The spine is one of the most common sites of lesions
in middle-aged and older individuals (>45 years of age)
(1). Conditions such as
intervertebral disc degeneration (IDD), ankylosing spondylitis (AS)
and spinal cord injury (SCI) are common non-infectious spinal
diseases that impair the quality of life of patients, and cause a
huge economic burden on families and society (2). In addition to causing financial
stress, IDD and other spinal disorders are important causes of pain
and disability in patients (3).
Notably, the incidence of spinal diseases such as IDD has
increased, with a trend towards younger individuals (13–20 years of
age) (4). Reportedly, ~80% of
individuals experience lower back pain due to IDD during their
lifetime (5). Unfortunately, the
current treatment for these spinal diseases encompasses pain
reduction and symptom improvement (6), and does not reverse or cure the
diseases (7). Therefore, a more
comprehensive understanding of the pathology of these diseases is
of great significance for developing new therapeutic drugs,
improving and optimising treatment programmes, and curing the
diseases.
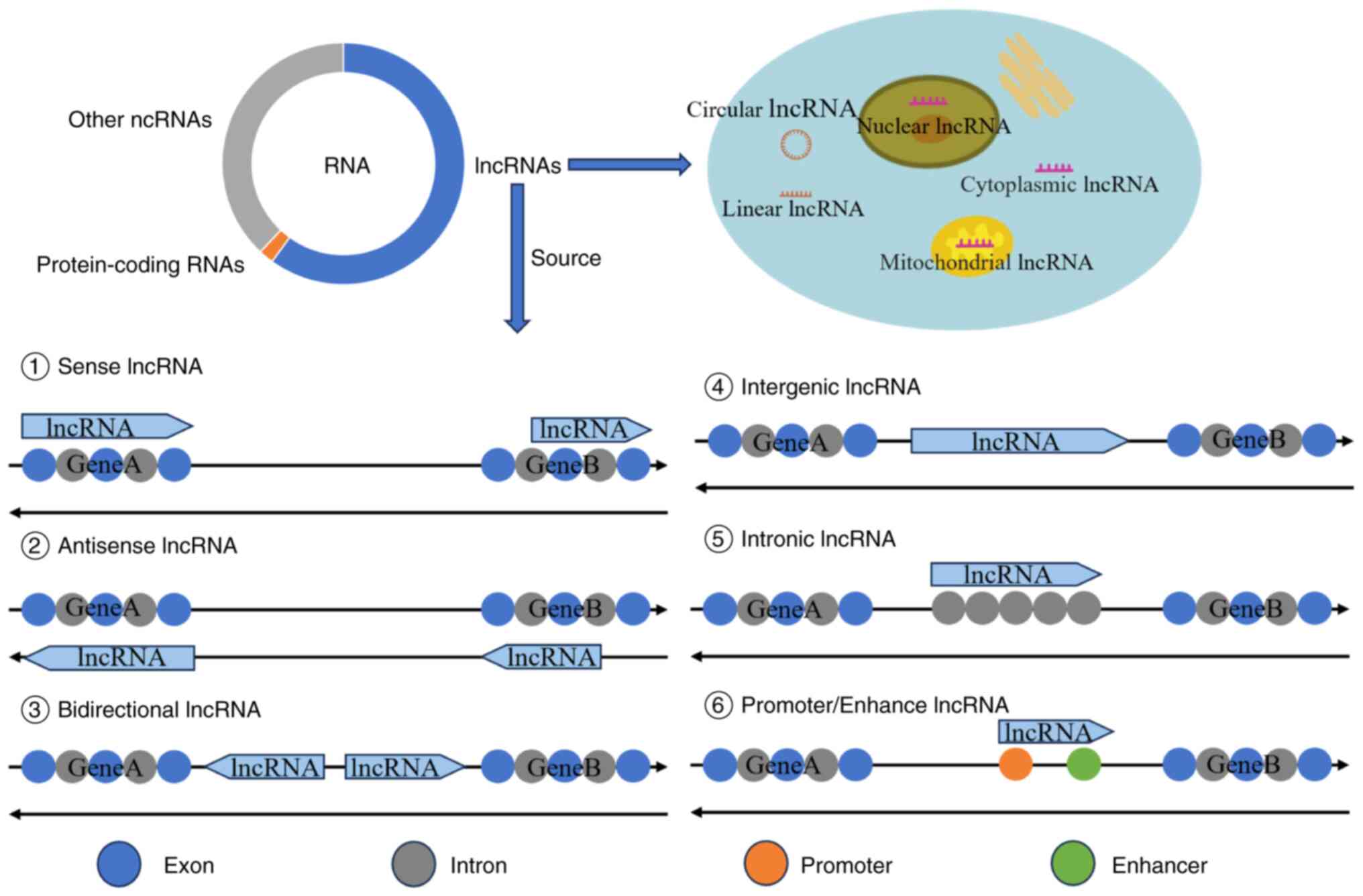
Although the human genome is expansive, only ~1% of
the genome is involved in coding proteins (5), with >98% not directly involved in
protein translation (8). RNAs
transcribed from these genes not directly involved in protein
translation is called non-coding RNAs (ncRNAs). Although ncRNAs are
not involved in protein synthesis, they play important roles in
regulating gene expression, cell function and development (9). NcRNAs can be divided into numerous
categories, including microRNAs (miRNAs/miRs), long non-coding RNAs
(lncRNAs), circular RNAs (circRNAs), small interfering RNAs and
PIWI-interacting RNA (10).
