Introduction
Male infertility is a complex, often multifactorial
pathological condition affecting ~7% of the global male population
(1). While the etiology of
infertility is wide-ranging, up to 15% of males with infertility
have an underlying causative genetic defect (1–3).
Mouse knockout models have identified >400 genes, including
ATM, SYCP1-3, SYCE1 and CADM, leading to monogenic
infertility in men (4). Studies
on the molecular mechanisms of cell-cell interactions have provided
a better understanding of the causes of fertility issues due to
fertilization defects (5,6). While there are established
guidelines and practices for diagnostic karyotyping, azoospermia
factor (AZF) deletion screening and cystic fibrosis transmembrane
conductance regulator testing, the overall diagnostic yield is as
low as 4% (7). However, rapid
advancements in genomic medicine achieved by next-generation
sequencing (NGS) technologies lack of results standardization,
creating a knowledge gap in the clinical area of male infertility
(8).
A literature review on monogenic forms of male
infertility identified 78 genes associated with male infertility in
1,337 publications in 2019 (9),
with this number rising by 33% to 104 genes by 2022 (10). In the era of genomics and
personalized medicine, this number is low compared with other
clinical fields, such as intellectual disability (8). To improve biological understanding
as well as diagnostic yield and clinical relevance of genetic
testing, further genes leading to male infertility must be
identified.
Our previous study outlined a possible cause for
asthenoteratozoospermia in a patient harboring a deletion of ~8-Mb
in the 5q22.2q23.1 locus, including the testis-specific serine
kinase 1B (TSSK1B) gene (11). Genes belonging to kinase and
phosphatase families are responsible for activation and
deactivation of intra- and extracellular transduction via
phosphorylation and dephosphorylation (12). These processes are key for correct
regulation and metabolic processes of the cell that are mediated by
different receptors and enzymes (13). TSSKs are part of the AMP-activated
protein kinase family (14). They
comprise six genes, which are almost exclusively present in the
testes (>1,000-fold concentration compared with other organs).
These genes serve a role in spermatogenesis and are responsible for
correct morphogenesis and differentiation following meiosis when
spermatid elongation occurs. This has been demonstrated using
recombinant mouse models, where sterile phenotypes have been
observed in double Tssk1 and Tssk2 knockout (KO) and
Tssk6 KO mice (15). A
sub-fertile phenotype with reduced TSSK1 and TSSK2
levels has also been observed in a Tssk4 KO model (16,17). These results highlight the
importance of TSSKs in research, as they can be targeted by both
drugs and inhibitors, suggesting that TSSKs may not only cause
infertility, but can also provide a route for the development of
male non-hormonal contraceptive drugs. Originally, TSSK
genes were found in mouse tissue using degenerate oligonucleotide
primers while searching for novel kinases, which led to the
discovery of TSSK1. Subsequently, TSSK2 was
identified via low stringency screening due to its close proximity
and genetic linkage to TSSK1 (18,19). Following the discovery of these
genes, yeast two-hybrid and co-immunoprecipitation experiments were
performed to detect proteins that may interact with these kinases.
A novel 65 kDa protein was identified, namely testis-specific
kinase substrate (TSKS), that interacts with both TSSK1 and TSSK2
(19).
