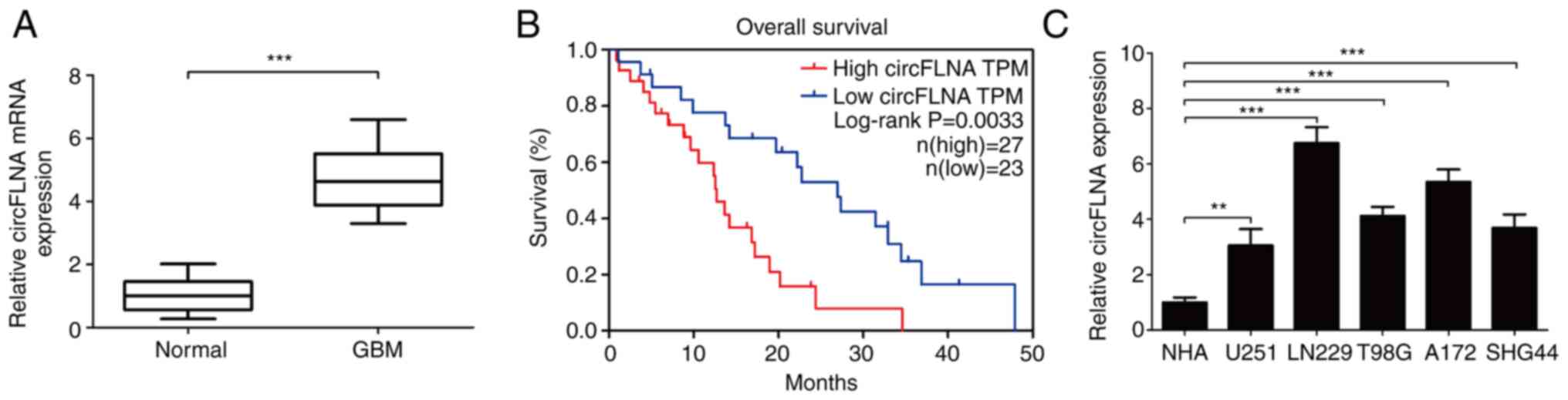
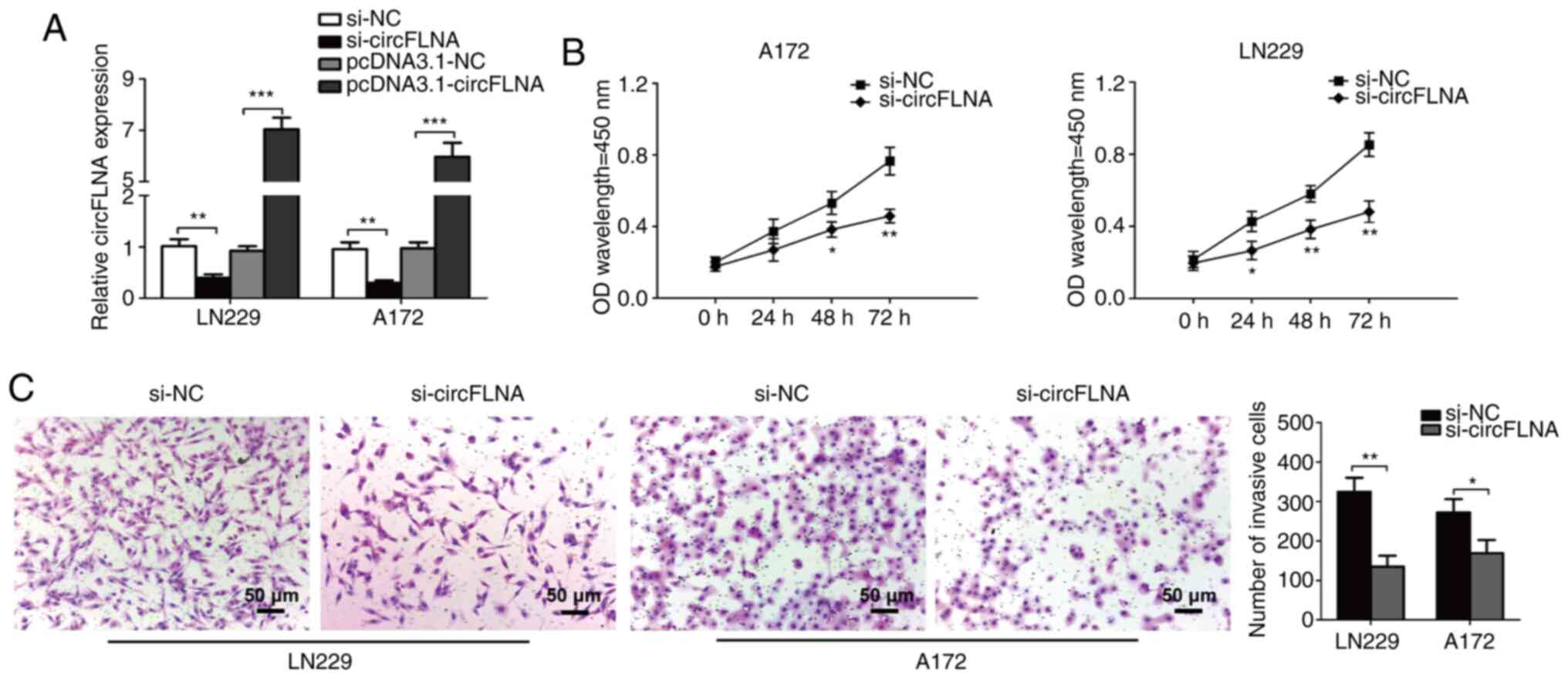
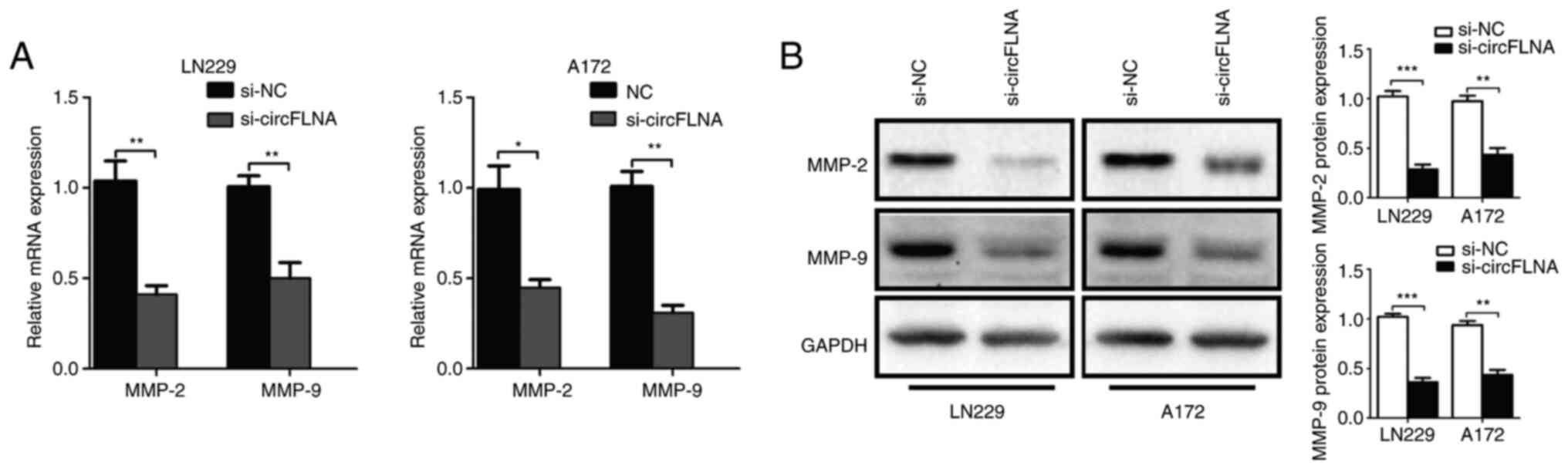