lncRNAs are RNAs with a length of >200 nucleotides that cannot
encode proteins and have various biological functions (11). The functions of lncRNAs include
gene regulation, RNAs processing and splicing regulation, cell
cycle and proliferation, cell signal transduction, chromosome
structure and nucleoplasmic transport (12). The present article reviews the
functional classification of lncRNAs and their roles in IDD, AS and
SCI, and discusses the potential of lncRNAs for treating spinal
diseases such as IDD.
The majority of lncRNAs are similar in origin to
mRNAs and are transcribed by RNA polymerase II. They also have the
same structure as mRNAs (13);
however, lncRNAs also have certain characteristics, although these
are not unique to lncRNAs. lncRNAs account for >60% of all
ncRNAs [certain studies report rates of >70% (14) or 70–90% (15)], and most lncRNAs contain >2
exons (16). Some lncRNAs, owing
to their reverse splicing or lack of a 5′ cap, are readily
connected with their own 3′ poly-A tail, generating circRNA
(17). Numerous lncRNA genes have
miRNA sequences embedded in their exons or introns, making them a
source of miRNAs (13). A number
of chemical modification methods exist for lncRNAs, including
adenosine methylation, cytosine modification, uridine
isomerisation, guanosine methylation and ribose modification
(18), resulting in more complex
ncRNA biogenesis.
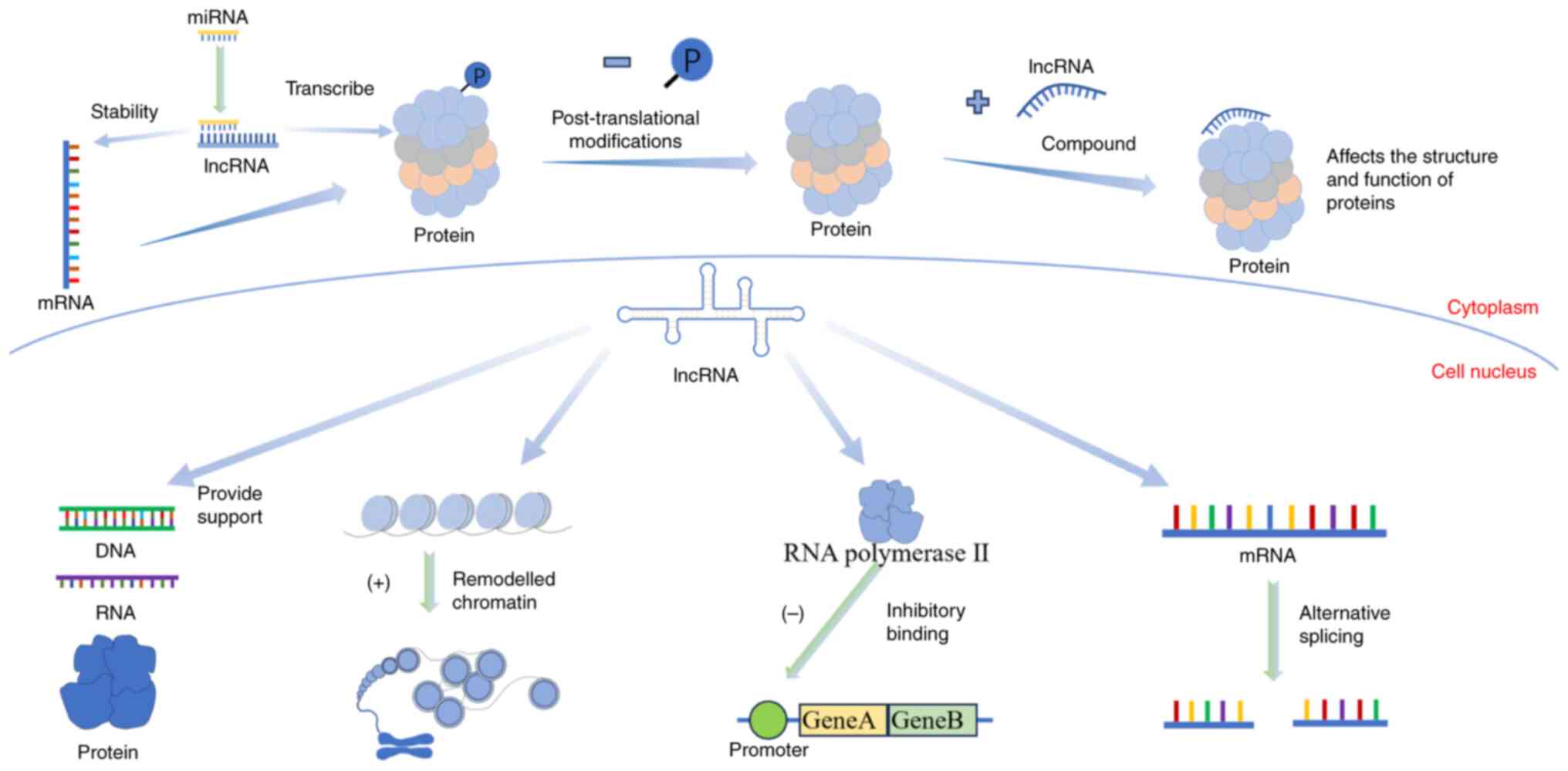