According to studies conducted on a diverse array of
species, it was proposed that the TSSK family originated with
Tssk5 in amphibians ~380 million years ago (MYA). Following
the Paleocene-Eocene radiation period, a novel gene appeared in
primates and humans, known as Tssk1b (Fig. 1). Moreover, in most species
Tssk1/2 are linked on one chromosome and their activity is
hypothesized to be parallel, as their combined absence has been
shown to cause infertility in KO mice (17). However, with the appearance of
Tssk1b (duplicated on another chromosome and not linked to
Tssk2), Tssk1 was inactivated through negative selection and
mutation to become a pseudogene known as Tssk1 (Fig. 1). Even though the genes diverge
from one another, they exhibit sequence conservation and
similarity. The N-terminal domain is represented by the 1–272 amino
acid sequence, which bears the kinase domain, whereas the
C-terminal is present in the 273-end sequence. In humans,
N-terminal similarity between Tssk1b and Tssk2 is
82.0%, which is similar to other mammalian species, while
C-terminal similarity between the two genes is notably lower at
14.7%. This may be due to the regulatory function of the C-terminal
domain, which is specific for each kinase. This could also mean
that each kinase serves specific functions, in addition to
exhibiting overlapping effects. With regards to evolutionary
conservation, the C-terminal of Tssk1 shows 65.7% mean
sequence conservation compared with the C-terminal of Tssk2,
which is 87.6%. When considering retrogenes and duplications, newer
genes seem to have the ability to evolve faster, meaning that their
conservation rate will be lower. Based on these observations on
conservation rates, it is hypothesized that Tssk2 was the
first gene to appear on the chromosome following the retroposition
of Tssk1 (20). Compared
with non-mammalian species, the C-terminal conservation of human
Tssk1b is lower (61.4%) than that of Tssk2 (86.8%),
whereas N-terminal conservation is similar (92.5 and 94.9%,
respectively) (20). This
suggests that the two domains are affected by different selective
evolutionary pressures.
Spermatogenesis is divided into three main phases:
Mitotic division, which generates a pool of spermatocytes; meiosis
to generate haploid spermatids and spermiogenesis, in which
spermatids differentiate into spermatozoa (21). Nayak et al (22) tested mouse tissue obtained from
maturing seminiferous tubules at different periods after birth. The
procedure included immuno-fluorescent staining to visualize
difference in expression of both Tssk1 and Tssk2. Tssk1 expression
was first observed at low levels in week 3, concurrent with the
appearance of round spermatids, predominantly in cells in meiotic
metaphase. By contrast, Tssk2 was detected in cells
undergoing spermiogenesis at weeks 4–5 and was not detected in
metaphase dividing cells (22).
This highlights the importance of TSSK1 (the first gene to
be expressed) in both early sperm development and mature sperm
formation (22).
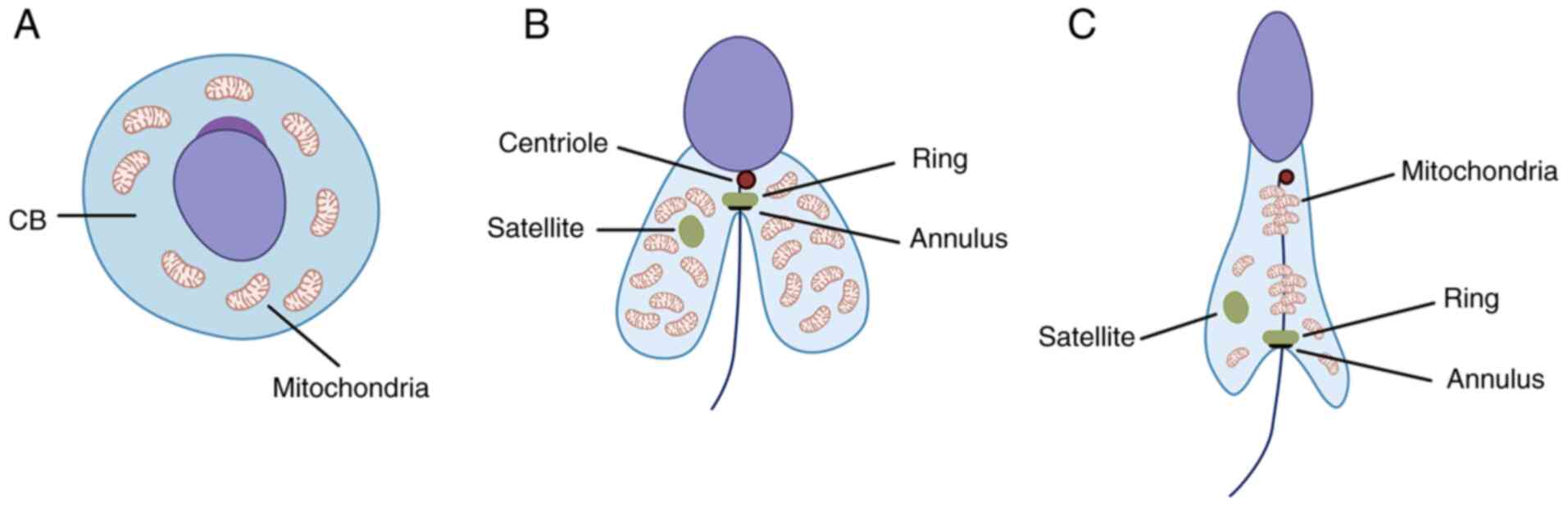
In spermatogenesis, following meiosis, a cloud-like
organelle (nuage) appears in the round spermatid known as
chromatoid body and is localized around the haploid nucleus
(23). The chromatoid body is the
RNA-controlling center responsible for organization and regulation
of mRNA and RNA pathways associated with the haploid genome of the
spermatid (Fig. 2A) (24). Its function is rather short-term
and during the transition from round to elongated spermatid, the
chromatoid body loses certain functions and enzymes (such as murine
P-element induced wimpy testis family member proteins) to split
into two well-defined structures, namely satellite and ring
(Fig. 2B) (25). It has been observed that
Tssk1 and Tssk2, as well as TSKS, accumulate in both
the ring around the flagellum and in the satellite in the
cytoplasm. Experiments in both wild-type and double Tssk1,2
КО mice revealed that КО results in disturbed mitochondrial sheath
formation, rendering the sperm cells non-functional (17).
