The intestinal tract contains a high concentration
of microorganisms and neurons, which have a profound influence on
both the physical and mental well-being of humans. The intestines
are also known as the ‘second brain’ of the human body, and while
they perform their normal functions, various intestinal diseases
are inevitably present (1–3). These diseases can be broadly
categorized into two groups: Functional diseases such as irritable
bowel syndrome (IBS), functional dyspepsia and functional
constipation, and organic diseases such as enterocolitis,
ulcerative colitis (UC), Crohn's disease (CD) and various
intestinal tumors (4–6). With the change in dietary habits and
lifestyles (such as irregular eating, oversatiety or overhunger) of
individuals, the incidence of IBS is on the rise, affecting ~20% of
the general population. This not only diminishes the quality of
life of individuals with the condition, but also causes a
substantial healthcare burden (7,8).
Furthermore, the prevalence of inflammatory bowel disease (IBD) has
been increasing worldwide over the last century. Due to the impact
of family history and weakened immune systems (9), there has been a notable rise in very
early onset IBD, which is identified before the age of 6 years
(10,11). This results in increased healthcare
expenses for patients and can also hinder their career aspirations
due to the enduring social stigma they face as they grow up.
Chronic inflammation of the colon and accelerated renewal of
epithelial cells can increase the risk of highly dysplastic growth,
leading to the further development of colorectal cancer (CRC)
(12). A previous study revealed
that individuals with UC or CD are at a higher risk of developing
both gastrointestinal and extraintestinal cancer (13,14).
The etiology, severity and advancement of this intricate set of
gastrointestinal disorders remain to be evaluated. For example, IBD
might initially be misdiagnosed as IBS, leading to a delay in early
treatment. The manifestations of IBD are varied and non-specific,
with a wide range of differential diagnoses. The possibility of
rapid growth and early metastasis of CRC is high, making early
detection particularly crucial (15–19).
Given the prevalence and challenges in diagnosing and treating
various gastrointestinal diseases, contemporary medical treatment
urgently needs more appropriate and optimized experimental methods
to explore and uncover the developmental origins of diseases in
order to effectively manage disease progression and provide
improved patient care.
The human immune system is a protective system
covering the whole body, which can maintain the immune homeostasis
of the body, while excluding exogenous antigenic substances
(20). The gastrointestinal tract,
as the initial line of defense for the immune system, is closely
associated with the occurrence of numerous diseases (21). The intestinal barrier has a
heterogeneous structure, including mechanical, chemical, microbial
and immune barriers (22,23). The intestinal mucosal barrier is
the first line of defense against penetration of luminal contents
and performs various biological functions (24), not only maintaining local
intestinal function, but also facilitating communication with the
nervous and endocrine systems via the bidirectional signaling
pathways of the brain-gut axis (25). Additionally, the barrier is
involved in immune, metabolic and emotional regulation, which is
closely linked to the function of intestinal microorganisms
(26). The human body, functioning
as a superorganism, coexists in symbiosis with trillions of
beneficial bacteria and eukaryotic cells. The advancement of the
Human Microbiome Project and METAgenomics (27,28)
of the human intestinal tract has offered novel technological
resources for the research of these microorganisms. There are
~500-1,000 different types of bacteria in the human body, which can
reproduce to about 100 trillion individual cells in an
adult-roughly 10 times the total number of a person's body cells.
These microorganisms benefit from the warm environment of the
intestines and serve a crucial role in digestion, vitamin
synthesis, immune system regulation, pathogen elimination, toxin
removal and maintenance of the normal functioning of the intestines
(29–32).
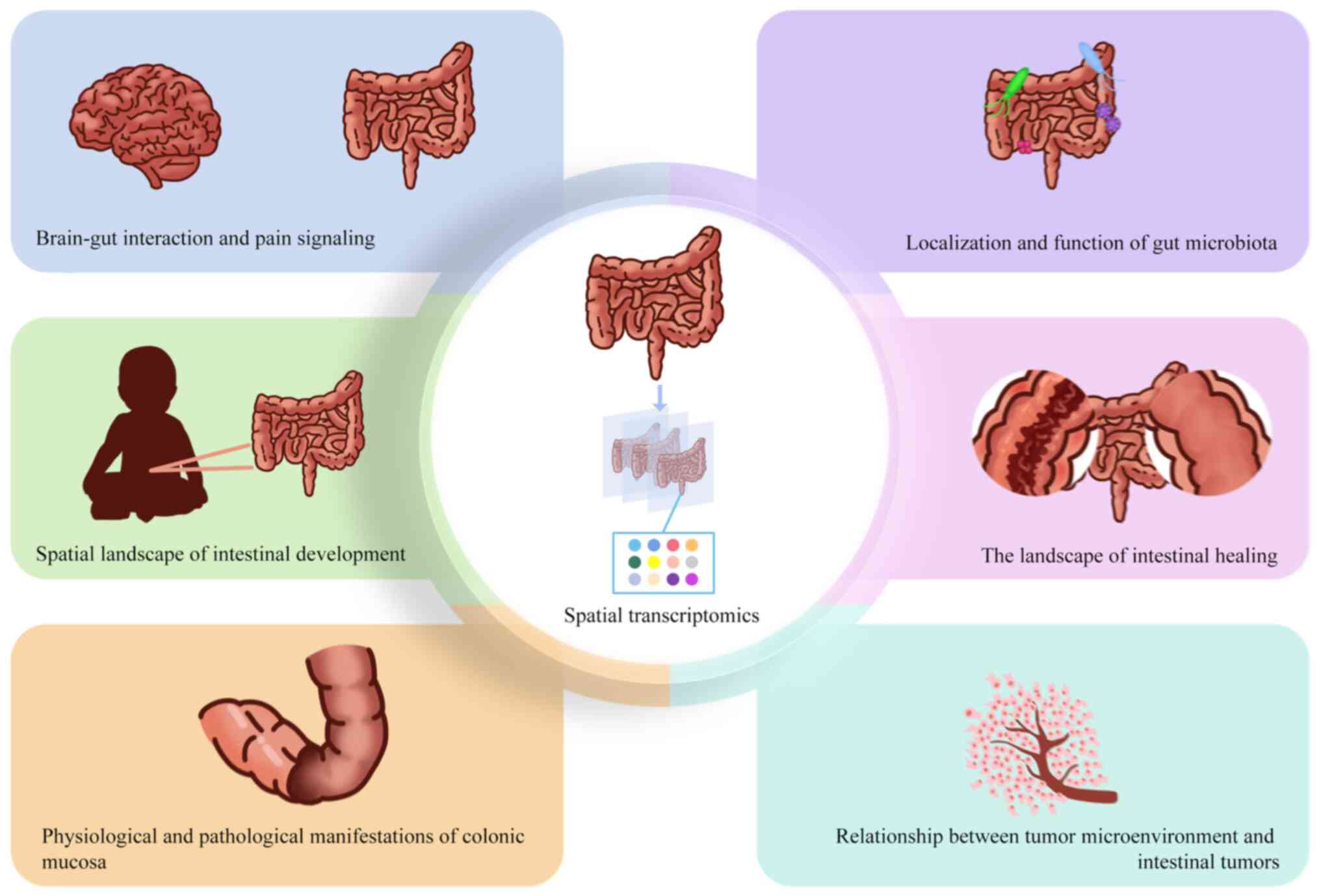
In summary, ST can help understand the biological
functions of the gut from multiple perspectives. First, generating
high-resolution maps of neurons or the gut using ST indicates the
development of the gut, the information exchange in the brain-gut
axis and pain signaling. Second, with the assistance of ST, the
physiopathological manifestations and healing of colonic tissues
can be observed dynamically, and accurate judgments can be made
about the development of intestinal diseases. Finally, spatially
specifying changes in the gut microbiota helps understand the
geographic features of the gut that have been altered by
pathogens.
Since the 1990s, with the rapid development of
high-throughput sequencing technology, transcriptomics has
gradually emerged as a discipline to study all transcribed RNAs in
cells. Researchers are starting to fully uncover gene expression
patterns and regulatory networks. However, conventional
transcriptomics techniques are unable to determine the effects of
different subcellular structures and environments on gene
expression. To more fully understand the regulatory processes of
gene expression, ST is rapidly emerging to reveal the spatial
distribution, subcellular location and interactions of genes within
the cell, providing novel perspectives for understanding the
spatial regulatory mechanisms of gene expression (39).
The development of transcriptome technology has gone
through three important stages: The first stage is transcriptome
sequencing (bulk RNA) of a large number of mixed cells, which
reveals the average expression level of a single gene in a large
cell population, but fails to demonstrate the transcriptional
expression level in a single cell (40). The second stage is scRNA-seq, which
constructs the expression profile of each cell at the single-cell
level, reflecting cell-to-cell heterogeneity (41). However, single-cell spatial
location information is lost during the preparation of the
suspension, resulting in changes in gene expression profiles of
some single cells during enzymatic digestion or after leaving a
specific microenvironment; this is one of the key features of organ
function. Driven by this need, the Joakim Lundeberg research group
proposed the concept of ST in 2016 and reported the first ST
technique based on in situ capture of RNA (42). This provides important research
tools in a number of fields, including tissue-cell function,
microenvironment interaction, lineage tracing of developmental
processes and disease pathology research. Subsequently, a series of
techniques capable of high-throughput in situ RNA detection
analysis have all been grouped into the category of ST techniques.
Although the principles of these techniques vary, they have a
common element with regard to recording information about the
spatial position of the detected RNA molecules (43).