The function of lncRNAs varies depending on their
spatial structure and location. lncRNAs have two functional forms
including spliced and unspliced (25). The biological genetic information
in mRNAs is mainly transmitted through ‘linear’ coding, while the
biological information in lncRNAs mainly works through its
different ‘spatial’ structural mechanisms (26). The main pathways through which
lncRNAs function include epigenetic, transcriptional and
post-transcriptional regulation (27). When lncRNAs perform their
functions, they usually combine with DNA, RNAs and proteins to form
the corresponding complexes. First, lncRNAs bind with DNA and play
a role in decompressing chromatin (28). They can also affect chromatin
accessibility (29), and the
R-loop can make lncRNAs an ideal regulatory centre (30). Next, lncRNAs bind to RNAs. The
lncRNAs regulate mRNA expression through competitive endogenous RNA
mechanisms (31). Finally, lncRNAs
bind with proteins to form complexes, regulating protein structure
and function (32). Some lncRNAs
directly bind to proteins, affecting cellular signal transduction
and gene expression.
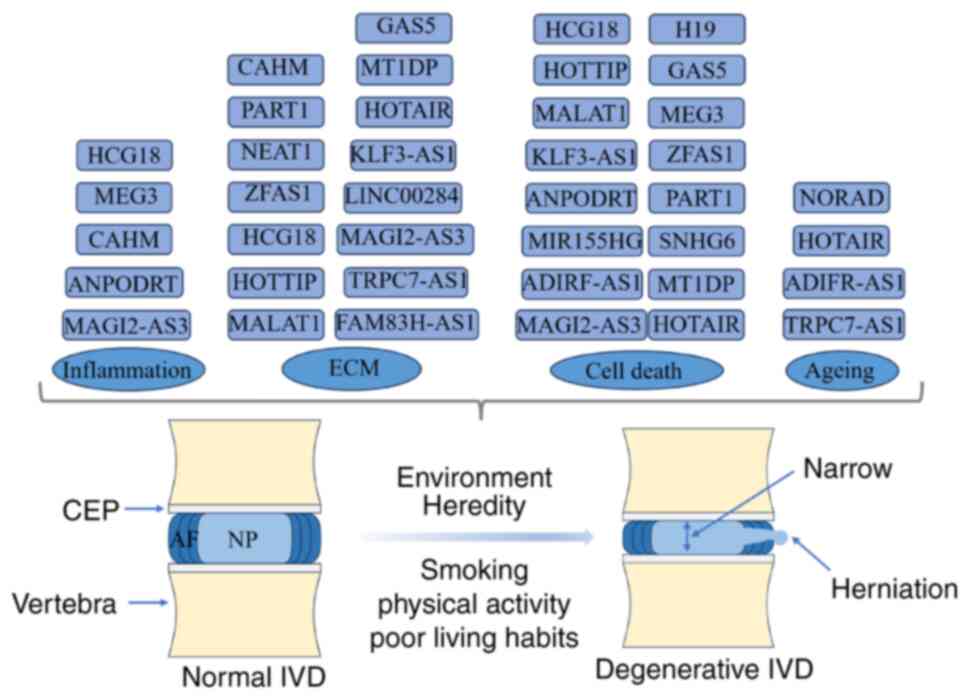
The intervertebral disc (IVD) is the largest
non-blood supply tissue in the human body (40). The IVD is composed of three parts:
i) The cartilage endplate (CEP) on the upper and lower sides; ii)
the surrounding annulus fibrosus (AF); and iii) the nucleus
pulposus (NP) in the middle (41).