The sheath-forming complex includes not only
TSSK1B, but also testis-specific phosphatase,
Ppp1cc2, which serves a key role in completion of
spermatogenesis in mice (26).
Pull-down assays using GST-Ppp1cc2 expressed and purified from
bacteria as bait against protein lysates from mouse testis tissue
have been performed to determine the interaction between Ppp1cc2
and other proteins (27). An
indirect interaction of Ppp1cc2 with Tssk1 was shown to be mediated
through TSKS via RVxF motif (at amino acid position 51–55), which
interacts with both proteins to form a complex. To test the
activity of Ppp1cc, deletion via mutagenesis was performed in male
mice; this resulted in germ cell reduction primarily occurring
during spermatid elongation (post-meiosis) phase at the time of
chromatoid body dissociation, causing the spermatogenic cycle to
halt (28). Thus, the testicular
kinase/phosphatase complex is key for formation of the
mitochondrial sheath during spermatogenesis.
Considering the literature and our previous
experience regarding the role of the TSSK1B gene in a
patient with asthenoteratozoospermia, the aim of the present study
was to perform TSSK1B genetic screening on a selected group
of mutation-negative male patients with infertility. The study
aimed to assess overall genomic variability within TSSK1B
compared with previously reported variants in population databases,
investigate the association between missense mutations in
TSSK1B and clinical phenotype with regards to semen quality
and to assess findings using in silico predictors and novel
protein folding algorithms.
Materials and methods
Patients
The examined patient cohort comprised 100 Bulgarian
male patient DNA samples obtained in a period of 3 years (January
2019 to December 2021) from the genetic biobank of Genetic
Medico-Diagnostic Laboratory ‘Genica’ (Sofia, Bulgaria). Patients
included in the study (age, 18–53; mean age, 34 years) had a
positive history of sperm dysmorphology according to European
Society of Human Reproduction and Embryology/Nordic Association for
Andrology criteria using Sperm Class Analyzer, a system of
quantitative and qualitative analysis of human sperm parameters
(29). Included patients were
selected based on negative results from Y-chromosome microdeletion
testing. Ethical approval was obtained from the ethical board of
Medical University (Sofia Bulgaria). For all subjects, written
informed consent was provided.
PCR and Sanger sequencing
Molecular testing for microdeletions in the AZF
region were performed as described in the guidelines and standards
of the European Academy of Andrology (30).
Genomic DNA was isolated from peripheral blood using
the QIAamp® DNA Blood Mini Kit (Qiagen GmbH) following
the manufacturer's recommendations. DNA was eluted in approximately
200 µl of buffer AE. Amplification was performed using forward
(5′-CTAGGAGGCAGGAACAGCAG-3′) and reverse
(5′-ACTGCCTTCCTTCTCTGGCT-3′) primers. Thermocycling conditions were
as follows: 5 min denaturation at 95°C, 35 cycles of 95°C for 45
sec, 60°C for 45 sec, 72°C for 90 sec and final extension for 5 min
at 72°C to obtain a 1,327 bp product. PCR products were verified by
3% agarose gel electrophoresis and visualized with ethidium
bromide. Sanger sequencing of the TSSK1B gene was performed
using BigDye®Terminator cycle sequencing kit v.3.1
(Applied Biosystems; Thermo Fisher Scientific, Inc.) on an ABI 3130
sequencer.