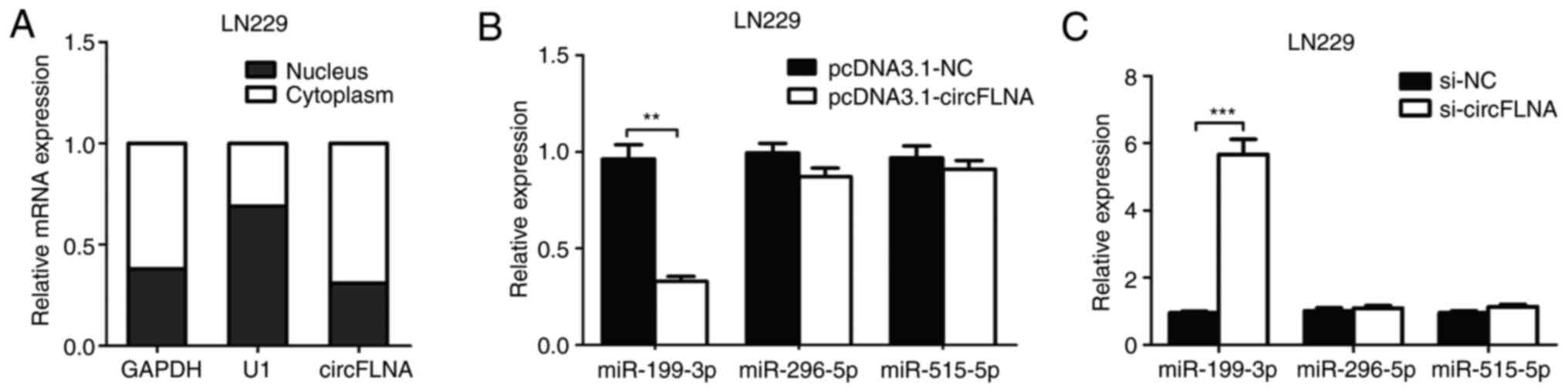
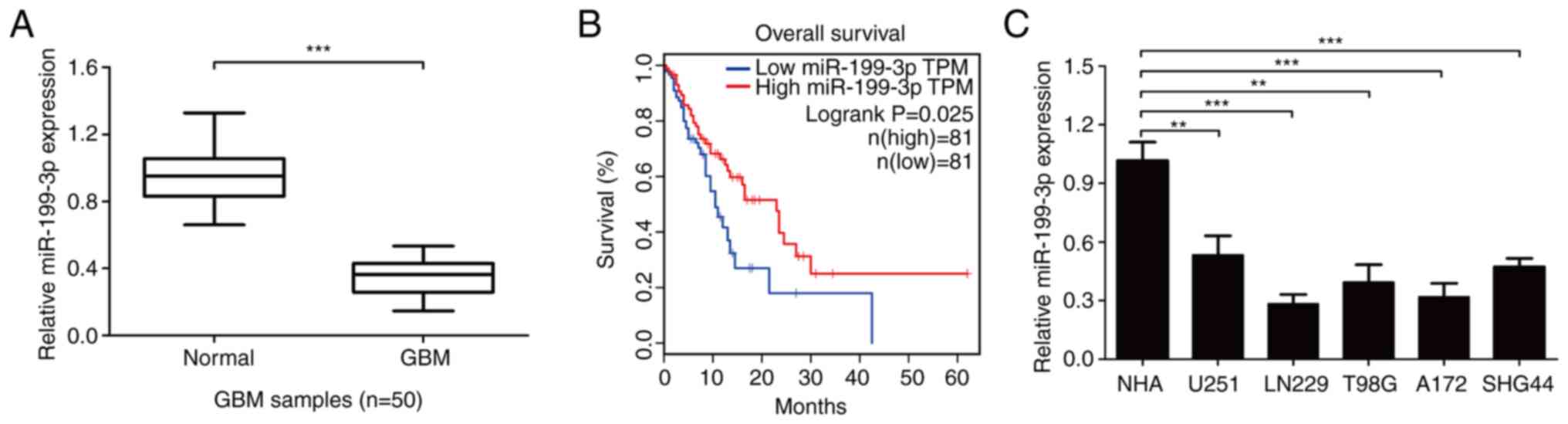
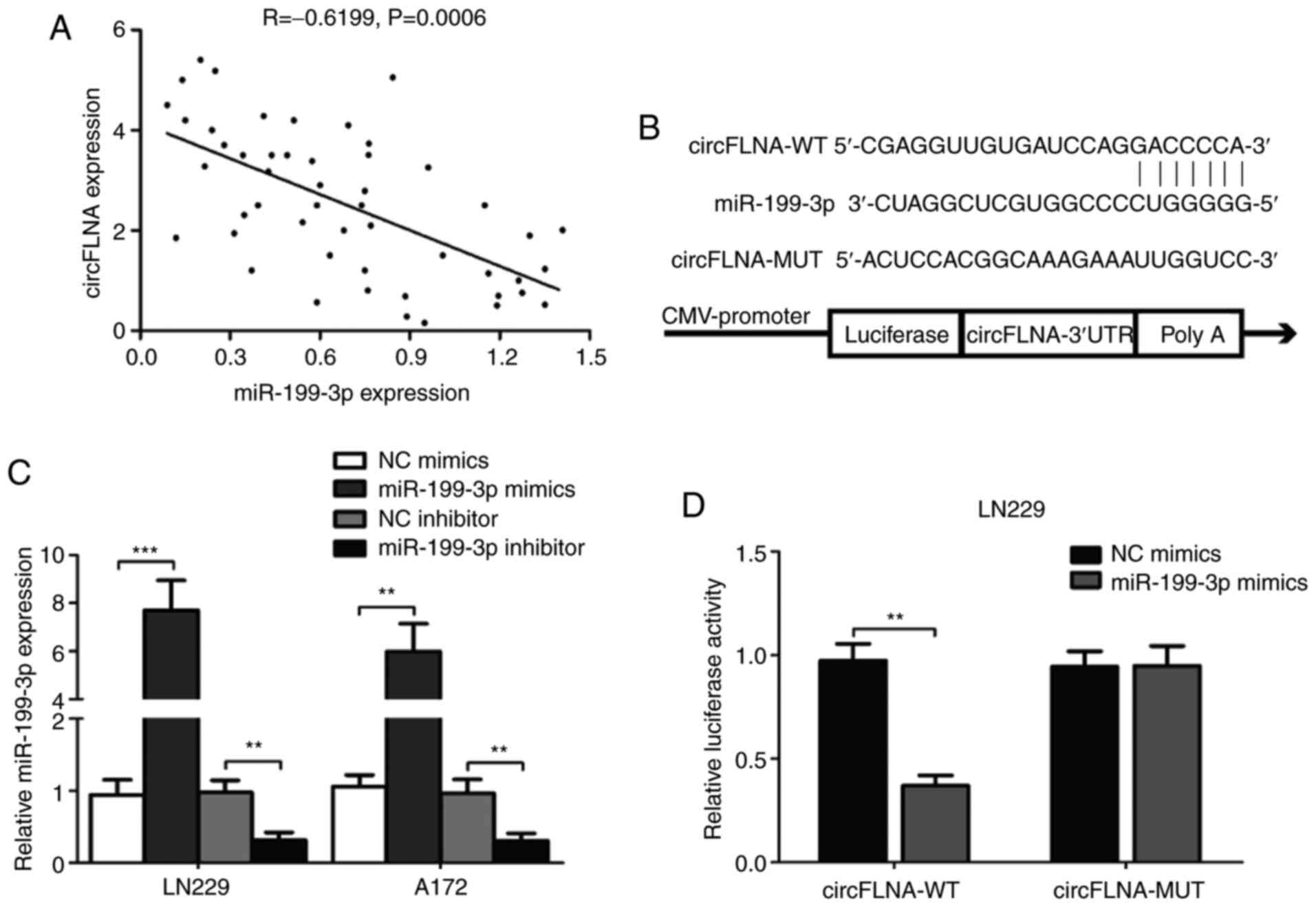