The CEP is ~1-mm thick and contains a number of micropores for
material exchange in the NP, but contains no nerve tissue. The AF
is divided into outer and inner rings, and the front and sides are
twice as thick as the back. Nerve endings are only distributed in
the outer ring (42). The AF is
very strong and plays a major role in supporting and stabilising
the spine (42). The NP is a
gel-like tissue with a high water content, containing collagen,
proteoglycans, NP cells and water (43). The composition and structure change
with age, accounting for 50–60% of the IVD cross-section (44). The toughness and elasticity of the
IVD enable it to perform important physiological functions, such as
supporting the spine, buffering pressure and providing spine
mobility (45).
As the IVD has no blood vessels, its self-repair
ability is poor, making it prone to degeneration (40). IDD initiates in NP cells (46), and this is hypothesised to be
primarily due to the joint action of genetic and environmental
factors. Smoking, physical activity and poor living habits are
contributing factors to IDD, although genetic factors are the most
important (47). It is estimated
that >70% of cases are caused by genetics and the programmed
cell death drives the occurrence of IDD (48). After the onset of IDD, the water
content in the IVD decreases, and the extracellular matrix (ECM),
including type II collagen and proteoglycans, degrades (49). After the CEP and AF are destroyed,
the barrier between the IVD, and the circulatory and immune systems
is broken, triggering an inflammatory reaction (50). Under the influence of these
pathogenic factors and changes in the microenvironment, the
proliferative ability of the NP cells decreases, causing cell
apoptosis, autophagy and senescence, ultimately resulting in IDD
(5). Surgical treatment of IDD
also has challenges, including infection (51), recurrence (52) and adjacent spondylosis (53). Therefore, understanding the
pathogenesis of IDD and exploring new therapeutic targets are
crucial.
Compared with normal tissues, IDD exhibits numerous
differentially expressed ncRNAs, including lncRNAs, which play an
important role in the occurrence and development of IDD. Wan et
al (54) used lncRNA-mRNA
microarrays to study lncRNA expression in NP specimens from human
degenerative NP tissue and normal groups. The results revealed that
the expression levels of 67 lncRNAs increased and those of 49
decreased. In another study, the same lncRNA-mRNA microarray
technology was used to analyse the lncRNA expression in NP
specimens of human degenerative NP tissue and normal groups, and
135 significantly upregulated and 170 downregulated lncRNAs were
found (55). A similar study found
that 2,418 mRNAs and 528 lncRNAs were differentially expressed in
the IDD group compared with the control group (56). Shi et al (57) recently constructed a ceRNA network
containing 15 lncRNAs, 9 miRNAs and 103 mRNAs, and discovered a
lncRNA/miRNA/mRNA axis, which affects the progression of IDD,
namely, the small nucleolar RNA host gene 5/miR-299-5p/activating
transcription factor 2 axis. However, further experimental
verification is still needed. In another study, a ceRNA network was
constructed using 15 lncRNAs and 21 miRNAs, and identified the two
hub genes MAPK8 and CAPN1 in this regulatory network as key
biomarkers of IDD (58). These
studies demonstrate that there are obvious differences in lncRNA
expression in IDD and that lncRNAs do not function independently
but form an extensive ceRNA network system with other ncRNAs.