Protein model
Protein models for the detected TSSK1B
missense mutations were simulated using AlphaFold2 v2.1.0 (29) for each altered TSSK1b protein
sequence specified by a missense mutation in the patient group
(Fig. 3). In silico
predictions were performed using Ensembl Variant Effect Predictor
(31) with pathogenicity scores
(from both SIFT and PolyPhen) classified according to the standards
of the American College of Medical Genetics (32).
Statistical analysis
Allele frequencies of each variant found in the
cohort were calculated and compared with those in the gnomAD v2.1.1
database (https://gnomad.broadinstitute.org/).
Results
Variant discovery in TSSK1
Screening of the TSSK1B gene within the
patient group revealed 11 nucleotide variations, with certain
patients presenting with up to four variants (three synonymous, one
missense), five of which were protein-altering missense mutations
and six of which were synonymous mutations (Table I). Two missense mutations,
p.3D>N and p.52F>L, are novel, without previous reports in
GnomAD (33). The other missense
mutations, p.66M>V, p.237R>C and p.293G>E, occur with a
higher allele frequency in the study cohort compared with global
databases (GnomAD v2.1.1).
 | Table I.Tssk, testis-specific serine kinase
1B variants in the patient cohort, with scores from SIFT, PolyPhen
and scoring according to the ACMG guidelines. |
Table I.
Tssk, testis-specific serine kinase
1B variants in the patient cohort, with scores from SIFT, PolyPhen
and scoring according to the ACMG guidelines.
| CDS position | Protein
position | Variant | Amino acids | SIFT | PolyPhen | ACMG | Cohort | gnomAD | Clinical
phenotype | Incidence rate,
% |
|---|
| 7 | 3 | Missense | D/N | Deleterious
(0.00) | Possibly damaging
(0.566) | VUS | 0.010 | - | N/A
Azoospermia | 2.56 (N/A) 3.84
(azoospermia) |
| 156 | 52 | Missense | F/L | Deleterious
(0.00) | Probably damaging
(0.925) | VUS | 0.005 | - | Azoospermia | 3.80 |
| 196 | 66 | Missense | M/V | Tolerated
(0.31) | Benign (0.000) | Likely benign | 0.005 |
7.37×10−4 | Asthenoteratozoo
spermia | 14.28 |
| 438 | 146 | Synonymous | K | - | - | - | 0.005 |
3.98×10−6 | - | - |
| 510 | 170 | Synonymous | A | - | - | - | 0.055 |
9.94×10−2 | - | - |
| 522 | 174 | Synonymous | T | - | - | - | 0.045 |
6.92×10−2 | - | - |
| 540 | 180 | Synonymous | A | - | - | - | 0.055 |
6.9×10−2 | - | - |
| 709 | 237 | Missense | R/C | Deleterious
(0.01) | Possibly damaging
(0.849) | Likely benign | 0.005 |
2.86×10−4 | Asthenoteratozoo
spermia | 14.28 |
| 878 | 293 | Missense | G/E | Tolerated low
confidence (0.89) | Benign (0.021) | Benign | 0.030 |
03.14×10−2 | N/A Azoospermia
Teratozoospermia | 7.69 (N/A) 3.84
(azoospermia) 50.00 (teratozoospermia) |
| 978 | 326 | Synonymous | T | - | - | - | 0.005 | - | - | - |
| 996 | 332 | Synonymous | A | - | - | - | 0.015 |
3.46×10−2 | - | - |
Correlations with sperm dysmorphology
phenotypes
Available clinical data were analyzed to demonstrate
a potential link between the mutations detected in the patient
cohort and clinical phenotype (Table
I). The mutations p.3D>N and p.52F>L were discovered in
patients with azoospermia. The p.66M>V and p.237R>C mutations
resulted in asthenoteratozoospermia, the same phenotype detected in
our initial TSSK1B case report (11). The missense variant p.293G>E
was seen in patients with azoospermia and teratozoospermia. Certain
patients harbored two or three synonymous variants in addition to a
protein-altering mutation.