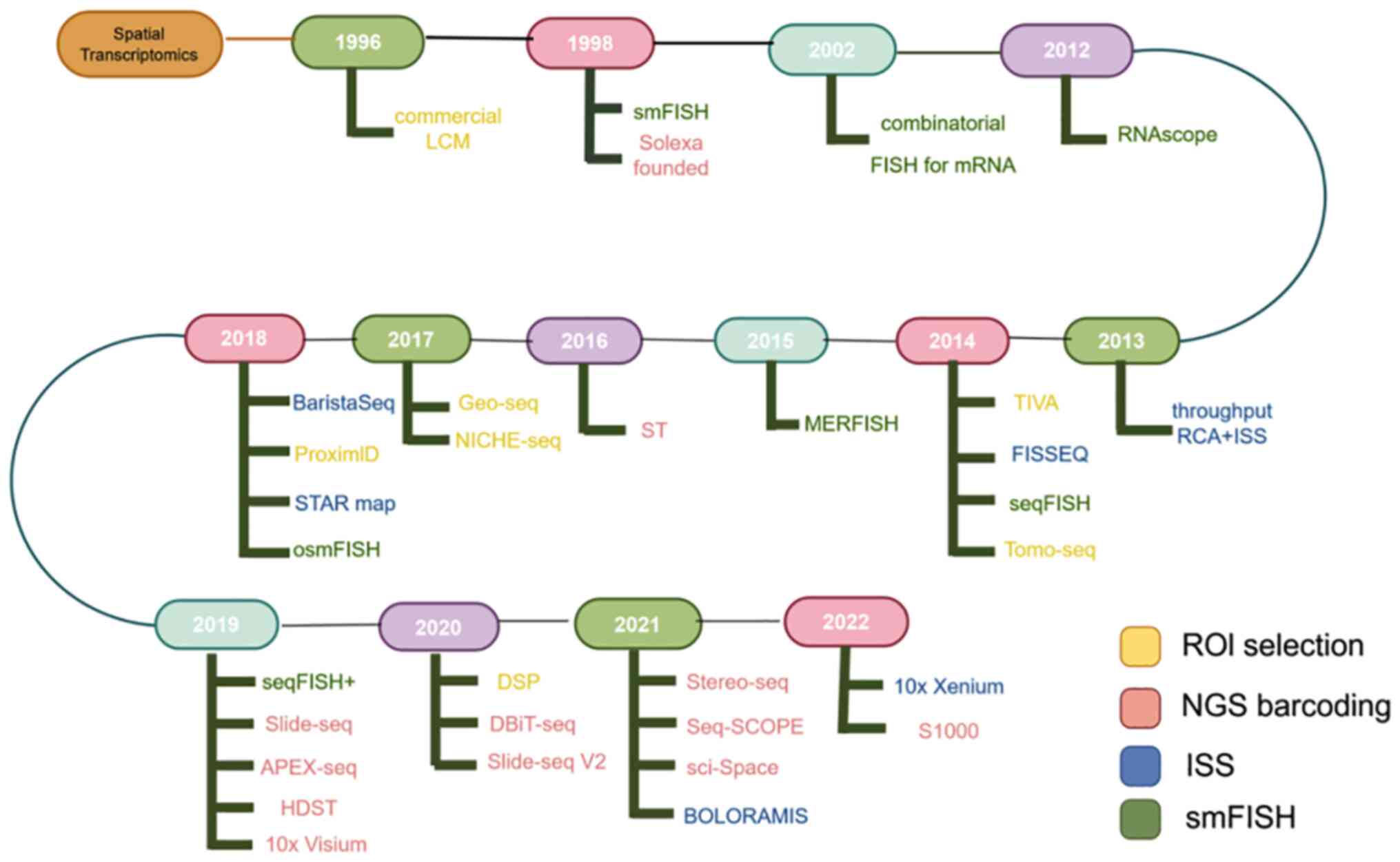
In the late 1990s, certain studies on ST started the
development of ST using technologies such as laser capture
microdissection (LCM), microarrays or RNA sequencing (RNA-seq) and
single-molecule FISH (smFISH). Some techniques from the 1980s,
although not called ‘spatial transcriptomics’, also acquired
transcriptional information in space. Moses and Pachter (42) refer to this part of the development
of ST technology as the ‘prequel era’ in their online ebook ‘Museum
of Spacial Transcriptomics’.
The prequel era technology depicts the general
technical characteristics of the spatial transcriptome: Imaging,
localization and expression level. At the same time, one of the
spatial transcriptomes that can see the ‘prequel era’ has been
working on imaging and single cells (resolution), and the
expression amount is often not obtained by high throughput.
Although spatial transcriptomics at that time was limited by
resolution, ease of operation, available software, rich databases
and other factors, particularly the high-throughput expression data
based on NGS not being available in the pre-transcriptomics era,
causing numerous technologies of that era to decline. However,
still, in the pre-transcriptomics era, numerous beneficial attempts
were made, and it can even be said that it provided a reference for
the spatial transcriptomics technology of the present era (39).
ST has undergone rapid development, with marked
improvements in resolution and flux. The spatial transcriptome was
declared to be the annual technical method by Nature Methods in
2020 and one of the seven noteworthy annual technologies by Nature
magazine in 2022. In 2023, space omics became one of the 10
technologies selected by the World Economic Forum to have the
highest potential and to have a positive impact on the world.
Currently, the commonly used spatial transcriptome
technologies are slide-DNA-seq, a method for capturing spatially
resolved DNA sequences from intact tissue sections (49), ST, sequential FISH, multiple
resistance error correction FISH (MERFISH), LCM-seq, geographical
position sequencing (Geo-seq) and Topographic Single Cell
Sequencing (Tomo-seq), which can be generally divided into four
different types of strategies, each with their own advantages and
disadvantages.
LCM-seq is a specific technique involving cutting of
frozen tissue or paraffin-embedded tissue, or single-cell cutting,
combined with subsequent chip or NGS to obtain the transcriptomic
data of single cells (59). In
2017, Chinese scientists established Geo-seq combined with LCM and
scRNA-seq technology, which overcomes the problem of insufficient
amounts of mRNA from a small number of cells and can achieve
cell-level accuracy; this can be used not only for 3D
reconstruction of a transcription map, but also to study the
transcriptomic information of a small number of tissues or cells
with special structures (60). In
2018, American scientists developed the TSCS technology. They used
lasers to capture single cells, amplified the whole genome of the
single cells, and attached independent label sequences. This
increased the experimental throughput. After mixed sequencing, they
matched the cell positions to tissues using labels and
computational methods (61).
Tomo-seq technology was inspired by clinical
multi-dimensional imaging technology. A research team sequenced all
zebrafish embryos in three different directions and reconstructed
different sections of the same axis to form complete 3D embryos by
computational methods. However, this method has high requirements
for computational methods, and necessitates three identical
samples, which is not applicable in clinical practice. The accuracy
also needs to be improved (62).
SmFISH uses a single probe sequence to bind the same
transcript, using the fluorescent color molecules on the probe to
display the expression level of the target molecule in the original
image. Although smFISH has high sensitivity and subcellular spatial
resolution, this method is susceptible to background influence and
low flux due to its weak light signal (65). In 2014, researchers improved the
invention of seqFISH, which uses multiple rounds of hybridization
to assign unique barcodes to individual transcripts within a single
cell. By performing consecutive rounds of hybridization, imaging
and probe stripping, a distinct barcode is generated for each mRNA
molecule, allowing for spatial transcriptomic mapping through color
recognition (66). However,
increasing the number of genes requires increasing the number of
hybridization rounds, which is expensive and time-consuming and
requires super-resolution microscopy (67).
The spatial barcode-based strategies can also enable
high-throughput detection of cells and are not limited to known
sequences. These strategies mainly consist of ST technology
(34) and Slide-seq technology
(68).
In 2016, Spatial transcriptomics (ST) printed 1,007
pre-defined points on a 6.2×6.6 mm array, each with a spatial
barcode, including paste part, sequencing fragments, position tags,
random unique molecular identifier and mRNA capture poly (T), and
each barcode was captured with mRNA molecules for library
construction and sequencing to obtain a transcriptome spatial map
(34). ST Technologies pioneered a
newera of combining in situ RNA amplification with spatial
barcode arrays. In 2018, 10× Genomics(https://www.10×genomics.com/) acquired ST
Technologies, and in 2019, they released 10× Visium, reducing the
distance from point to point from 200 to 100 microns for higher
precision. ST indicates genome-wide spatial expression at the
micrometer scale, but does not achieve single-cell resolution.
In 2019, Professors Alice Y. Ting and Howard Y.
Chang from Stanford University introduced a new RNA localization
detection technique called APEX-seq (69). The use of peroxidase APEX2 can
produce covalent biotinylation of RNA molecules and has different
sensitivities in different organelle regions, which allows the
realization of transcriptome studies on different organelles.
However, as cells are required to express APEX2, which is an
engineered soybean ascorbate peroxidase, it cannot be used in
clinical practice.
In 2020, NanoString Technologies released the GeoMx
Digital Spatial Profiler, which is conjugated to DNA Oligo on
antibodies or RNA (70). Each DNA
Oligo corresponds to one target, with multiple reactions achieved
through multiple targets. When the antibody or RNA is bound to the
target on the tissue, ultraviolet light is used to cut off the
linker between DNA Oligo and the antibody or RNA, thus releasing
DNA Oligo for the next quantification of the transcriptome and
proteome.
High-definition ST (HDST) and Slide-seq can
implement spatial barcodes on microbeads (68,71).
Slide-seq links spatial barcodes on a batch of 10-µm microbeads,
and mRNA molecules in tissue sections are captured by microbead
binding, which are then processed using ‘sequencing by oligo
ligation detection’ sequencing to map tissue space by spatial
barcode. HDST is a microwell arranged with 2 micron, each
containing a microbead, which links a spatial barcode on the bead,
bound to the mRNA in the tissue, and then allows Illumina, Inc.
Sequencing.
In 2021, space omics technology was a research
focus. The group of Professor Rong Fan from Yale University
invented DBiT-seq spatial transcriptomics, which is a method of
adding barcodes in a different way (72). Tissues are first attached to
slides, with a minimum inter-hole distance of 10 micrometers
achieved using microfluidic technology. Two sets of liquid
containing barcodes are then flowed through the tissue sequentially
from two axes, binding to mRNA and protein molecules in the tissue
to form a 2D spatial barcode. Reverse transcription is performed
inside the cells, and cDNA is collected for subsequent sequencing
(73). Although the method can
achieve simultaneous capture of the transcriptome and proteome,
there may be barcode contamination, and the pore spacing can only
be 10 µm, limiting the accuracy of the technique (74).
Chinese scientists established Stereo-seq space
omics technology on a BGI sequencing platform. The technique is
based on the DNA Nanoballs (DNB) sequencing technology. It works by
depositing DNB containing random barcode sequences on a lithoetched
modified chip. The nanosphere spacing can be 500 or 715 nm, using a
rolling ring to amplify the barcode pool, and obtain the coordinate
code of the DNB after the first round of sequencing. Then, by
hybridization, connecting the molecular coding and polyT sequence,
the tissue was loaded onto the chip and the barcode was captured
with mRNA for a second round of sequencing to finally obtain the
transcriptome map (75).
American scientists have released the Seq-Scope
spatial group on the Illumina platform. This method involves two
rounds of sequencing. In the first round, sequencing is done on the
Illumina sequencing platform to amplify clusters on a chip with
fixed labels. The distance between clusters can be achieved at
0.6–0.8 micrometers. After amplification, a restriction
endonuclease is used to expose the Oligo-dT sequence. Following the
capture of mRNA molecules in the tissue, a second round of
sequencing is conducted to obtain the transcriptome map (76).
Numerous research studies have used scRNA-seq
techniques to identify the diversity of intestinal epithelial,
mesenchymal and immune cell types in the adult intestines (95–97).