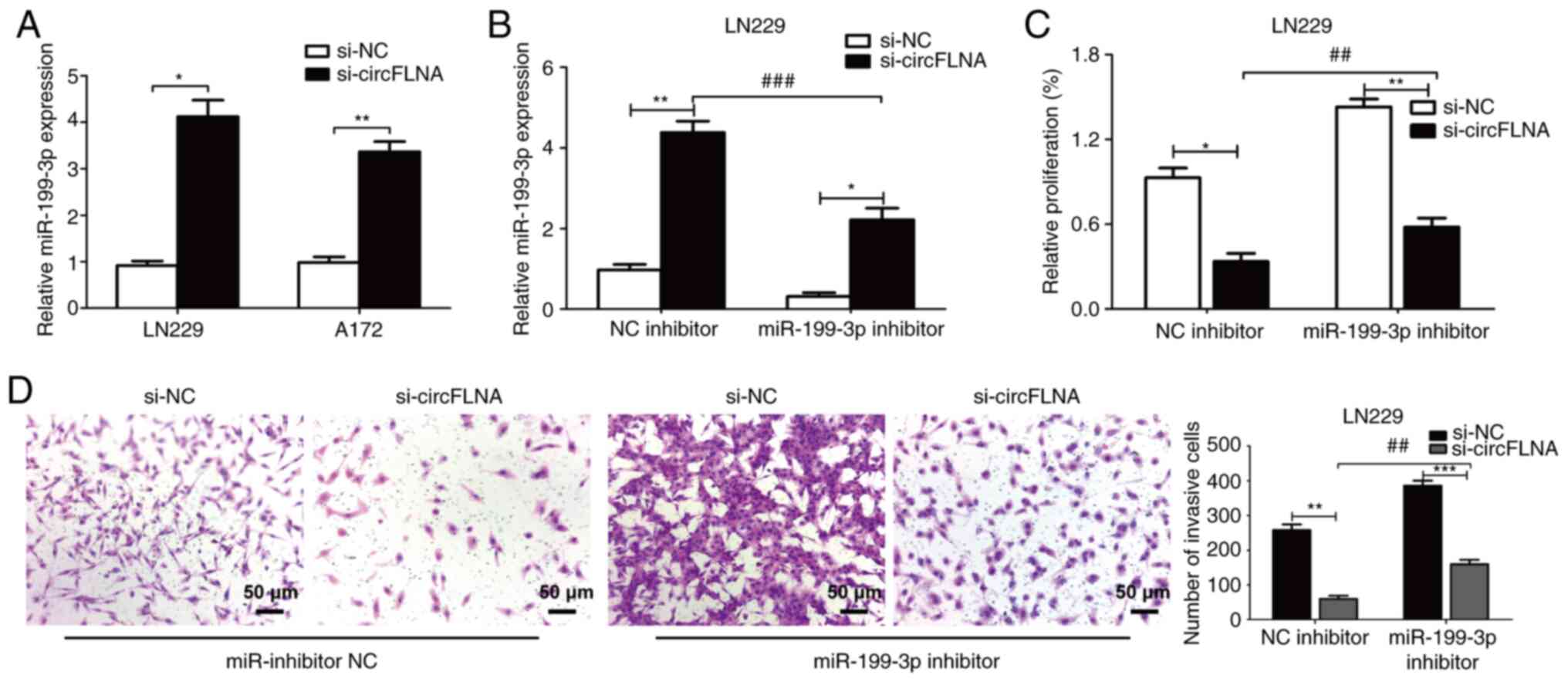
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bernstock JD, Mooney JH, Ilyas A, Chagoya
G, Estevez-Ordonez D, Ibrahim A and Nakano I: Molecular and
cellular intratumoral heterogeneity in primary glioblastoma:
Clinical and translational implications. J Neurosurg. 23:1–9.