Structure alterations caused by
missense variants
Alignments to canonical TSSK1B protein revealed
potential misfolding of the protein in four of the models
(p.3D>N, p.66M>V, p.237R>C and p.293G>E). This occurred
within the kinase domain of the protein, between positions 45–47,
creating a small Arg-Lys-Lys helix immediately after a predicted
β-sheet structure. Additionally, the mutation p.66M>V created a
small helix motif of Glu-Ile-Leu at position 340–342. From in
silico prediction methods for mutations p.3D>N, p.66M>V
and p.237R>C, pathogenic scoring from Sorting Intolerant from
Tolerant (SIFT) and PolyPhen was observed.
Discussion
Our previous report presented the first known case
of a TSSK1B deletion associated with a clinical phenotype of
asthenoteratozoospermia and male infertility (11). Since then, the TSSK family has
emerged as a potential cause for male infertility and may be a
target for the development of novel male contraceptive solutions
through targeted inhibition of TSSK1B (15). By performing comprehensive
targeted patient screening to investigate the role of TSSK1B
in male infertility, the present study discovered both known and
novel missense variants that should be evaluated in vivo to
determine the extent of their clinical manifestation. Mutations
discovered toward the C-terminus of TSSK1B, namely p.3D>N
and p.52F>L, did not exhibit the same phenotype as in our
initial report (asthenoteratozoospermia), yet yielded pathogenic
scores via in silico prediction methods. Of note, p.66M>V
and p.237R>C mutations were associated with the clinical
phenotype discovered in our initial study. Finally, the p.293G>E
mutation manifested as two clinical phenotypes, azoospermia and
teratozoospermia. While classification via ACMG guidelines shows
either that mutations are of unknown significance or possibly
benign, this may be due to >0.01 population frequency in
reference databases (34).
Infertility-causing mutations might not be functionally
investigated because of their high representation in a population.
Such sequence alterations may not directly affect the quality of
life of the individual; hence they are treated as benign.
While in silico predictors such as PolyPhen
and SIFT (31) have been used to
determine the potential outcome of a missense mutation, the next
step in the assessment of novel pathological variants is to build
and compare protein models. Although AlphaFold2 does not provide a
solution to the protein folding problem (34), it has an accuracy level comparable
to that of X-ray crystallography (35). This novel artificial intelligence
program achieved a milestone level of accuracy in the biannual
Critical Assessment of Protein Structure Prediction experiment in
2020 and is used to predict protein structures for databases such
as the European Bioinformatics Institute. While it is still not
validated for clinical use, the results produced are notable, with
a margin of error of only 1.6 angstroms when simulating the folding
of the protein (36). Using this
method, altered amino acid sequence caused by the missense
mutations identified in our patient cohort were simulated;
AlphaFold2 predicted a small helical motif in four of the altered
sequences. This fold occurred seemingly with no regard to the
location of the change yet it was predicted to affect the kinase
domain of TSSK1B. As previous studies show, damaging or removing
one of the copies of this gene can cause infertility (14,17), which indicates that small
structure alterations lead to a pathological changes.
To support the present findings, the evolutionary
background of TSSKs and integration of the Ppp1cc2-Tssk1 complex
were investigated, which suggested that this gene is key for sperm
maturity. TSSK1B was the first of the family to evolve in
humans and the complex with Ppp1cc2 is essential for proceeding
with the spermatid elongation stage and completing spermatogenesis.
Furthermore, TSSK1B is the earliest kinase of this family to
be expressed, at 3 weeks opposed to other TSSKs expressed at 3.5
weeks (22,37), which further outlines its
importance in spermatogenesis.
In the present cohort, 11 of 100 patients carried a
missense mutation in TSSK1B; although this frequency is
biased by the cohort size and patient selection, an 11% carrier
rate warrants further study. Not all mutations are equal: In
silico predictors showed that 3 of the 5 protein-altering
variants have scores that are interpreted as damaging. As the
N-terminus of TSSK1 is highly conserved, this suggests a
potential damaging outcome caused by the mutations reported in the
present study. Further functional studies are required to verify
the pathological consequences of these alterations.