This has led to the exploration of several important questions
using this information. Initially, the process of crypt-villus axis
formation was elucidated. By scoring the activity of all modules
(in the field of transcriptomics, modules can be understood as
collections of genes with similar expression patterns), it was
found that modules derived from endothelial cells, fibroblasts and
pericytes are located deep in the tissue; epithelial-specific
modules expressing Hedgehog pathway genes (such as the convolution
donor receptor LDL receptor related protein 5) (94), are located in the vicinity of the
canaliculus, and genes expressed by modules containing
myofibroblasts (such as WNT2B) emerge later and are abundant at PCW
12, indicating that the ISC-myofibroblast signaling circuit is
established only after crypt formation. These findings suggest that
ST may partially restore the disrupted morphological gradients and
increased physical distances during intestinal development
(93). Furthermore, it can
determine the hierarchical structure and differentiation functions
of fibroblasts and myofibroblast subtypes. After classifying
fibroblasts (98), intestinal
fibroblasts were divided into different functional zones according
to the characteristics of different phenotypes along the crypt
structure to the villus axis (99). According to such anatomical zoning,
as the main determinant of intestinal fibroblast heterogeneity, one
of the original studies identified human colonic fibroblasts with
numerous fibroblast subcharacteristics, Wnt differential expression
and bone morphogenic protein signaling genes, which reflect the
specific location along the anatomical axis of crypt villi
(100). Antibodies in colon
mesenchymal cells of UC patients were further investigated using
scRNA-Seq and flow sorting to enrich CD90 cells. This study
identified four populations of colonic stromal fibroblasts (S1-S4).
After classifying fibroblasts, the analysis of various morphogens
with different cells and genes expression revealed that the
submucosal structural cells were mainly composed of S1 cells. S2
fibroblasts mainly contribute to maintaining the epithelial crypt
ecological niche and promoting epithelial formation (101,102), whereas S3 fibroblasts are more
commonly found in large vessels, emphasizing their role in forming
the intestinal vascular support ecotone (93). S4 fibroblasts are closely
associated with immune follicular cells in the adult colon and
exhibit characteristics of follicular reticulocytes, which are
crucial for the formation of lymphoid structures and are closely
linked to the pathogenesis of UC (98). The development of blood vessels and
nerves in the intestines, the formation of Peyer's patches (PP),
gut-associated lymphoid tissue (GALT), and the immune system
processes have been established (94). PPs are secondary lymphoid organs
that interact with the external environment via the intestinal
lumen. Spatial transcriptome analysis showed that fetal type 3
lymphocytes (innate lymphoid cells) express IL7RA and inhibitor of
DNA binding 2, crucial genes for PP formation in the mouse uterus
(103). Additionally,
deficiencies in GDNF family receptor α3 also result in
developmental disorders of PP. A study confirmed that GALT
formation occurs in the small intestine prenatally and in the colon
after birth (104), and lymphoid
structures can be visualized by mapping factors enriched for
B-cell-related genes onto colonic tissue. Enrichment of the NF-κB
and TNF-α pathways has been detected in regions of immune and
inflammatory activity, closely matching the spatial distribution of
lymphoid clusters (105).
The intestinal epithelium maintains the normal
physiology of the intestinal tract through continuous regeneration,
and disruption of the regeneration process of intestinal epithelium
can lead to pathogen translocation, which in turn mediates various
chronic intestinal diseases (106). scRNA-seq technology has broadened
the understanding of the cell types, cell subpopulations and cell
states present in physiological and pathological states (107). scRNA-seq has also enabled the
detection of subpopulations of cells abnormally driven (activation
of KRAS in lung cancer is a poor prognostic indicator) by disease
conditions, which is of considerable importance in the study of
disease mechanisms (108,109). One study revealed that even
though mice given dextran sodium sulfate (DSS) showed improvement
in symptoms and body weight after a period of DSS discontinuation,
the inflammatory changes in the colon did not fully return to
normal levels (110). Therefore,
it is crucial to analyze the transcriptomic landscape of the
mucosal healing process in detail. Parigi et al (111) used ST to reveal previously
unrecognized molecular regionalization in the colon of healthy
mice. The study revealed a spatially organized transcriptional
program for regionalized mucosal healing, demonstrating that
factors in the proximal and distal colon have distinct functions
and are involved in different regulatory processes, which was
determined through dataset integration and associated pathway
analyses. At the same time, the integration of longitudinal and ST
data to observe the expression status of different modules at
different stages can reveal the dynamic timing of colon tissue
healing (110,111). Studies have revealed that
clusterin accumulated during the repair phase following intestinal
injury. Clusterin is seldom present in a healthy state, and further
verification is needed to determine its association with infection.
Multimodal spatial analysis will help identify important events
related to tissue damage repair (111,112).
Macrophages are resident immune cells that exist in
two primary states following activation: M1 macrophages and M2
macrophages. The functions and appearances of these macrophages
vary depending on the microenvironment and location, and there is
ongoing debate with regard to their roles and characteristics
(113). What has been
demonstrated is a higher expression of the M1 population in IBD
samples, and that the phenotype and function of intestinal
macrophages are related to their spatial distribution in the gut
(114). ST has revealed the
transcriptional status of macrophages in healthy and inflammatory
states, defined specific markers for M2 macrophages, and clarified
cell types and differences in the spatial distribution of different
cells during intestinal inflammation (99). For example, ST showed that M1
macrophages and neutrophils were located near the mucosal surface
in areas with intestinal ulcers. Spatial analysis has verified the
diversity of macrophage populations and highlighted their
interaction with inflammatory fibroblasts (115). This interaction enhances colony
stimulating factor 2 expression and stimulates macrophage
activation, leading to the release of IL-6 and TNF (115), which serve a role in the
inflammatory process, and this spatial analysis technique is
essential for comprehending the interactions between several
specific cells.
The gut microbiota is an integral part of the human
body, and although a portion is highly conserved, the dynamic
microbiota changes in response to age, and physiological and
pathological states, and participates in processes such as
neuromodulation, immunity and metabolism (116,117). The human gut consists mainly of
Bacteroidota and Bacillota bacterial phyla, and an
imbalance between these two phyla is considered to be a sign of
microbial dysbiosis, which is closely linked to the pathogenesis of
IBD (118). Dysfunction of the
microbiota, such as an increase in facultative anaerobes and a
decrease in obligate anaerobes, occurs during periods of active
intestinal inflammation (119).
Currently, the study of gut microbes is dominated by the use of
genomics, which provides a comprehensive understanding of microbial
categories and certain functions (120,121), but it does not offer spatial
information about the microbes. The spatial organization of gut
microbes can affect various properties such as colonization,
metabolism and stability of the internal environment (122,123). It has been found that the
distribution and abundance of different phyla is heterogeneous in
different regions of the gut and that the microbial composition
differs considerably between species (124).
The stomach, being the most acidic part of the
digestive system, has fewer bacterial species. In the transition to
the duodenum and ileum, the pH markedly increases and the oxygen
pressure gradient decreases, creating a more favorable environment
for bacterial growth. Although study results have indicated that
the bacterial diversity in this area is either lower or equal to
that of the stomach, there is a notable increase in the abundance
of specific microbiota (125,126). This low diversity environment
protects the intestinal tract to a certain extent, while bacterial
overgrowth can potentially contribute to the onset of conditions
such as IBS or functional dyspepsia (127). By the time it reaches the cecum
and proximal colon, the intestinal lumen becomes intensely
anaerobic and transit time is slowed due to the decreased pH, as a
result of fermentation of fibers and complex polysaccharides.
Dietary fibers provide ample nutrients for the growth of microbes,
leading to a marked increase in both the quantity and variety of
microbial species (128,129). In the human colon, there are
higher concentrations of Bacteroidota, Pseudomonadota and
Bacillota, and processes that shape the gut microbiota are
thought to be mostly niche-driven (122). The intestinal epithelium has
folds, villi and invaginations on its surface, known as crypts,
which offer specialized protection to the intestine, but the crypts
are heterogeneous in their response to injury. Paneth cells
colonize the crypts of the small intestine and secrete
antimicrobial peptides to achieve multi-level management of the
intestinal flora and to maintain a stable intestinal flora
structure. Since the large intestine lacks Paneth cells, it is
guarded at the entrance of the colonic crypts by cup cells, which
secrete mucin and form a mucus barrier to protect the epithelial
cells (130). In addition, it has
been previously reported that chemokines with antimicrobial
activity against Escherichia coli and Salmonella
enterica are found in colonic crypt compartments, which serve
an important role in coordinating the normal influx of immune cells
and inflammation in the intestine (131). Various microorganisms inhabit
crypts, and some also extend their colonization beyond these areas,
including into the PP, a specific region where bacteria directly
interact with host tissues. This site serves as a sampling location
for the microbiota of the immune system. Here, specialized cells
access microbial and environmental antigens, passing them to
antigen-presenting dendritic cells, participating in the immune
process (132).
The degree of bacterial spatial organization in the
gut microbiota varies, and the peristaltic contractions of the
intestines can promote the fusion of bacteria in the gut.
Therefore, even in the colon where the content flow rate is low,
there is a high degree of mixing in the gut microbiota (133,134). A sample block sampling technique
used to characterize the spatial organization of microbial
communities at the 10–30-µm scale found well-mixed sites scattered
with micron-scale clusters of specific taxa (126), which may be related to the
heterogeneity of food and plant particles within the lumen
(131,135). Bifidobacterium
pseudolongum has been proved to colonize starch granules and
plant granules, and Bacteroides colonizes undigested plant
granules and is spatially segregated into multiple small colonies,
suggesting that mucosal communities based on different intestinal
regions could be useful in determining the role of mucosal
geography in driving colonization of epithelial surfaces (136,137). Alterations in the microbiota are
strongly associated with various human illnesses, such as IBD and
CRC, as well as skin and mental health conditions (138,139). Clarifying the geographical
distribution of gut microbiota affected by pathogens will provide
novel insights for the understanding of host health and disease
(140).
IBD and IBS are commonly viewed as gastrointestinal
conditions resulting from heightened sensitivity in the colon,
along with dysfunction in other visceral and somatic organ
(141,142). The interaction of sensory signals
between multiple organs, known as ‘cross-organ sensitization’, is
caused by the dorsal root ganglia transmitting sensory data for
integrated processing (143).
Injury receptors are located in the dorsal root ganglion and
trigeminal ganglion, where they receive sensory signals related to
bodily injury. These receptors are the first neurons in the pain
pathway and express a range of receptors, allowing them to respond
to various stimuli (144). In
cases of cross-organ sensitization linked to intestinal
dysfunction, the sensitization of colonic afferent neurons can lead
to the cross-activation of reflex pathways in other uninjured
organs, with the release of substance P, calcitonin gene-related
peptide and excitatory amino acids to the peripheral organs, which
activate the injury receptors and lower their thresholds (143). The excitability of these neurons
increases in both acute and chronic pain states, and their excited
phenotype changes are directly associated with the chronic pain
state, so these neurons are key targets for the treatment of pain
(144). ST enables researchers to
draw high-resolution maps of human sensory neurons in the dorsal
root ganglion, which can help identify more effective drug targets,
therefore opening up novel avenues for pain management (145).