As aforementioned, IDD originates primarily from NP
cells. As early as 2006, researchers found that transplantation of
human NP cells into degenerated IVDs in rabbits could improve the
degree of disc degeneration (59).
A subsequent similar study showed that transplantation of NP stem
cells into degenerated IVDs can more effectively improve IDD than
transplantation of NP cells (60).
These studies indicate that NP cells play a crucial role in the
occurrence and development of IDD. The state of NP cells is
influenced by the ECM, inflammation, apoptosis, necrosis and ageing
factors. Under a series of negative influences, this ultimately
leads to dysregulation of the NP cell phenotype, thereby
participating in the occurrence and development of IDD (61). Jiang et al (62) reported that miR-365 can alleviate
the development of IDD by regulating the synthesis and degradation
of the ECM in NP cells. Propionibacterium acnes has been
shown to induce the apoptosis of NP cells through the TLR2/JNK
pathway, which can alter the process of IDD (63). Inflammation in NP cells also
affects IDD, whereby extracellular lactate can promote activation
of the NLR family pyrin domain containing 3 (NLRP3) inflammasome,
increasing inflammatory levels in the NP cells and thereby
promoting IDD (64). Experimental
results from Du et al (65)
suggested that activation of cannabinoid receptor type 2 can delay
the ageing of NP cells, restore the balance of ECM metabolism and
attenuate IDD. Overall, changes in the ECM, cell death, cell
inflammation and cell ageing are all part of the complex mechanisms
underlying the occurrence and development of IDD. They interact
with each other, collectively influencing the process of IDD.
The ECM, type II collagen and proteoglycan
(aggrecan) are the main components of the NP. The ECM is produced
by NP cells, and the balance between ECM synthesis and degradation
contributes to the biomechanical balance and structural stability
of the IVD, which is also considered a key indicator for evaluating
NP cell function (66). Subsequent
studies indicate that lncRNAs can affect the synthesis and
degradation of the ECM through a ceRNA mechanism and can directly
regulate hub genes or signalling pathways. Gao et al
(67) studied degenerated human
tissues and cells, and found that prostate androgen-regulated
transcript 1 (PART1) was upregulated in degenerated NP tissues. By
contrast, PART1 knockdown led to increased ECM synthesis, reduced
degradation and enhanced proliferation of NP cells. Growth arrest
specific 5 has been shown to be expressed at abnormally high levels
in human degenerative NP tissues (68). After downregulation, angiopoietin 2
has been shown to be inhibited in a miR-17-3p-dependent manner,
promoting ECM remodelling, inhibiting NP cell apoptosis and
improving IDD (68). A recent
study showed that the transcription factor FOXO3 may enhance the
competitive binding of HOXA transcript at the distal tip (HOTTIP)
and miR-615-3p by activating HOTTIP transcription, increasing the
expression of the target gene collagen type II α1 of miR-615-3p,
inducing NP cell proliferation, and reducing apoptosis to prevent
ECM degradation (69).
Krüppel-like factor 3 antisense RNA 1 (KLF3-AS1), like HOTTIP, was
also shown to be expressed at low levels in degenerative NP
tissues. Overexpression of KLF3-AS1 can increase NP cell viability,
prevent cell apoptosis and increase ECM synthesis (70).
Owing to different damage factors, NP cells die
through a number of the same mechanisms as other cells, such as
apoptosis, pyroptosis, autophagy, ferroptosis and necrosis
(76). Currently, the most studied
process is NP cell apoptosis. As aforementioned, PART1 not only
affects the synthesis and degradation of the ECM through the
miR-93/matrix metalloproteinase-2 signalling axis, but increased
PART1 expression in degenerated tissues can also promote the
apoptosis of NP cells and promote IDD (67). Similarly, HOTTIP, while preventing
ECM degradation through a ceRNA mechanism, can also inhibit the
apoptosis of NP cells and promote proliferation, improving the
progression of IDD (69). In
degenerated NP cells, small nucleolar RNA host gene 6 (SNHG6)
expression has been reported to be upregulated and SNHG6 can induce
NP cell apoptosis by targeting miR-101-3p (77). Chen et al (70) illustrated the possible impact of
KLF3-AS1 on IDD. Oe-KLF3-AS1 enhanced NP cell viability and
prevented apoptosis. The experimental results confirmed that
KLF3-AS1 overexpression improved degenerative changes in NP cells
through the miR-10a-3p/zinc finger and BTB domain-containing
protein 20 (ZBTB20) axis. Yu and Li (78) detected low expression of membrane
associated guanylate kinase, WW and PDZ domain containing 2
(MAGI2)-AS3 and IL-10, and high expression of miR-374b-5p in
lipopolysaccharide (LPS)-induced NP cells. Notably, miR-374b-5p is
a target of MAGI2-AS3 and IL-10. MAGI2-AS3 can increase the
expression of IL-10 by competing with miR-374b-5p, thereby reducing
the inflammatory response and cell apoptosis. Thus, the regulation
of NP cell apoptosis by lncRNAs is mainly based on the ceRNA
mechanism.