Multiple studies have highlighted the need for novel
male contraceptives (38,39). Current methods are limited to
condoms and vasectomy, compared with the variety of available
female contraceptives; the limitations of these methods are high
failure rate of condoms and irreversibility of surgical vasectomy
(39). While trials of hormonal
methods are successful, they come with side effects, such as weight
gain, acne, mood changes and changes in libido (39,40). Previous studies demonstrate
promising results in targeting TSSKs using kinase inhibitors and
the potential application of TSSK allosteric inhibitors (15,41), thereby emphasizing not only the
important role of this family of kinases in spermatogenesis, but
also how they can be targeted pharmaceutically.
The only putative pathological manifestation of
TSSK1B has been studied in mice, where haploinsufficiency
results in decreased sperm maturation and offspring carrying
exclusively the wild-type TSSK1B allele (14). The present patient cohort was
referred for a number of clinical phenotypes of infertility and
some individuals carried a heterozygous variant of TSSK1B.
Functional study investigating TSSK1B function in human
spermatozoids is warranted. Second, through the advent of NGS,
carrier screening can be performed in cases with previously
unidentified genetic causes. Lastly, research into this gene may
lead to novel male contraceptive solutions.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
TK and IT confirm the authenticity of all the raw
data. TK and IT conceptualized the study. TK, VZ and DAS designed
the study. KD, DM and ST performed the experiments and wrote the
manuscript. IT, VZ and DAS performed the statistical analysis. TK,
VZ and DAS supervised the study. KD visualized data. TK, IT, VZ and
DAS reviewed and edited the manuscript. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study was conducted according to the
guidelines of the Declaration of Helsinki. The study was approved
by the Ethics Committee of Sofia Medical University. All patients
provided signed informed consent forms for participation in the
study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests. DAS is the Editor-in-Chief for the journal, but had no
personal involvement in the reviewing process, or any influence in
terms of adjudicating on the final decision, for this article.
References
|
1
|
Krausz C and Riera-Escamilla A: Genetics
of male infertility. Nat Rev Urol. 15:369–384. 2018. View Article : Google Scholar
|
|
2
|
Tournaye H, Krausz C and Oates RD: Novel
concepts in the aetiology of male reproductive impairment. Lancet
Diabetes Endocrinol. 5:544–553. 2017. View Article : Google Scholar
|
|
3
|
Xavier MJ, Salas-Huetos A, Oud MS, Aston
KI and Veltman JA: Disease gene discovery in male infertility:
Past, present and future. Hum Genet. 140:7–19. 2021. View Article : Google Scholar
|
|
4
|
Jamsai D and O'Bryan MK: Mouse models in
male fertility research. Asian J Androl. 13:139–151. 2011.
View Article : Google Scholar
|
|
5
|
Cardona Barberán A, Boel A, Vanden
Meerschaut F, Stoop D and Heindryckx B: Diagnosis and treatment of
male infertility-related fertilization failure. J Clin Med.
9:38992020. View Article : Google Scholar
|
|
6
|
Georgadaki K, Khoury N, Spandidos DA and
Zoumpourlis V: The molecular basis of fertilization (Review). Int J
Mol Med. 38:979–986. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tüttelmann F, Simoni M, Kliesch S, Ledig
S, Dworniczak B, Wieacker P and Röpke A: Copy number variants in
patients with severe oligozoospermia and sertoli-cell-only
syndrome. PLoS One. 6:e194262011. View Article : Google Scholar
|
|
8
|
Alhathal N, Maddirevula S, Coskun S, Alali
H, Assoum M, Morris T, Deek HA, Hamed SA, Alsuhaibani S, Mirdawi A,
et al: A genomics approach to male infertility. Genet Med.