The mental, nervous, endocrine and immune regulation
via the brain-gut axis serves an important role in the onset of
certain intestinal diseases (146). The hypothalamus serves a leading
role in regulating the basic social behaviors of the body and
homeostatic functions, and is involved in the regulation of
anxiety, depression, immune inflammation and gut microbiota
(147,148). Studies have found that the volume
of the hypothalamus in patients with IBD was larger than that of
patients in the control group, and that it is also more strongly
connected to other brain functional areas (149,150). This excessive connectivity can
lead to the activation of abnormal anxiety circuits, mediating
emotional disorders (151).
However, the molecular characteristics of the development of the
human hypothalamus are still unclear, and there is limited
knowledge about the structure, spatial organization and function of
the hypothalamic nucleus. The imaging-based single-cell
transcriptomics method combines gene expression profiles with
activity markers to directly image and analyze multiple RNAs in
their natural cellular environment, thereby mapping out their
spatial structure (152). By
uniformly slicing the preoptic area, researchers have created a
cellular map of the mouse hypothalamic preoptic area, identifying
rare cell types and describing the spatial organization of specific
neuronal cell types. This revealed their functions in different
behaviors, providing potential insight into how different cell
types communicate in physiological and pathological states
(63). At the same time, ST can
reveal the spatiotemporal transcriptional profile and cell type
characteristics of human hypothalamic development. Mapping the
spatiotemporal transcriptome of human hypothalamic development has
demonstrated the asynchrony of spatial development of different
neurons and neuroglial cells, identified key regulatory genes that
neural progenitor cells and neural epithelial cells, and provided a
deeper understanding of cellular network organization, circuit
formation and the mechanisms of hypothalamic dysfunction (63,153). Analyzing the spatial
transcription of key areas of the brain and gaining a comprehensive
understanding of the connectivity patterns of the nervous system of
the brain can provide valuable insights for studying the brain-gut
axis, potentially offering novel therapeutic targets for the
treatment of brain-gut-related diseases (153,154).
During the progression of tumors, the interactions
between cancer cells and other cell populations promote the
heterogeneity of tumors. Experimental findings have demonstrated
that the tumor microenvironment (TME) serves an important role in
the development of cancer, and its spatial distribution and
specific host-microbe cell interactions to some extent influence
the progression of tumors (155,156). Single-cell sequencing of the
genome, transcriptome and epigenome has contributed to the
understanding of the internal structure of tumors, and more
advanced ST is expected to decipher the complex principles and
mechanisms of gene activity in three dimensions, which will have
profound implications for life science research (157).
CRC is the third most common malignant tumor
globally, and due to its high mortality rate due to metastasis,
nearly 900,000 individuals die from CRC each year (158). Since CRC only presents with
noticeable symptoms in the late stages, it is particularly
important to identify relevant oncogenes and improve the early
detection rate and early treatment of CRC (159,160). The combined application of
multi-omics is important in studying the biological characteristics
of the early local spread and distant metastasis of CRC (161). Cancer-associated fibroblasts
(CAFs) are a major component of the stromal cells in numerous
malignant tumors (109,162). In order to determine the
interaction between the TME and CAFs in the pathogenesis of CRC,
researchers have combined ST and bulk RNA-seq. Two types of CAFs
were identified: Myo-CAFs and inflammatory CAFs. The latter not
only promoted the progression and metastasis of tumors, but were
also associated with the poor prognosis of the tumor (163). Certain studies have also found
that spatial analysis can reveal the molecular and immunological
characteristics of early-stage CRC, identify biomarkers associated
with disease progression and prognosis, and reveal genes related to
CRC invasion, providing a deeper understanding of the biological
processes associated with tumor malignancy (164,165). The ST and GeoMx digital spatial
profiling technologies of 10× Visium can identify the identity and
location of the internal microbial community within tumors. The
technologies can also detect areas with low vascularization, high
levels of immune suppression and association with malignant tumors.
Research has shown that the degree of vascularization in
bacteria-colonized regions is lower than in bacteria-negative
regions, with decreased expression of smooth muscle actin,
decreased proliferation levels and downregulation of Ki-67 and p90
RSK. The bacteria-colonized microniches significantly increases the
phosphorylation levels of JNK, ERK1, ERK2 and P38 in CRC tumors,
revealing signal pathways that are activated in response to
bacteria. The results indicated that the infected areas of CRC
tumor tissue have lower proliferative potential compared to
uninfected areas (166–168).
The liver metastasis of CRC poses a significant
challenge for clinicians, with 80% of CRC cases experiencing tumor
metastasis to the liver before the primary tumor can be detected
clinically, which is closely associated with low survival rates
(169,170). There is controversy regarding
whether or not aging cancer cells serve a positive role in the
progression and metastasis of tumors. Researchers have used ST and
multi-database integration to comprehensively map the whole
transcriptome cell atlas of CRC and related liver metastasis. They
analyzed the molecular specificity within the TME and the
transcriptional heterogeneity of senescent cancer cells, which
could identify the aging-dependent cancer ecosystem in CRC liver
metastasis and potentially provide therapeutic targets for
CRC-related cancer types (171,172).
As one of the eight regions of the digestive
system, the gastrointestinal tract serves an important role in
absorption, digestion, excretion, endocrine, nutritional
metabolism, immune balance and other functions. Due to the
diversity of intestinal cells, dissecting the characteristics,
functions and internal operation of the cells in the intestinal
mucosa has been a technical challenge (173,174). The advent of ST provides a
higher-resolution research tool for developmental and
cancer-related studies of the gut, adding new dimensions to the
understanding of multiple gastrointestinal diseases (94). Table
I shows that different ST technologies are applied to different
intestinal diseases, and the corresponding conclusions are drawn
from different materials, indicating that they are widely used in
intestinal diseases (111,175–181).
The present review discusses the applications of ST
technologies related to the gut. The rapid development of ST and
its combined application with multi-omics has provided powerful
technical support for studying the mechanisms involved in gut
development and physiological pathology.
Due to the complex structure of the intestines,
accurately mapping their spatial organization and molecular
characteristics is crucial for understanding the progression of
diseases. Various sequencing technologies have been continuously
evolving, from scRNA-Seq to ST based on scRNA-Seq, enabling the
visualization of the spatial positioning of tissues and cells.
Given that low-resolution ST cannot meet higher imaging
requirements, researchers have developed high-definition spatial
genomics. HDST has been combined with powerful imaging
modularization techniques through the modification of microbead
arrays and the development of polynomial Bayesian classifiers,
resulting in a marked increase in the resolution of the technique
to 1,400 times higher than that of normal ST. Together with the
highly specific nature of HDST signals, which can be interpreted
through computational integration with morphological features and
single-cell profiles, this has opened up novel avenues for
high-resolution spatial analyses of cells and tissues (70,182). With the continuous development of
sequencing technology, phenotypic information at the protein level
on the cell surface has been gradually and well presented by novel
technologies, and the use of DNA-barcoded antibodies allows for
simultaneous analysis of the single-cell transcriptome and
proteome, whereas NGS-based ST allows for the analysis of spatial
multi-omics data. The single-cell multi-omics approach permits a
more comprehensive understanding of intestinal biology and disease
mechanisms, which facilitates the prediction of drug targets and
clinical responses (183,184). More in-depth studies are already
underway, including the use of ST to study epigenomic, proteomic or
chromosomal structures (185),
which can define neuroscience- and cancer-related cell types,
provide spatial analyses that can present epigenetic traits at the
single-cell level, and provide insight into the spatial state of
protein molecules in tissues (186–188).
Numerous discoveries in the life sciences are
closely related to the biological function of tissue and cell
interactions, where spatial relationships between cells determine
cell fate and diseases emerge as abnormalities in spatial
structures within tissues (189).
ST reconstructs spatial organization based on single-cell
sequencing and combines it with other methods, such as different
sequencing techniques, computer programming and visualization
technology to present biological processes more intuitively. These
technologies have facilitated new discoveries in various fields
ranging from intestinal development to various diseases of the
intestines.
Efficient research methods serving research should
also serve the clinic, and the clinical application of ST is one of
the important issues in translational medicine (190). Complete and efficient sampling
and testing protocols, as well as the analysis and interpretation
of data, are key to applying ST in clinical settings. A number of
transcriptome-based commercial platforms are progressively being
developed for applications that utilize artificial intelligence
(AI) to provide open datasets and accurate data processing methods
(echnologies such as SOMDE, SEDR and STAGATE), leveraging the
potentials of AI, which will contribute to the clinical application
of ST (68,191–193).
Not applicable.
The present study was supported by Chongqing Natural Science
Foundation (grant no. cstc2021jcyj-msxmX0858).
Not applicable.
YH and YG discussed the premise of the study and
conceived the format. YG drafted the initial manuscript based on
this discussion. CR completed the revisions of the manuscript. YH
and YW generated the figures. YW revised the content of the
manuscript. XY proposed the main idea of the article and
coordinated the work to ensure the accuracy or completeness of the
manuscript and address any issues that arise during the writing
process. All authors have read and approved the final version of
the manuscript. Data authentication is not applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Caruso R, Lo BC and Núñez G:
Host-microbiota interactions in inflammatory bowel disease. Nat Rev
Immunol. 20:411–426. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Round JL and Mazmanian SK: The gut
microbiota shapes intestinal immune responses during health and
disease. Nat Rev Immunol. 9:313–323. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sardinha-Silva A, Alves-Ferreira EVC and
Grigg ME: Intestinal immune responses to commensal and pathogenic
protozoa. Front Immunol. 13:9637232022. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Longstreth GF, Thompson WG, Chey WD,
Houghton LA, Mearin F and Spiller RC: Functional bowel disorders.
Gastroenterology. 130:1480–1491. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mayer EA, Ryu HJ and Bhatt RR: The
neurobiology of irritable bowel syndrome. Mol Psychiatry.
28:1451–1465. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Flynn S and Eisenstein S: Inflammatory
bowel disease presentation and diagnosis. Surg Clin North Am.
99:1051–1062. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bonetto S, Fagoonee S, Battaglia E,
Grassini M, Saracco GM and Pellicano R: Recent advances in the
treatment of irritable bowel syndrome. Pol Arch Intern Med.
131:709–715. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Saha L: Irritable bowel syndrome:
Pathogenesis, diagnosis, treatment, and evidence-based medicine.