2019.
|
|
4
|
Aldape K, Zadeh G, Mansouri S,
Reifenberger G and von Deimling A: Glioblastoma: Pathology,
molecular mechanisms and markers. Acta Neuropathol. 129:829–848.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hara A, Kanayama T, Noguchi K, Niwa A,
Miyai M, Kawaguchi M, Ishida K, Hatano Y, Niwa M and Tomita H:
Treatment strategies based on histological targets against invasive
and resistant glioblastoma. J Oncol. 2019:29647832019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Garton ALA, Kinslow CJ, Rae AI, Mehta A,
Pannullo SC, Magge RS, Ramakrishna R, McKhann GM, Sisti MB, Bruce
JN, et al: Extent of resection, molecular signature, and survival
in 1p19q-codeleted gliomas. J Neurosurg. 134:1357–1367. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang X, Zhang W, Mao XG, Cao WD, Zhen HN
and Hu SJ: Malignant intracranial high grade glioma and current
treatment strategy. Curr Cancer Drug Targets. 19:101–108. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Qu S, Yang X, Li X, Wang J, Gao Y, Shang
R, Sun W, Dou K and Li H: Circular RNA: A new star of noncoding
RNAs. Cancer Lett. 365:141–148. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong
F, Ren D, Ye X, Li C, Wang Y, et al: Circular RNAs function as
ceRNAs to regulate and control human cancer progression. Mol
Cancer. 17:792018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yin Y, Long J, He Q, Li Y, Liao Y, He P
and Zhu W: Emerging roles of circRNA in formation and progression
of cancer. J Cancer. 10:5015–5021. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hao Z, Hu S, Liu Z, Song W, Zhao Y and Li
M: Circular RNAs: Functions and prospects in glioma. J Mol
Neurosci. 67:72–81. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cheng J, Meng J, Zhu L and Peng Y:
Exosomal noncoding RNAs in Glioma: Biological functions and
potential clinical applications. Mol Cancer. 19:662020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hingorani DV, Lippert CN, Crisp JL,
Savariar EN, Hasselmann JPC, Kuo C, Nguyen QT, Tsien RY, Whitney MA
and Ellies LG: Impact of MMP-2 and MMP-9 enzyme activity on wound
healing, tumor growth and RACPP cleavage. PLoS One.
13:e01984642018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Qu J, Yang J, Chen M, Wei R and Tian J:
CircFLNA Acts as a sponge of miR-646 to facilitate the
proliferation, metastasis, glycolysis, and apoptosis inhibition of
gastric cancer by targeting PFKFB2. Cancer Manag Res. 12:8093–8103.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang N, Gao L, Ren W, Li S, Zhang D, Song
X, Zhao C and Zhi K: Fucoidan affects oral squamous cell carcinoma
cell functions in vitro by regulating FLNA derived circular RNA.
Ann N Y Acad Sci. 1462:65–78. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lu C, Shi X, Wang AY, Tao Y, Wang Z, Huang
C, Qiao Y, Hu H and Liu L: RNA-Seq profiling of circular RNAs in
human laryngeal squamous cell carcinomas. Mol Cancer. 17:862018.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang JX, Liu Y, Jia XJ, Liu SX, Dong JH,
Ren XM, Xu O, Zhang HZ, Duan HJ and Shan CG: Upregulation of
circFLNA contributes to laryngeal squamous cell carcinoma migration
by circFLNA-miR-486-3p-FLNA axis. Cancer Cell Int. 19:1962019.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 11:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen Z, Li J, Tian L, Zhou C, Gao Y, Zhou
F, Shi S, Feng X, Sun N, Yao R, et al: MiRNA expression profile
reveals a prognostic signature for esophageal squamous cell
carcinoma. Cancer Lett. 350:34–42. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang Q, Ye B, Wang P, Yao F, Zhang C and
Yu G: Overview of microRNA-199a regulation in cancer. Cancer Manag
Res. 11:10327–10335. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chi GN, Yang FW, Xu DH and Liu WM:
Silencing hsa_circ_PVT1 (circPVT1) suppresses the growth and
metastasis of glioblastoma multiforme cells by up-regulation of
miR-199a-5p. Artif Cells Nanomed Biotechnol. 48:188–196. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhu J, Ye J, Zhang L, Xia L, Hu H, Jiang
H, Wan Z, Sheng F, Ma Y, Li W, et al: Differential expression of
circular RNAs in glioblastoma multiforme and its correlation with
prognosis. Transl Oncol. 10:271–279. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yuan Y, Jiaoming L, Xiang W, Yanhui L, Shu
J, Maling G and Qing M: Analyzing the interactions of mRNAs,
miRNAs, lncRNAs and circRNAs to predict competing endogenous RNA
networks in glioblastoma. J Neurooncol. 137:493–502. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate-A practical and powerful approach to
multiple testing. J R Stat Soc. 1:289–300. 1995.
|
|
26
|
Zhang HD, Jiang LH, Sun DW, Hou JC and Ji
ZL: CircRNA: A novel type of biomarker for cancer. Breast Cancer.
