protein (0.7), which may participate in the processing of
endocytosed antigens, reactive oxygen species production, the
innate immune response, and antigen processing and presentation,
are suppressed by A. fumigatus infection (Tables SI–SIV). Notably, the above proteins were
associated with Toll-like receptor signaling pathways and
RIG-I-like receptor signaling pathways, which indicated that these
pathways were involved in A. fumigatus infection. The
findings of the present study were consistent with the
transcriptome and metabolome of A. fumigatus-infected human
dendritic cells (22). Thus, the
function of non-C type lectin-like pattern recognition receptors
deserves more attention in studies of the interaction between A.
fumigatus conidia and lung epithelial cells.
NDRG1, a member of the NDRG family (NDRG 1–4), is
expressed in epithelial cells (23). NDRG1 is involved in multiple cell
biological processes, such as cell proliferation, differentiation
and development (24). However,
the role of NDRG1 in viral infection is ambiguous. NDRG1 limits the
life cycle of hepatitis C virus (25) but promotes the replication of
influenza A virus (23). The
present study identified that the infection of A549 cells with
A. fumigatus significantly increased the protein expression
of NDRG1, which serves an important role in maintaining the
integrity of the lung epithelial cell barrier (26). However, whether elevated NDRG1
expression on epithelial cells is helpful for A. fumigatus
conidia invasion or the body's defense remains to be elucidated.
The internalization efficiency was reduced when the NDRG1
gene was silenced, but NDRG1 silencing had little effect on
the expression levels of cytokines. This indicated that NDRG1 may
have a positive regulatory effect on the rearrangement of
cytoskeletal proteins, thereby promoting A. fumigatus
conidia internalization.
CD44 is a transmembrane adhesion molecule with has
hyaluronic glucosaminidase activity and cytokine receptor activity.
It is widely expressed in a number of cells and can recognize a
variety of ligands, including hyaluronan (HA), collagen, laminin,
fibronectin, osteopontin and other extracellular proteins involved
in cell adhesion, growth, proliferation and motility (27–29).
Studies have demonstrated that CD44 can promote or inhibit
microbial infection depending on the type of microorganism and the
initial infection site (30,31).
CD44-deficient mice have prolonged survival rates but elevated lung
inflammation compared with wild-type mice following infection with
a lethal dose of Klebsiella pneumoniae by intranasal
administration (31). However, the
effect of Streptococcus equi subsp. Zooepidemicus
infection is in contrast to that of Klebsiella pneumoniae
infection (31). CD44-knockout
mice survive longer and have a lower fungal burden in the brain and
cerebrospinal fluid than wild-type mice following lateral venule
injection of Cryptococcus neoformans (32). This may be because HA on the
surface of Cryptococcus neoformans binds CD44 on the surface
of human brain microvascular endothelial cells (33). It is known that CD44 promotes cell
adhesion and invasion (32,33).
In the present study, the inhibition of CD44 expression may be
caused by the absence of CD44-related ligands on the cell wall
surface of A. fumigatus. Qadri et al (34) demonstrated that CD44-knockdown
reduces NF-κB nuclear translocation and downstream proinflammatory
cytokines production. Therefore, the present study assayed the
expression levels of proinflammatory cytokines following
transfection with CD44 siRNA. This demonstrated that the expression
levels of IL-6 and IL-8 were reduced, which indicated that A.
fumigatus conidia could inhibit the inflammatory reaction in
epithelial cells via CD44. However, CD44 does not affect the
internalization of A. fumigatus conidia, which is in
contrast to Cryptococcus neoformans (32). This phenomenon might be related to
the surface content of the fungal cell wall. The study of CD44 and
its role in the extracellular matrix in fungal infection would be
of great interest.
In summary, to the best of our knowledge, proteomic
data on the interaction between A. fumigatus conidia and
A549 alveolar epithelial cells have been acquired for the first
time and a number of differentially expressed proteins, including
NDRG1 and CD44, were identified. In addition, NDRG1 and CD44 were
found to affect endocytosis and cytokine expression in A549 cells,
respectively, and these proteins may serve as diagnostic targets
and may be involved in treatment regimens for invasive pulmonary
aspergillosis.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
The authors would like to thank Professor Koji
Yokoyama, from Medical Mycology Research Center of Chiba University
(Chiba, Japan), for providing the A. fumigatus strain for
the current research.
Funding
The present study was supported in part by the
National Natural Science Foundation of China (grant no. 81271802)
and the National Science and Technology Major Project of the
Ministry of Science and Technology of China (grant no.