World J Gastroenterol. 20:6759–6773. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Singh N and Bernstein CN: Environmental
risk factors for inflammatory bowel disease. United European
Gastroenterol J. 10:1047–1053. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rosen MJ, Dhawan A and Saeed SA:
Inflammatory bowel disease in children and adolescents. JAMA
Pediatr. 169:1053–1060. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang YZ and Li YY: Inflammatory bowel
disease: Pathogenesis. World J Gastroenterol. 20:91–99. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Neurath MF: IL-23 in inflammatory bowel
diseases and colon cancer. Cytokine Growth Factor Rev. 45:1–8.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nadeem MS, Kumar V, Al-Abbasi FA, Kamal MA
and Anwar F: Risk of colorectal cancer in inflammatory bowel
diseases. Semin Cancer Biol. 64:51–60. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Brackmann S, Andersen SN, Aamodt G,
Langmark F, Clausen OPF, Aadland E, Fausa O, Rydning A and Vatn MH:
Relationship between clinical parameters and the colitis-colorectal
cancer interval in a cohort of patients with colorectal cancer in
inflammatory bowel disease. Scand J Gastroenterol. 44:46–55. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Borowitz SM: The epidemiology of
inflammatory bowel disease: Clues to pathogenesis? Front Pediatr.
10:11037132023. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kaplan GG: The global burden of IBD: From
2015 to 2025. Nat Rev Gastroenterol Hepatol. 12:720–727. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zheng HB, de la Morena MT and Suskind DL:
The growing need to understand very early onset inflammatory bowel
disease. Front Immunol. 12:6751862021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Taylor S and Lobo AJ: Diagnosis and
treatment of inflammatory bowel disease. Practitioner. 260:19–23.
2016.PubMed/NCBI
|
|
19
|
Chachu KA and Osterman MT: How to diagnose
and treat IBD mimics in the refractory IBD patient who does not
have IBD. Inflamm Bowel Dis. 22:1262–1274. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Parkin J and Cohen B: An overview of the
immune system. Lancet. 357:1777–1789. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kayama H, Okumura R and Takeda K:
Interaction between the microbiota, epithelia, and immune cells in
the intestine. Annu Rev Immunol. 38:23–48. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Groschwitz KR and Hogan SP: Intestinal
barrier function: Molecular regulation and disease pathogenesis. J
Allergy Clin Immunol. 124:3–22. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wu S, Yang L, Fu Y, Liao Z, Cai D and Liu
Z: Intestinal barrier function and neurodegenerative disease. CNS
Neurol Disord Drug Targets. Nov 24–2023.(Epub ahead of print).
|
|
24
|
Wu Y, Tang L, Wang B, Sun Q, Zhao P and Li
W: The role of autophagy in maintaining intestinal mucosal barrier.
J Cell Physiol. 234:19406–19419. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ding JH, Jin Z, Yang XX, Lou J, Shan WX,
Hu YX, Du Q, Liao QS, Xie R and Xu JY: Role of gut microbiota via
the gut-liver-brain axis in digestive diseases. World J
Gastroenterol. 26:6141–6162. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rutsch A, Kantsjö JB and Ronchi F: The
gut-brain axis: How microbiota and host inflammasome influence
brain physiology and pathology. Front Immunol. 11:6041792020.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Integrative HMP (iHMP) Research Network
Consortium, . The integrative human microbiome project. Nature.
569:641–648. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gueimonde M and Collado MC: Metagenomics
and probiotics. Clin Microbiol Infect. 18 (Suppl 4):S32–S34. 2012.
View Article : Google Scholar
|
|
29
|
Adak A and Khan MR: An insight into gut
microbiota and its functionalities. Cell Mol Life Sci. 76:473–493.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Patterson E, Ryan PM, Cryan JF, Dinan TG,
Ross RP, Fitzgerald GF and Stanton C: Gut microbiota, obesity and
diabetes. Postgrad Med J. 92:286–300. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu X, Chen Y, Zhang S and Dong L: Gut
microbiota-mediated immunomodulation in tumor. J Exp Clin Cancer
Res. 40:2212021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Qin J, Li R, Raes J, Arumugam M, Burgdorf
KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al: A
human gut microbial gene catalogue established by metagenomic
sequencing. Nature. 464:59–65. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Marx V: Method of the year: Spatially
resolved transcriptomics. Nat Methods. 18:9–14. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ståhl PL, Salmén F, Vickovic S, Lundmark
A, Navarro JF, Magnusson J, Giacomello S, Asp M, Westholm JO, Huss
M, et al: Visualization and analysis of gene expression in tissue
sections by spatial transcriptomics. Science. 353:78–82. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang Y, Liu B, Zhao G, Lee Y, Buzdin A, Mu
X, Zhao J, Chen H and Li X: Spatial transcriptomics: Technologies,
applications and experimental considerations. Genomics.
115:1106712023. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shah S, Lubeck E, Zhou W and Cai L: In
situ transcription profiling of single cells reveals spatial
organization of cells in the mouse hippocampus. Neuron. 92:342–357.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lee JH, Daugharthy ER, Scheiman J, Kalhor
R, Yang JL, Ferrante TC, Terry R, Jeanty SS, Li C, Amamoto R, et
al: Highly multiplexed subcellular RNA sequencing in situ. Science.
343:1360–1363. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang X, Allen WE, Wright MA, Sylwestrak
EL, Samusik N, Vesuna S, Evans K, Liu C, Ramakrishnan C, Liu J, et
al: Three-dimensional intact-tissue sequencing of single-cell
transcriptional states. Science. 361:eaat56912018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Asp M, Bergenstråhle J and Lundeberg J:
Spatially Resolved transcriptomes-Next generation tools for tissue
exploration. Bioessays. 42:e19002212020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Shi Y, Wu X, Zhou J, Cui W, Wang J, Hu Q,
Zhang S, Han L, Zhou M, Luo J, et al: Single-nucleus RNA sequencing
reveals that decorin expression in the amygdala regulates
perineuronal nets expression and fear conditioning response after
traumatic brain injury. Adv Sci (Weinh). 9:e21041122022. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li J, Wu C, Hu H, Qin G, Wu X, Bai F,
Zhang J, Cai Y, Huang Y, Wang C, et al: Remodeling of the immune
and stromal cell compartment by PD-1 blockade in mismatch
repair-deficient colorectal cancer. Cancer Cell. 41:1152–1169.e7.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Moses L and Pachter L: Museum of spatial
transcriptomics. Nat Methods. 19:534–546. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chen KH, Boettiger AN, Moffitt JR, Wang S
and Zhuang X: RNA imaging. Spatially resolved, highly multiplexed
RNA profiling in single cells. Science. 348:aaa60902015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Schena M, Shalon D, Davis RW and Brown PO:
Quantitative monitoring of gene expression patterns with a
complementary DNA microarray. Science. 270:467–470. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Luo L, Salunga RC, Guo H, Bittner A, Joy
KC, Galindo JE, Xiao H, Rogers KE, Wan JS, Jackson MR and Erlander
MG: Gene expression profiles of laser-captured adjacent neuronal
subtypes. Nat Med. 5:117–122. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
46
|
Bhatia HS, Brunner AD, Öztürk F, Kapoor S,
Rong Z, Mai H, Thielert M, Ali M, Al-Maskari R, Paetzold JC, et al:
Spatial proteomics in three-dimensional intact specimens. Cell.
185:5040–5058.e19. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Alfieri CM, Mattinzoli D, Ikehata M,
Cresseri D, Moroni G, Vaira V, Ferri G, Ferrero S and Messa P:
Laser capture microdissection on formalin-fixed and
paraffin-embedded renal transplanted biopsies: Technical
perspectives for clinical practice application. Exp Mol Pathol.
116:1045162020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Achanta S, Gorky J, Leung C, Moss A,
Robbins S, Eisenman L, Chen J, Tappan S, Heal M, Farahani N, et al:
A comprehensive integrated anatomical and molecular atlas of rat
intrinsic cardiac nervous system. iScience. 23:1011402020.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhao T, Chiang ZD, Morriss JW, LaFave LM,
Murray EM, Del Priore I, Meli K, Lareau CA, Nadaf NM, Li J, et al:
Spatial genomics enables multi-modal study of clonal heterogeneity
in tissues. Nature. 601:85–91. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Liao J, Lu X, Shao X, Zhu L and Fan X:
Uncovering an organ's molecular architecture at single-cell
resolution by spatially resolved transcriptomics. Trends
Biotechnol. 39:43–58. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Satija R, Farrell JA, Gennert D, Schier AF
and Regev A: Spatial reconstruction of single-cell gene expression
data. Nat Biotechnol. 33:495–502. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Sun YM and Chen YQ: Principles and
innovative technologies for decrypting noncoding RNAs: From
discovery and functional prediction to clinical application. J
Hematol Oncol. 13:1092020. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhang L, Yu X, Zheng L, Zhang Y, Li Y,
Fang Q, Gao R, Kang B, Zhang Q, Huang JY, et al: Lineage tracking
reveals dynamic relationships of T cells in colorectal cancer.
Nature. 564:268–272. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zhang Q, He Y, Luo N, Patel SJ, Han Y, Gao
R, Modak M, Carotta S, Haslinger C, Kind D, et al: Landscape and
dynamics of single immune cells in hepatocellular carcinoma. Cell.
179:829–845.e20. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Emmert-Buck MR, Bonner RF, Smith PD,
Chuaqui RF, Zhuang Z, Goldstein SR, Weiss RA and Liotta LA: Laser
capture microdissection. Science. 274:998–1001. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Liew LC, Narsai R, Wang Y, Berkowitz O,
Whelan J and Lewsey MG: Temporal tissue-specific regulation of
transcriptomes during barley (Hordeum vulgare) seed germination.
Plant J. 101:700–715. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Shaw R, Tian X and Xu J: Single-cell
transcriptome analysis in plants: Advances and challenges. Mol
Plant. 14:115–126. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Guo W, Hu Y, Qian J, Zhu L, Cheng J, Liao
J and Fan X: Laser capture microdissection for biomedical research:
Towards high-throughput, multi-omics, and single-cell resolution. J
Genet Genomics. 50:641–651. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Nichterwitz S, Chen G, Aguila Benitez J,
Yilmaz M, Storvall H, Cao M, Sandberg R, Deng Q and Hedlund E:
Laser capture microscopy coupled with Smart-seq2 for precise
spatial transcriptomic profiling. Nat Commun. 7:121392016.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Chen J, Suo S, Tam PP, Han JDJ, Peng G and
Jing N: Spatial transcriptomic analysis of cryosectioned tissue
samples with Geo-seq. Nat Protoc. 12:566–580. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Casasent AK, Schalck A, Gao R, Sei E, Long
A, Pangburn W, Casasent T, Meric-Bernstam F, Edgerton ME and Navin
NE: Multiclonal invasion in breast tumors identified by topographic
single cell sequencing. Cell. 172:205–217.e12. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Junker JP, Noël ES, Guryev V, Peterson KA,
Shah G, Huisken J, McMahon AP, Berezikov E, Bakkers J and van
Oudenaarden A: Genome-wide RNA Tomography in the zebrafish embryo.