22:1967–1975. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Oud MS, Volozonoka L, Smits RM, Vissers
LELM, Ramos L and Veltman JA: A systematic review and standardized
clinical validity assessment of male infertility genes. Hum Reprod
Oxf Engl. 34:932–941. 2019. View Article : Google Scholar
|
|
10
|
Houston BJ, Riera-Escamilla A, Wyrwoll MJ,
Salas-Huetos A, Xavier MJ, Nagirnaja L, Friedrich C, Conrad DF,
Aston KI, Krausz C, et al: A systematic review of the validated
monogenic causes of human male infertility: 2020 update and a
discussion of emerging gene-disease relationships. Hum Reprod
Update. 28:15–29. 2022. View Article : Google Scholar
|
|
11
|
Kadiyska T, Tourtourikov I, Petrov A,
Chavoushian A, Antalavicheva M, König EM, Klopocki E, Vessela N and
Stanislavov R: Interstitial deletion of 5q22.2q23.1 including
APC and TSSK1B in a patient with adenomatous
polyposis and asthenoteratozoospermia. Mol Syndromol. 9:235–240.
2019. View Article : Google Scholar
|
|
12
|
Bononi A, Agnoletto C, De Marchi E, Marchi
S, Patergnani S, Bonora M, Giorgi C, Missiroli S, Poletti F,
Rimessi A and Pinton P: Protein kinases and phosphatases in the
control of cell fate. Enzyme Res. 2011:3290982011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ardito F, Giuliani M, Perrone D, Troiano G
and Lo Muzio L: The crucial role of protein phosphorylation in cell
signaling and its use as targeted therapy (Review). Int J Mol Med.
40:271–280. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xu B, Hao Z, Jha KN, Zhang Z, Urekar C,
Digilio L, Pulido S, Strauss JF III, Flickinger CJ and Herr JC:
Targeted deletion of Tssk1 and 2 causes male infertility due to
haploinsufficiency. Dev Biol. 319:211–222. 2008. View Article : Google Scholar
|
|
15
|
Salicioni AM, Gervasi MG, Sosnik J,
Tourzani DA, Nayyab S, Caraballo DA and Visconti PE:
Testis-specific serine kinase protein family in male fertility and
as targets for non-hormonal male contraception†. Biol Reprod.
103:264–274. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Spiridonov NA, Wong L, Zerfas PM, Starost
MF, Pack SD, Paweletz CP and Johnson GR: Identification and
characterization of SSTK, a serine/threonine protein kinase
essential for male fertility. Mol Cell Biol. 25:4250–4261. 2005.
View Article : Google Scholar
|
|
17
|
Shang P, Baarends WM, Hoogerbrugge J, Ooms
MP, van Cappellen WA, de Jong AA, Dohle GR, van Eenennaam H, Gossen
JA and Grootegoed JA: Functional transformation of the chromatoid
body in mouse spermatids requires testis-specific serine/threonine
kinases. J Cell Sci. 123:331–339. 2010. View Article : Google Scholar
|
|
18
|
Bielke W, Blaschke RJ, Miescher GC,
Zürcher G, Andres AC and Ziemiecki A: Characterization of a novel
murine testis-specific serine/threonine kinase. Gene. 139:235–239.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kueng P, Nikolova Z, Djonov V, Hemphill A,
Rohrbach V, Boehlen D, Zuercher G, Andres AC and Ziemiecki A: A
novel family of serine/threonine kinases participating in
spermiogenesis. J Cell Biol. 139:1851–1859. 1997. View Article : Google Scholar
|
|
20
|
Shang P, Hoogerbrugge J, Baarends WM and
Grootegoed JA: Evolution of testis-specific kinases TSSK1B and
TSSK2 in primates. Andrology. 1:160–168. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sharpe R: Regulation of spermatogenesis.
The Physuiology of Reproduction. Knobil E and Neil JD: Raven Press;
New York, NY: pp. 1363–434. 1994
|
|
22
|
Nayak S, Galili N and Buck CA:
Immunohistochemical analysis of the expression of two
serine-threonine kinases in the maturing mouse testis. Mech Dev.
74:171–174. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yokota S: Historical survey on chromatoid
body research. Acta Histochem Cytochem. 41:65–82. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kotaja N, Bhattacharyya SN, Jaskiewicz L,
Kimmins S, Parvinen M, Filipowicz W and Sassone-Corsi P: The
chromatoid body of male germ cells: Similarity with processing
bodies and presence of Dicer and microRNA pathway components. Proc
Natl Acad Sci USA. 103:2647–2652. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Grivna ST, Pyhtila B and Lin H: MIWI
associates with translational machinery and PIWI-interacting RNAs
(piRNAs) in regulating spermatogenesis. Proc Natl Acad Sci.