2013ZX10004612-006).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
XZ performed the laboratory experiments and drafted
the manuscript. DH analyzed the experimental data and revised the
manuscript. SG and YW participated in laboratory experiments. LW
designed the project and supervised the present study. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Latgé JP: The pathobiology of
Aspergillus fumigatus. Trends Microbiol. 9:382–389. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ogorman CM: Airborne Aspergillus
fumigatus conidia: A risk factor for aspergillosis. Fungal Biol
Rev. 25:151–157. 2011. View Article : Google Scholar
|
|
3
|
Croft CA, Culibrk L, Moore MM and Tebbutt
SJ: Interactions of Aspergillus fumigatus Conidia with
airway epithelial cells: A critical review. Front Microbiol.
7:4722016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bassetti M, Garnacho-Montero J, Calandra
T, Kullberg B, Dimopoulos G, Azoulay E, Chakrabarti A, Kett D, Leon
C, Ostrosky-Zeichner L, et al: Intensive care medicine research
agenda on invasive fungal infection in critically ill patients.
Intensive Care Med. 43:1225–1238. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Toor A, Culibrk L, Singhera GK, Moon KM,
Prudova A, Foster LJ, Moore MM, Dorscheid DR and Tebbutt SJ:
Transcriptomic and proteomic host response to Aspergillus
fumigatus conidia in an air-liquid interface model of human
bronchial epithelium. PLoS One. 13:e02096522018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jia X, Chen F, Pan W, Yu R, Tian S, Han G,
Fang H, Wang S, Zhao J, Li X, et al: Gliotoxin promotes
Aspergillus fumigatus internalization into type II human
pneumocyte A549 cells by inducing host phospholipase D activation.
Microbes Infect. 16:491–501. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Paris S, Boisvieux-Ulrich E, Crestani B,
Houcine O, Taramelli D, Lombardi L and Latgé JP: Internalization of
Aspergillus fumigatus conidia by epithelial and endothelial
cells. Infect Immun. 65:1510–1514. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ye F, Zhang H, Yang YX, Hu HD, Sze SK,
Meng W, Qian J, Ren H, Yang BL, Luo MY, et al: Comparative proteome
analysis of 3T3-L1 adipocyte differentiation using iTRAQ-coupled 2D
LC-MS/MS. J Cell Biochem. 112:3002–3014. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Vuong NQ, Goegan P, Mohottalage S, Breznan
D, Ariganello M, Williams A, Elisma F, Karthikeyan S, Vincent R and
Kumarathasan P: Proteomic changes in human lung epithelial cells
(A549) in response to carbon black and titanium dioxide exposures.
J Proteomics. 149:53–63. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
She X, Zhang P, Gao Y, Zhang L, Wang Q,
Chen H, Calderone R, Liu W and Li D: A mitochondrial proteomics
view of complex I deficiency in Candida albicans.
Mitochondrion. 38:48–57. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pizzatti L, Binato R, Cofre J, Gomes BE,
Dobbin J, Haussmann ME, D'Azambuja D, Bouzas LF and Abdelhay E:
SUZ12 is a candidate target of the non-canonical WNT pathway in the
progression of chronic myeloid leukemia. Genes Chromosomes Cancer.
49:107–118. 2010.PubMed/NCBI
|
|
12
|
Zhang M, Cheng ST, Wang HY, Wu JH, Luo YM,
Wang Q, Wang FX and Xia GX: iTRAQ-based proteomic analysis of
defence responses triggered by the necrotrophic pathogen
Rhizoctonia solani in cotton. J Proteomics. 152:226–235.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li W, Cohen A, Sun Y, Squires J, Braas D,
Graeber TG, Du L, Li G, Li Z, Xu X, et al: The role of CD44 in
glucose metabolism in prostatic small cell neuroendocrine
carcinoma. Mol Cancer Res. 14:344–353. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bao Z, Han X, Chen F, Jia X, Zhao J, Zhang
C, Yong C, Tian S, Zhou X and Han L: Evidence for the involvement
of cofilin in Aspergillus fumigatus internalization into
type II alveolar epithelial cells. BMC Microbiol. 15:1612015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Seddigh P, Bracht T, Molinier-Frenkel V,
Castellano F, Kniemeyer O, Schuster M, Weski J, Hasenberg A, Kraus
A, Poschet G, et al: Quantitative analysis of proteome modulations
in alveolar epithelial type II cells in response to pulmonary
Aspergillus fumigatus Infection. Mol Cell Proteomics.