Cell. 159:662–675. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Moffitt JR, Bambah-Mukku D, Eichhorn SW,
Vaughn E, Shekhar K, Perez JD, Rubinstein ND, Hao J, Regev A, Dulac
C and Zhuang X: Molecular, spatial, and functional single-cell
profiling of the hypothalamic preoptic region. Science.
362:eaau53242018. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Xia C, Fan J, Emanuel G, Hao J and Zhuang
X: Spatial transcriptome profiling by MERFISH reveals subcellular
RNA compartmentalization and cell cycle-dependent gene expression.
Proc Natl Acad Sci USA. 116:19490–19499. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Femino AM, Fay FS, Fogarty K and Singer
RH: Visualization of single RNA transcripts in situ. Science.
280:585–590. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Wang F, Flanagan J, Su N, Wang LC, Bui S,
Nielson A, Wu X, Vo HT, Ma XJ and Luo Y: RNAscope: A novel in situ
RNA analysis platform for formalin-fixed, paraffin-embedded
tissues. J Mol Diagn. 14:22–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Lubeck E, Coskun AF, Zhiyentayev T, Ahmad
M and Cai L: Single-cell in situ RNA profiling by sequential
hybridization. Nat Methods. 11:360–361. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Rodriques SG, Stickels RR, Goeva A, Martin
CA, Murray E, Vanderburg CR, Welch J, Chen LM, Chen F and Macosko
EZ: Slide-seq: A scalable technology for measuring genome-wide
expression at high spatial resolution. Science. 363:1463–1467.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Fazal FM, Han S, Parker KR, Kaewsapsak P,
Xu J, Boettiger AN, Chang HY and Ting AY: Atlas of subcellular RNA
localization revealed by APEX-seq. Cell. 178:473–490.e26. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Vickovic S, Eraslan G, Salmén F,
Klughammer J, Stenbeck L, Schapiro D, Äijö T, Bonneau R,
Bergenstråhle L, Navarro JF, et al: High-definition spatial
transcriptomics for in situ tissue profiling. Nat Methods.
16:987–990. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Srivatsan SR, Regier MC, Barkan E, Franks
JM, Packer JS, Grosjean P, Duran M, Saxton S, Ladd JJ, Spielmann M,
et al: Embryo-scale, single-cell spatial transcriptomics. Science.
373:111–117. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Liu Y, Yang M, Deng Y, Su G, Enninful A,
Guo CC, Tebaldi T, Zhang D, Kim D, Bai Z, et al:
High-spatial-resolution multi-omics sequencing via deterministic
barcoding in tissue. Cell. 183:1665–1681.e18. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Su G, Qin X, Enninful A, Bai Z, Deng Y,
Liu Y and Fan R: Spatial multi-omics sequencing for fixed tissue
via DBiT-seq. STAR Protoc. 2:1005322021. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Dixon EE, Wu H, Sulvarán-Guel E, Guo J and
Humphreys BD: Spatially resolved transcriptomics and the kidney:
Many opportunities. Kidney Int. 102:482–491. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Chen A, Liao S, Cheng M, Ma K, Wu L, Lai
Y, Qiu X, Yang J, Xu J, Hao S, et al: Spatiotemporal transcriptomic
atlas of mouse organogenesis using DNA nanoball-patterned arrays.
Cell. 185:1777–1792.e21. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Cho CS, Xi J, Si Y, Park SR, Hsu JE, Kim
M, Jun G, Kang HM and Lee JH: Microscopic examination of spatial
transcriptome using Seq-Scope. Cell. 184:3559–3572.e22. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Levsky JM, Shenoy SM, Pezo RC and Singer
RH: Single-cell gene expression profiling. Science. 297:836–840.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Lubeck E and Cai L: Single-cell systems
biology by super- resolution imaging and combinatorial labeling.
Nat Methods. 9:743–748. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Ke R, Mignardi M, Pacureanu A, Svedlund J,
Botling J, Wählby C and Nilsson M: In situ sequencing for RNA
analysis in preserved tissue and cells. Nat Methods. 10:857–860.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Lee JH, Daugharthy ER, Scheiman J, Kalhor
R, Ferrante TC, Terry R, Turczyk BM, Yang JL, Lee HS, Aach J, et
al: Fluorescent in situ sequencing (FISSEQ) of RNA for gene
expression profiling in intact cells and tissues. Nat Protoc.
10:442–458. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Olin A, Henckel E, Chen Y, Lakshmikanth T,
Pou C, Mikes J, Gustafsson A, Bernhardsson AK, Zhang C, Bohlin K
and Brodin P: Stereotypic immune system development in newborn
children. Cell. 174:1277–1292.e14. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Soderholm AT and Pedicord VA: Intestinal
epithelial cells: At the interface of the microbiota and mucosal
immunity. Immunology. 158:267–280. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Schreurs RRCE, Baumdick ME, Sagebiel AF,
Kaufmann M, Mokry M, Klarenbeek PL, Schaltenberg N, Steinert FL,
van Rijn JM, Drewniak A, et al: Human fetal
TNF-α-cytokine-producing CD4+ effector memory T cells
promote intestinal development and mediate inflammation early in
life. Immunity. 50:462–476.e8. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Olsen TK and Baryawno N: Introduction to
single-cell RNA sequencing. Curr Protoc Mol Biol. 122:e572018.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Rodaway A and Patient R: Mesendoderm. An
ancient germ layer? Cell. 105:169–172. 2001.PubMed/NCBI
|
|
86
|
Zorn AM and Wells JM: Molecular basis of
vertebrate endoderm development. Int Rev Cytol. 259:49–111. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Zorn AM and Wells JM: Vertebrate endoderm
development and organ formation. Annu Rev Cell Dev Biol.
25:221–251. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Kimelman D and Griffin KJ: Vertebrate
mesendoderm induction and patterning. Curr Opin Genet Dev.
10:350–356. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Spence JR, Lauf R and Shroyer NF:
Vertebrate intestinal endoderm development. Dev Dyn. 240:501–520.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Que J, Okubo T, Goldenring JR, Nam KT,
Kurotani R, Morrisey EE, Taranova O, Pevny LH and Hogan BL:
Multiple dose-dependent roles for Sox2 in the patterning and
differentiation of anterior foregut endoderm. Development.
134:2521–2531. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Sherwood RI, Chen TYA and Melton DA:
Transcriptional dynamics of endodermal organ formation. Dev Dyn.
238:29–42. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Walton KD, Whidden M, Kolterud Å, Shoffner
SK, Czerwinski MJ, Kushwaha J, Parmar N, Chandhrasekhar D, Freddo
AM, Schnell S and Gumucio DL: Villification in the mouse: Bmp
signals control intestinal villus patterning. Development.
143:427–436. 2016.PubMed/NCBI
|
|
93
|
Grey RD: Morphogenesis of intestinal
villi. I. Scanning electron microscopy of the duodenal epithelium
of the developing chick embryo. J Morphol. 137:193–213. 1972.
View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Fawkner-Corbett D, Antanaviciute A, Parikh
K, Jagielowicz M, Gerós AS, Gupta T, Ashley N, Khamis D, Fowler D,
Morrissey E, et al: Spatiotemporal analysis of human intestinal
development at single-cell resolution. Cell. 184:810–826.e23. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Martin JC, Chang C, Boschetti G, Ungaro R,
Giri M, Grout JA, Gettler K, Chuang LS, Nayar S, Greenstein AJ, et
al: Single-cell analysis of Crohn's disease lesions identifies a
pathogenic cellular module associated with resistance to anti-TNF
therapy. Cell. 178:1493–1508.e20. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Parikh K, Antanaviciute A, Fawkner-Corbett
D, Jagielowicz M, Aulicino A, Lagerholm C, Davis S, Kinchen J, Chen
HH, Alham NK, et al: Colonic epithelial cell diversity in health
and inflammatory bowel disease. Nature. 567:49–55. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Smillie CS, Biton M, Ordovas-Montanes J,
Sullivan KM, Burgin G, Graham DB, Herbst RH, Rogel N, Slyper M,
Waldman J, et al: Intra- and inter-cellular rewiring of the human
colon during ulcerative colitis. Cell. 178:714–730.e22. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Fenderico N, van Scherpenzeel RC, Goldflam
M, Proverbio D, Jordens I, Kralj T, Stryeck S, Bass TZ, Hermans G,
Ullman C, et al: Anti-LRP5/6 VHHs promote differentiation of
Wnt-hypersensitive intestinal stem cells. Nat Commun. 10:3652019.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Kinchen J, Chen HH, Parikh K,
Antanaviciute A, Jagielowicz M, Fawkner-Corbett D, Ashley N, Cubitt
L, Mellado-Gomez E, Attar M, et al: Structural remodeling of the
human colonic mesenchyme in inflammatory bowel disease. Cell.
175:372–386.e17. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Powell DW, Pinchuk IV, Saada JI, Chen X
and Mifflin RC: Mesenchymal cells of the intestinal lamina propria.
Annu Rev Physiol. 73:213–237. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
McCarthy N, Manieri E, Storm EE,
Saadatpour A, Luoma AM, Kapoor VN, Madha S, Gaynor LT, Cox C,
Keerthivasan S, et al: Distinct mesenchymal cell populations
generate the essential intestinal BMP signaling gradient. Cell Stem
Cell. 26:391–402.e5. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Degirmenci B, Valenta T, Dimitrieva S,
Hausmann G and Basler K: GLI1-expressing mesenchymal cells form the
essential Wnt-secreting niche for colon stem cells. Nature.
558:449–453. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
van Es JH, Sato T, van de Wetering M,
Lyubimova A, Yee Nee AN, Gregorieff A, Sasaki N, Zeinstra L, van
den Born M, Korving J, et al: Dll1+ secretory progenitor cells
revert to stem cells upon crypt damage. Nat Cell Biol.
14:1099–1104. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
van de Pavert SA and Mebius RE: New
insights into the development of lymphoid tissues. Nat Rev Immunol.
10:664–674. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Scheich S, Chen J, Liu J, Schnütgen F,
Enssle JC, Ceribelli M, Thomas CJ, Choi J, Morris V, Hsiao T, et
al: Targeting N-linked glycosylation for the therapy of aggressive
lymphomas. Cancer Discov. 13:1862–1883. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Guan Q: A comprehensive review and update
on the pathogenesis of inflammatory bowel disease. J Immunol Res.
2019:72472382019. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Tang F, Barbacioru C, Wang Y, Nordman E,
Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, et al: mRNA-Seq
whole-transcriptome analysis of a single cell. Nat Methods.
6:377–382. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Yu X, Abbas-Aghababazadeh F, Chen YA and
Fridley BL: Statistical and bioinformatics analysis of data from
bulk and single-cell RNA sequencing experiments. Methods Mol Biol.