103:13415–13420. 2006. View Article : Google Scholar
|
|
26
|
Varmuza S, Jurisicova A, Okano K, Hudson
J, Boekelheide K and Shipp EB: Spermiogenesis is impaired in mice
bearing a targeted mutation in the protein phosphatase 1cgamma
gene. Dev Biol. 205:98–110. 1999. View Article : Google Scholar
|
|
27
|
MacLeod G, Shang P, Booth GT, Mastropaolo
LA, Manafpoursakha N, Vogl AW and Varmuza S: PPP1CC2 can form a
kinase/phosphatase complex with the testis-specific proteins TSSK1
and TSKS in the mouse testis. Reprod Camb Engl. 147:1–12. 2014.
View Article : Google Scholar
|
|
28
|
Forgione N, Vogl AW and Varmuza S: Loss of
protein phosphatase 1c(gamma) (PPP1CC) leads to impaired
spermatogenesis associated with defects in chromatin condensation
and acrosome development: An ultrastructural analysis.
Reproduction. 139:1021–1029. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
World Health Organization (WHO), . WHO
laboratory manual for the examination of human semen and
sperm-cervical mucus interaction. 4th edition. Published on behalf
of the World Health Organization (by). Cambridge University Press;
Cambridge: pp. p1281999
|
|
30
|
Krausz C, Hoefsloot L, Simoni M and
Tüttelmann F; European Academy of Andrology and European Molecular
Genetics Quality Network, : EAA/EMQN best practice guidelines for
molecular diagnosis of Y-chromosomal microdeletions:
State-of-the-art 2013. Andrology. 2:5–19. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
McLaren W, Gil L, Hunt SE, Riat HS,
Ritchie GR, Thormann A, Flicek P and Cunningham F: The ensembl
variant effect predictor. Genome Biol. 17:1222016. View Article : Google Scholar
|
|
32
|
Richards S, Aziz N, Bale S, Bick D, Das S,
Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al:
Standards and guidelines for the interpretation of sequence
variants: A joint consensus recommendation of the American college
of medical genetics and genomics and the association for molecular
pathology. Genet Med. 17:405–424. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Karczewski KJ, Francioli LC, Tiao G,
Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A,
Birnbaum DP, et al: The mutational constraint spectrum quantified
from variation in 141,456 humans. Nature. 581:434–443. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jumper J, Evans R, Pritzel A, Green T,
Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A,
Potapenko A, et al: Highly accurate protein structure prediction
with AlphaFold. Nature. 596:583–589. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Callaway E: ‘It will change everything’:
DeepMind's AI makes gigantic leap in solving protein structures.
Nature. 588:203–204. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jumper J, Evans R, Pritzel A, Green T,
Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A,
Potapenko A, et al: Applying and improving AlphaFold at CASP14.
Proteins Struct Funct Bioinforma. 89:1711–1721. 2021. View Article : Google Scholar
|
|
37
|
Li Y, Sosnik J, Brassard L, Reese M,
Spiridonov NA, Bates TC, Johnson GR, Anguita J, Visconti PE and
Salicioni AM: Expression and localization of five members of the
testis-specific serine kinase (Tssk) family in mouse and human
sperm and testis. Mol Hum Reprod. 17:42–56. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Dorman E and Bishai D: Demand for male
contraception. Expert Rev Pharmacoecon Outcomes Res. 12:605–613.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Abbe CR, Page ST and Thirumalai A: Male
contraception. Yale J Biol Med. 93:603–613. 2020.PubMed/NCBI
|
|
40
|
Gava G and Meriggiola MC: Update on male
hormonal contraception. Ther Adv Endocrinol Metab.
10:20420188198348462019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hawkinson JE, Sinville R, Mudaliar D,
Shetty J, Ward T, Herr JC and Georg GI: Potent pyrimidine and
pyrrolopyrimidine inhibitors of testis-specific serine/threonine
kinase 2 (TSSK2). ChemMedChem. 12:1857–1865. 2017. View Article : Google Scholar : PubMed/NCBI
|