16:2184–2198. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rabinovitch M: Professional and
non-professional phagocytes: An introduction. Trends Cell Biol.
5:85–87. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Crapo JD, Barry BE, Gehr P, Bachofen M and
Weibel ER: Cell number and cell characteristics of the normal human
lung. Am Rev Respir Dis. 126:332–337. 1982.PubMed/NCBI
|
|
20
|
Escobar N, Ordonez SR, Wosten HA, Haas PJ,
de Cock H and Haagsman HP: Hide, keep quiet, and keep low:
Properties that make Aspergillus fumigatus a successful lung
pathogen. Front Microbiol. 7:4382016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen F, Zhang C, Jia X, Wang S, Wang J,
Chen Y, Zhao J, Tian S, Han X and Han L: Transcriptome profiles of
human lung epithelial cells A549 interacting with Aspergillus
fumigatus by RNA-Seq. PLoS One. 10:e01357202015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Srivastava M, Bencurova E, Gupta SK, Weiss
E, Loffler J and Dandekar T: Aspergillus fumigatus
challenged by human dendritic cells: Metabolic and regulatory
pathway responses testify a tight battle. Front Cell Infect
Microbiol. 9:1682019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen L, Xing C, Ma G, Luo J, Su W, Li M,
Shi Q and He H: N-myc downstream-regulated gene 1 facilitates
influenza A virus replication by suppressing canonical NF-kappaB
signaling. Virus Res. 252:22–28. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Melotte V, Qu X, Ongenaert M, van
Criekinge W, de Bruïne AP, Baldwin HS and van Engeland M: The N-myc
downstream regulated gene (NDRG) family: Diverse functions,
multiple applications. FASEB J. 24:4153–4166. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Schweitzer CJ, Zhang F, Boyer A, Valdez K,
Cam M and Liang TJ: N-Myc downstream-regulated gene 1 restricts
hepatitis C virus propagation by regulating lipid droplet
biogenesis and viral assembly. J Virol. 92:e01166–1792.
2018.PubMed/NCBI
|
|
26
|
Gon Y, Maruoka S, Kishi H, Kozu Y,
Kazumichi K, Nomura Y, Takeshita I, Oshima T and Hashimoto S: NDRG1
is important to maintain the integrity of airway epithelial barrier
through claudin-9 expression. Cell Biol Int. 41:716–725. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ouhtit A, Rizeq B, Saleh HA, Rahman MM and
Zayed H: Novel CD44-downstream signaling pathways mediating breast
tumor invasion. Int J Biol Sci. 14:1782–1790. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Naor D, Nedvetzki S, Golan I, Melnik L and
Faitelson Y: CD44 in cancer. Crit Rev Clin Lab Sci. 39:527–579.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Babasola O, Rees-Milton KJ, Bebe S, Wang J
and Anastassiades TP: Chemically modified N-acylated hyaluronan
fragments modulate proinflammatory cytokine production by
stimulated human macrophages. J Biol Chem. 289:24779–24791. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fu Q, Wei Z, Xiao P, Chen Y and Liu X:
CD44 enhances macrophage phagocytosis and plays a protective role
in Streptococcus equi subsp. zooepidemicus infection. Vet
Microbiol. 198:121–126. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
van der Windt GJ, Florquin S, de Vos AF,
van't Veer C, Queiroz KC, Liang J, Jiang D, Noble PW and van der
Poll T: CD44 deficiency is associated with increased bacterial
clearance but enhanced lung inflammation during Gram-negative
pneumonia. Am J Pathol. 177:2483–2494. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jong A, Wu CH, Gonzales-Gomez I,
Kwon-Chung KJ, Chang YC, Tseng HK, Cho WL and Huang SH: Hyaluronic
acid receptor CD44 deficiency is associated with decreased
Cryptococcus neoformans brain infection. J Biol Chem.
287:15298–15306. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chang YC, Stins MF, McCaffery MJ, Miller
GF, Pare DR, Dam T, Paul-Satyaseela M, Kim KS and Kwon-Chung KJ:
Cryptococcal yeast cells invade the central nervous system via
transcellular penetration of the blood-brain barrier. Infect Immun.
72:4985–4995. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Qadri M, Almadani S, Jay GD and Elsaid KA:
Role of CD44 in regulating TLR2 activation of human macrophages and
downstream expression of proinflammatory cytokines. J Immun.
200:758–767. 2018. View Article : Google Scholar : PubMed/NCBI
|