2194:143–175. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Nath A and Bild AH: Leveraging single-cell
approaches in cancer precision medicine. Trends Cancer. 7:359–372.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Czarnewski P, Parigi SM, Sorini C, Diaz
OE, Das S, Gagliani N and Villablanca EJ: Conserved transcriptomic
profile between mouse and human colitis allows unsupervised patient
stratification. Nat Commun. 10:28922019. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Parigi SM, Larsson L, Das S, Ramirez
Flores RO, Frede A, Tripathi KP, Diaz OE, Selin K, Morales RA, Luo
X, et al: The spatial transcriptomic landscape of the healing mouse
intestine following damage. Nat Commun. 13:8282022. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Ayyaz A, Kumar S, Sangiorgi B, Ghoshal B,
Gosio J, Ouladan S, Fink M, Barutcu S, Trcka D, Shen J, et al:
Single-cell transcriptomes of the regenerating intestine reveal a
revival stem cell. Nature. 569:121–125. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Smythies LE, Sellers M, Clements RH,
Mosteller-Barnum M, Meng G, Benjamin WH, Orenstein JM and Smith PD:
Human intestinal macrophages display profound inflammatory anergy
despite avid phagocytic and bacteriocidal activity. J Clin Invest.
115:66–75. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Martinez FO and Gordon S: The M1 and M2
paradigm of macrophage activation: Time for reassessment.
F1000Prime Rep. 6:132014. View
Article : Google Scholar : PubMed/NCBI
|
|
115
|
Garrido-Trigo A, Corraliza AM, Veny M,
Dotti I, Melón-Ardanaz E, Rill A, Crowell HL, Corbí Á, Gudiño V,
Esteller M, et al: Macrophage and neutrophil heterogeneity at
single-cell spatial resolution in human inflammatory bowel disease.
Nat Commun. 14:45062023. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Schoeler M and Caesar R: Dietary lipids,
gut microbiota and lipid metabolism. Rev Endocr Metab Disord.
20:461–472. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Milani C, Duranti S, Bottacini F, Casey E,
Turroni F, Mahony J, Belzer C, Delgado Palacio S, Arboleya Montes
S, Mancabelli L, et al: The first microbial colonizers of the human
gut: Composition, activities, and health implications of the infant
gut microbiota. Microbiol Mol Biol Rev. 81:e00036–17. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Human Microbiome Project Consortium, .
Structure, function and diversity of the healthy human microbiome.
Nature. 486:207–214. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Lloyd-Price J, Arze C, Ananthakrishnan AN,
Schirmer M, Avila-Pacheco J, Poon TW, Andrews E, Ajami NJ, Bonham
KS, Brislawn CJ, et al: Multi-omics of the gut microbial ecosystem
in inflammatory bowel diseases. Nature. 569:655–662. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Huang H, Ren Z, Gao X, Hu X, Zhou Y, Jiang
J, Lu H, Yin S, Ji J, Zhou L and Zheng S: Integrated analysis of
microbiome and host transcriptome reveals correlations between gut
microbiota and clinical outcomes in HBV-related hepatocellular
carcinoma. Genome Med. 12:1022020. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Zhang SL, Cheng LS, Zhang ZY, Sun HT and
Li JJ: Untangling determinants of gut microbiota and tumor
immunologic status through a multi-omics approach in colorectal
cancer. Pharmacol Res. 188:1066332023. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Pereira FC and Berry D: Microbial nutrient
niches in the gut. Environ Microbiol. 19:1366–1378. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Kim HJ, Boedicker JQ, Choi JW and
Ismagilov RF: Defined spatial structure stabilizes a synthetic
multispecies bacterial community. Proc Natl Acad Sci USA.
105:18188–18193. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Rausch P, Basic M, Batra A, Bischoff SC,
Blaut M, Clavel T, Gläsner J, Gopalakrishnan S, Grassl GA, Günther
C, et al: Analysis of factors contributing to variation in the
C57BL/6J fecal microbiota across German animal facilities. Int J
Med Microbiol. 306:343–355. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Seekatz AM, Schnizlein MK, Koenigsknecht
MJ, Baker JR, Hasler WL, Bleske BE, Young VB and Sun D: Spatial and
temporal analysis of the stomach and small-intestinal microbiota in
fasted healthy humans. mSphere. 4:e00126–19. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Sheth RU, Li M, Jiang W, Sims PA, Leong KW
and Wang HH: Spatial metagenomic characterization of microbial
biogeography in the gut. Nat Biotechnol. 37:877–883. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Choi CH and Chang SK: Role of small
intestinal bacterial overgrowth in functional gastrointestinal
disorders. J Neurogastroenterol Motil. 22:3–5. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Evans DF, Pye G, Bramley R, Clark AG,
Dyson TJ and Hardcastle JD: Measurement of gastrointestinal pH
profiles in normal ambulant human subjects. Gut. 29:1035–1041.
1988. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Macfarlane GT, Gibson GR and Cummings JH:
Comparison of fermentation reactions in different regions of the
human colon. J Appl Bacteriol. 72:57–64. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Birchenough GM, Johansson ME, Gustafsson
JK, Bergström JH and Hansson GC: New developments in goblet cell
mucus secretion and function. Mucosal Immunol. 8:712–719. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Kotarsky K, Sitnik KM, Stenstad H,
Kotarsky H, Schmidtchen A, Koslowski M, Wehkamp J and Agace WW: A
novel role for constitutively expressed epithelial-derived
chemokines as antibacterial peptides in the intestinal mucosa.
Mucosal Immunol. 3:40–48. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
McCallum G and Tropini C: The gut
microbiota and its biogeography. Nat Rev Microbiol. 22:105–118.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Cremer J, Arnoldini M and Hwa T: Effect of
water flow and chemical environment on microbiota growth and
composition in the human colon. Proc Natl Acad Sci USA.
114:6438–6443. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Cremer J, Segota I, Yang CY, Arnoldini M,
Sauls JT, Zhang Z, Gutierrez E, Groisman A and Hwa T: Effect of
flow and peristaltic mixing on bacterial growth in a gut-like
channel. Proc Natl Acad Sci USA. 113:11414–11419. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Nagara Y, Takada T, Nagata Y, Kado S and
Kushiro A: Microscale spatial analysis provides evidence for
adhesive monopolization of dietary nutrients by specific intestinal
bacteria. PLoS One. 12:e01754972017. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Walker AW, Duncan SH, Harmsen HJ, Holtrop
G, Welling GW and Flint HJ: The species composition of the human
intestinal microbiota differs between particle-associated and
liquid phase communities. Environ Microbiol. 10:3275–3283. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Leitch ECMW, Walker AW, Duncan SH, Holtrop
G and Flint HJ: Selective colonization of insoluble substrates by
human faecal bacteria. Environ Microbiol. 9:667–679. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Sobhani I, Bergsten E, Couffin S, Amiot A,
Nebbad B, Barau C, de'Angelis N, Rabot S, Canoui-Poitrine F,
Mestivier D, et al: Colorectal cancer-associated microbiota
contributes to oncogenic epigenetic signatures. Proc Natl Acad Sci
USA. 116:24285–24295. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
David LA, Maurice CF, Carmody RN,
Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y,
Fischbach MA, et al: Diet rapidly and reproducibly alters the human
gut microbiome. Nature. 505:559–563. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Nguyen J, Pepin DM and Tropini C: Cause or
effect? The spatial organization of pathogens and the gut
microbiota in disease. Microbes Infect. 23:1048152021. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Borghini R, Donato G, Alvaro D and
Picarelli A: New insights in IBS-like disorders: Pandora's box has
been opened; a review. Gastroenterol Hepatol Bed Bench. 10:79–89.
2017.PubMed/NCBI
|
|
142
|
Ng QX, Soh AYS, Loke W, Lim DY and Yeo WS:
The role of inflammation in irritable bowel syndrome (IBS). J
Inflamm Res. 11:345–349. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Qiao LY and Tiwari N: Spinal
neuron-glia-immune interaction in cross-organ sensitization. Am J
Physiol Gastrointest Liver Physiol. 319:G748–G760. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
North RY, Li Y, Ray P, Rhines LD, Tatsui
CE, Rao G, Johansson CA, Zhang H, Kim YH, Zhang B, et al:
Electrophysiological and transcriptomic correlates of neuropathic
pain in human dorsal root ganglion neurons. Brain. 142:1215–1226.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Tavares-Ferreira D, Shiers S, Ray PR,
Wangzhou A, Jeevakumar V, Sankaranarayanan I, Cervantes AM, Reese
JC, Chamessian A, Copits BA, et al: Spatial transcriptomics of
dorsal root ganglia identifies molecular signatures of human
nociceptors. Sci Transl Med. 14:eabj81862022. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Bonaz BL and Bernstein CN: Brain-gut
interactions in inflammatory bowel disease. Gastroenterology.
144:36–49. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Tsigos C and Chrousos GP:
Hypothalamic-pituitary-adrenal axis, neuroendocrine factors and
stress. J Psychosom Res. 53:865–871. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Kitaoka S: Inflammation in the brain and
periphery found in animal models of depression and its behavioral
relevance. J Pharmacol Sci. 148:262–266. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
De Bellis MD, Casey BJ, Dahl RE, Birmaher
B, Williamson DE, Thomas KM, Axelson DA, Frustaci K, Boring AM,
Hall J and Ryan ND: A pilot study of amygdala volumes in pediatric
generalized anxiety disorder. Biol Psychiatry. 48:51–57. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Schienle A, Ebner F and Schäfer A:
Localized gray matter volume abnormalities in generalized anxiety
disorder. Eur Arch Psychiatry Clin Neurosci. 261:303–307. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Qiao J, Li A, Cao C, Wang Z, Sun J and Xu
G: Aberrant functional network connectivity as a biomarker of
generalized anxiety disorder. Front Hum Neurosci. 11:6262017.
View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Moffitt JR, Hao J, Bambah-Mukku D, Lu T,
Dulac C and Zhuang X: High-performance multiplexed fluorescence in
situ hybridization in culture and tissue with matrix imprinting and
clearing. Proc Natl Acad Sci USA. 113:14456–14461. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Zhou X, Lu Y, Zhao F, Dong J, Ma W, Zhong
S, Wang M, Wang B, Zhao Y, Shi Y, et al: Deciphering the
spatial-temporal transcriptional landscape of human hypothalamus
development. Cell Stem Cell. 29:328–343.e5. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Oh SW, Harris JA, Ng L, Winslow B, Cain N,
Mihalas S, Wang Q, Lau C, Kuan L, Henry AM, et al: A mesoscale
connectome of the mouse brain. Nature. 508:207–214. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Qian J, Olbrecht S, Boeckx B, Vos H, Laoui
D, Etlioglu E, Wauters E, Pomella V, Verbandt S, Busschaert P, et
al: A pan-cancer blueprint of the heterogeneous tumor
microenvironment revealed by single-cell profiling. Cell Res.