25:1–7. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kristensen LS, Hansen TB, Venø MT and
Kjems J: Circular RNAs in cancer: Opportunities and challenges in
the field. Oncogene. 37:555–565. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao
L, Zhang Y, Wu YM, Dhanasekaran SM, Engelke CG, Cao X, et al: The
landscape of circular RNA in cancer. Cell. 176:869–881. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhao W, Dong M, Pan J, Wang Y, Zhou J, Ma
J and Liu S: Circular RNAs: A novel target among non-coding RNAs
with potential roles in malignant tumors (Review). Mol Med Rep.
20:3463–3474. 2019.PubMed/NCBI
|
|
30
|
Ng WL, Mohd Mohidin TB and Shukla K:
Functional role of circular RNAs in cancer development and
progression. RNA Biol. 15:995–1005. 2018.PubMed/NCBI
|
|
31
|
Liu Z, Zhou Y, Liang G, Ling Y, Tan W, Tan
L, Andrews R, Zhong W, Zhang X, Song E and Gong C: Circular RNA
hsa_circ_001783 regulates breast cancer progression via sponging
miR-200c-3p. Cell Death Dis. 10:552019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dong W, Bi J, Liu H, Yan D, He Q, Zhou Q,
Wang Q, Xie R, Su Y, Yang M, et al: Circular RNA ACVR2A suppresses
bladder cancer cells proliferation and metastasis through
miR-626/EYA4 axis. Mol Cancer. 18:952019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lu J, Wang YH, Yoon C, Huang XY, Xu Y, Xie
JW, Wang JB, Lin JX, Chen QY, Cao LL, et al: Circular RNA
circ-RanGAP1 regulates VEGFA expression by targeting miR-877-3p to
facilitate gastric cancer invasion and metastasis. Cancer Lett.
471:38–48. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Oliveto S, Mancino M, Manfrini N and Biffo
S: Role of microRNAs in translation regulation and cancer. World J
Biol Chem. 8:45–56. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wu M, Wang G, Tian W, Deng Y and Xu Y:
miRNA-based therapeutics for lung cancer. Curr Pharm Des.
23:5989–5996. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shin VY and Chu KM: miRNA as potential
biomarkers and therapeutic targets for gastric cancer. World J
Gastroenterol. 20:10432–10439. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Fridrichova I and Zmetakova I: MicroRNAs
contribute to breast cancer invasiveness. Cells. 8:13612019.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Deng JH, Deng Q, Kuo CH, Delaney SW and
Ying SY: miRNA targets of prostate cancer. Methods Mol Biol.
936:357–369. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang HY, Li CH, Wang XC, Luo YQ, Cao XD
and Chen JJ: miR-199 inhibits EMT and invasion of hepatoma cells
through inhibition of Snail expression. Eur Rev Med Pharmacol Sci.
23:7884–7891. 2019.PubMed/NCBI
|
|
40
|
Koshizuka K, Hanazawa T, Kikkawa N, Arai
T, Okato A, Kurozumi A, Kato M, Katada K, Okamoto Y and Seki N:
Regulation of ITGA3 by the anti-tumor miR-199 family inhibits
cancer cell migration and invasion in head and neck cancer. Cancer
Sci. 108:1681–1692. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Percie du Sert N, Hurst V, Ahluwalia A,
Alam S, Avey MT, Baker M, Browne WJ, Clark A, Cuthill IC, Dirnagl
U, et al: The ARRIVE guidelines 2.0: Updated guidelines for
reporting animal research. PLOS Biol. 18:e30004102020. View Article : Google Scholar : PubMed/NCBI
|