30:745–762. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Galeano Niño JL, Wu H, LaCourse KD,
Kempchinsky AG, Baryiames A, Barber B, Futran N, Houlton J, Sather
C, Sicinska E, et al: Effect of the intratumoral microbiota on
spatial and cellular heterogeneity in cancer. Nature. 611:810–817.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Ozato Y, Kojima Y, Kobayashi Y, Hisamatsu
Y, Toshima T, Yonemura Y, Masuda T, Kagawa K, Goto Y, Utou M, et
al: Spatial and single-cell transcriptomics decipher the cellular
environment containing HLA-G+ cancer cells and SPP1+ macrophages in
colorectal cancer. Cell Rep. 42:1119292023. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Dekker E, Tanis PJ, Vleugels JLA, Kasi PM
and Wallace MB: Colorectal cancer. Lancet. 394:1467–1480. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Zhao W, Dai S, Yue L, Xu F, Gu J, Dai X
and Qian X: Emerging mechanisms progress of colorectal cancer liver
metastasis. Front Endocrinol (Lausanne). 13:10815852022. View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Brouwer NP, Webbink L, Haddad TS, Rutgers
N, van Vliet S, Wood CS, Jansen PW, Lafarge MW; imCMS Consortium;
de Wilt JH, ; et al: Transcriptomics and proteomics reveal distinct
biology for lymph node metastases and tumour deposits in colorectal
cancer. J Pathol. 261:401–412. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Kobayashi H, Gieniec KA, Lannagan TRM,
Wang T, Asai N, Mizutani Y, Iida T, Ando R, Thomas EM, Sakai A, et
al: The origin and contribution of cancer-associated fibroblasts in
colorectal carcinogenesis. Gastroenterology. 162:890–906. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Peng Z, Ye M, Ding H, Feng Z and Hu K:
Spatial transcriptomics atlas reveals the crosstalk between
cancer-associated fibroblasts and tumor microenvironment components
in colorectal cancer. J Transl Med. 20:3022022. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Qi J, Sun H, Zhang Y, Wang Z, Xun Z, Li Z,
Ding X, Bao R, Hong L, Jia W, et al: Single-cell and spatial
analysis reveal interaction of FAP+ fibroblasts and
SPP1+ macrophages in colorectal cancer. Nat Commun.
13:17422022. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Valdeolivas A, Amberg B, Giroud N,
Richardson M, Gálvez EJC, Badillo S, Julien-Laferrière A, Túrós D,
Voith von Voithenberg L, Wells I, et al: Profiling the
heterogeneity of colorectal cancer consensus molecular subtypes
using spatial transcriptomics. NPJ Precis Oncol. 8:102024.
View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Roelands J, van der Ploeg M, Ijsselsteijn
ME, Dang H, Boonstra JJ, Hardwick JCH, Hawinkels LJAC, Morreau H
and de Miranda NFCC: Transcriptomic and immunophenotypic profiling
reveals molecular and immunological hallmarks of colorectal cancer
tumourigenesis. Gut. 72:1326–1339. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Zwing N, Failmezger H, Ooi CH, Hibar DP,
Cañamero M, Gomes B, Gaire F and Korski K: Analysis of spatial
organization of suppressive myeloid cells and effector T cells in
colorectal cancer-A potential tool for discovering prognostic
biomarkers in clinical research. Front Immunol. 11:5502502020.
View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Liu HT, Chen SY, Peng LL, Zhong L, Zhou L,
Liao SQ, Chen ZJ, Wang QL, He S and Zhou ZH: Spatially resolved
transcriptomics revealed local invasion-related genes in colorectal
cancer. Front Oncol. 13:10890902023. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Manfredi S, Lepage C, Hatem C, Coatmeur O,
Faivre J and Bouvier AM: Epidemiology and management of liver
metastases from colorectal cancer. Ann Surg. 244:254–259. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Hu Z, Ding J, Ma Z, Sun R, Seoane JA,
Scott Shaffer J, Suarez CJ, Berghoff AS, Cremolini C, Falcone A, et
al: Quantitative evidence for early metastatic seeding in
colorectal cancer. Nat Genet. 51:1113–1122. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Wang F, Long J, Li L, Wu ZX, Da TT, Wang
XQ, Huang C, Jiang YH, Yao XQ, Ma HQ, et al: Single-cell and
spatial transcriptome analysis reveals the cellular heterogeneity
of liver metastatic colorectal cancer. Sci Adv. 9:eadf54642023.
View Article : Google Scholar : PubMed/NCBI
|
|
172
|
Garbarino O, Lambroia L, Basso G, Marrella
V, Franceschini B, Soldani C, Pasqualini F, Giuliano D, Costa G,
Peano C, et al: Spatial resolution of cellular senescence dynamics
in human colorectal liver metastasis. Aging Cell. 22:e138532023.
View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Cheng LK, O'Grady G, Du P, Egbuji JU,
Windsor JA and Pullan AJ: Gastrointestinal system. Wiley
Interdiscip Rev Syst Biol Med. 2:65–79. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
174
|
Peterson LW and Artis D: Intestinal
epithelial cells: Regulators of barrier function and immune
homeostasis. Nat Rev Immunol. 14:141–153. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
175
|
Pelka K, Hofree M, Chen JH, Sarkizova S,
Pirl JD, Jorgji V, Bejnood A, Dionne D, Ge WH, Xu KH, et al:
Spatially organized multicellular immune hubs in human colorectal
cancer. Cell. 184:4734–4752.e20. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
176
|
Frede A, Czarnewski P, Monasterio G,
Tripathi KP, Bejarano DA, Ramirez Flores RO, Sorini C, Larsson L,
Luo X, Geerlings L, et al: B cell expansion hinders the
stroma-epithelium regenerative cross talk during mucosal healing.
Immunity. 55:2336–2351.e12. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
177
|
Zhang X, Hu C, Huang C, Wei Y, Li X, Hu M,
Li H, Wu J, Czajkowsky DM, Guo Y and Shao Z: Robust Acquisition of
spatial transcriptional programs in tissues with
immunofluorescence-guided laser capture microdissection. Front Cell
Dev Biol. 10:8531882022. View Article : Google Scholar : PubMed/NCBI
|
|
178
|
Ghaddar B and De S: Reconstructing
physical cell interaction networks from single-cell data using
Neighbor-seq. Nucleic Acids Res. 50:e822022. View Article : Google Scholar : PubMed/NCBI
|
|
179
|
Zhu Z, Cai J, Hou W, Xu K, Wu X, Song Y,
Bai C, Mo YY and Zhang Z: Microbiome and spatially resolved
metabolomics analysis reveal the anticancer role of gut Akkermansia
muciniphila by crosstalk with intratumoral microbiota and
reprogramming tumoral metabolism in mice. Gut Microbes.
15:21667002023. View Article : Google Scholar : PubMed/NCBI
|
|
180
|
Sun C, Wang A, Zhou Y, Chen P, Wang X,
Huang J, Gao J, Wang X, Shu L, Lu J, et al: Spatially resolved
multi-omics highlights cell-specific metabolic remodeling and
interactions in gastric cancer. Nat Commun. 14:26922023. View Article : Google Scholar : PubMed/NCBI
|
|
181
|
Tun HM, Peng Y, Massimino L, Sin ZY,
Parigi TL, Facoetti A, Rahman S, Danese S and Ungaro F: Gut virome
in inflammatory bowel disease and beyond. Gut. 73:350–360. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
182
|
Zhao E, Stone MR, Ren X, Guenthoer J,
Smythe KS, Pulliam T, Williams SR, Uytingco CR, Taylor SEB, Nghiem
P, et al: Spatial transcriptomics at subspot resolution with
BayesSpace. Nat Biotechnol. 39:1375–1384. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
183
|
Saviano A, Henderson NC and Baumert TF:
Single-cell genomics and spatial transcriptomics: Discovery of
novel cell states and cellular interactions in liver physiology and
disease biology. J Hepatol. 73:1219–1230. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
184
|
Peterson VM, Zhang KX, Kumar N, Wong J, Li
L, Wilson DC, Moore R, McClanahan TK, Sadekova S and Klappenbach
JA: Multiplexed quantification of proteins and transcripts in
single cells. Nat Biotechnol. 35:936–939. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
185
|
Thornton CA, Mulqueen RM, Torkenczy KA,
Nishida A, Lowenstein EG, Fields AJ, Steemers FJ, Zhang W,
McConnell HL, Woltjer RL, et al: Spatially mapped single-cell
chromatin accessibility. Nat Commun. 12:12742021. View Article : Google Scholar : PubMed/NCBI
|
|
186
|
Stoeckius M, Hafemeister C, Stephenson W,
Houck-Loomis B, Chattopadhyay PK, Swerdlow H, Satija R and Smibert
P: Simultaneous epitope and transcriptome measurement in single
cells. Nat Methods. 14:865–868. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
187
|
Vickovic S, Lötstedt B, Klughammer J,
Mages S, Segerstolpe Å, Rozenblatt-Rosen O and Regev A: SM-omics is
an automated platform for high-throughput spatial multi-omics. Nat
Commun. 13:7952022. View Article : Google Scholar : PubMed/NCBI
|
|
188
|
Park HE, Jo SH, Lee RH, Macks CP, Ku T,
Park J, Lee CW, Hur JK and Sohn CH: Spatial transcriptomics:
Technical aspects of recent developments and their applications in
neuroscience and cancer research. Adv Sci (Weinh). 10:e22069392023.
View Article : Google Scholar : PubMed/NCBI
|
|
189
|
Rao A, Barkley D, França GS and Yanai I:
Exploring tissue architecture using spatial transcriptomics.
Nature. 596:211–220. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
190
|
Zhang L, Chen D, Song D, Liu X, Zhang Y,
Xu X and Wang X: Clinical and translational values of spatial
transcriptomics. Signal Transduct Target Ther. 7:1112022.
View Article : Google Scholar : PubMed/NCBI
|
|
191
|
Li Y, Stanojevic S and Garmire LX:
Emerging artificial intelligence applications in spatial
transcriptomics analysis. Comput Struct Biotechnol J. 20:2895–2908.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
192
|
Hao M, Hua K and Zhang X: SOMDE: A
scalable method for identifying spatially variable genes with
self-organizing map. Bioinformatics. 37:4392–4398. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
193
|
Dries R, Zhu Q, Dong R, Eng CL, Li H, Liu
K, Fu Y, Zhao T, Sarkar A, Bao F, et al: Giotto: A toolbox for
integrative analysis and visualization of spatial expression data.
Genome Biol. 22:782021. View Article : Google Scholar : PubMed/NCBI
|