LncRNAs regulate the apoptosis of NP cells and
affect pyroptosis and autophagy. For instance, MIR155 host gene
(MIR155HG) has been shown to be upregulated in degenerated human NP
tissues, and further experiments revealed that MIR155HG sponges
miR-223-3p and promotes NLRP3 expression, thereby inducing
pyroptosis in NP cells (79).
Furthermore, platelet-rich plasma-derived extracellular vesicles
may inhibit tert-butyl hydroperoxide-induced NP cell injury by
upregulating metastasis-associated lung adenocarcinoma transcript 1
(MALAT1) expression; MALAT1 regulates the miR-217/silent
information regulator sirtuin 1 (SIRT1) signalling axis, and its
overexpression can alleviate NP cell pyroptosis (80). As aforementioned, HOTAIR can affect
the synthesis and decomposition of the ECM in NP cells and the
apoptosis of NP cells. Simultaneously, HOTAIR can also affect
autophagy and alter the progression of IDD (75). As the first lncRNA discovered
(81), H19 imprinted maternally
expressed transcript (H19) is highly conserved and widely expressed
in mammals. Sun et al (82)
found that H19 was highly expressed in degenerated NP tissues, and
experimentally verified that H19 can promote autophagy and
apoptosis in NP cells through the miR-139-3p/C-X-C motif chemokine
receptor type 4/nuclear factor-κB (NF-κB) signalling axis, thereby
affecting IDD (Table I and
Fig. 3).
Inflammation can accelerate the process of IDD and
cause damage and dehydration of the IVD tissue, resulting in loss
of its original elasticity and function. Therefore, preventing and
controlling inflammation is crucial for treating IDD (83). LncRNAs are important molecules that
regulate inflammatory responses. Jiang et al (84) found that IDD tissues showed
decreased expression of family with sequence similarity 83 member H
(FAM83H)-AS1 compared with normal tissues, and Oe-FAM83H-AS1 could
promote NP-cell proliferation, and reduce inflammatory responses
and ECM degradation. In addition, miR-22-3p mediated the effect of
FAM83H-AS1 on degenerated NP tissue (84). In IL-1β-stimulated NP cells, the
expression of HLA complex group 18 (HCG18) and follistatin-like
protein 1 (FSTL1) has been shown to be increased. Subsequent
overexpression experiments demonstrated the impact of the
HCG18/miR-495-3p/FSTL1 signalling axis on NP tissue inflammation
and apoptosis (85). Zhang et
al (86) showed that melatonin
can reduce inflammation and apoptosis in NP cells while
upregulating maternally expressed gene 3 (MEG3) expression, whereas
MEG3 expression was shown to be downregulated in the degenerated
NP. The study revealed that melatonin can reduce NP cell
inflammation and apoptosis through the MEG3/miR-15a-5p/peroxisome
proliferator-activated receptor γ coactivator 1α/SIRT1 pathway.
Similar to most lncRNAs described previously, MAGI2-AS3 also
affects IDD progression through a ceRNA mechanism. MAGI2-AS3 has
been reported to be poorly expressed in degenerated NP cells,
whereas oe-MAGI2-AS3 can downregulate miR-374b-5p and upregulate
IL-10 expression to improve inflammation and ECM degradation in NP
cells (78). Exosomes derived from
bone marrow mesenchymal stem cells (BMSC-Exos) have therapeutic
effects on IDD. In co-culture experiments, BMSC-Exos delivered
colon adenocarcinoma hypermethylated (CAHM) to inhibit the
polarisation of M1 macrophages, thereby reducing the apoptosis of
degenerated NP cells, ECM degradation and the expression of
inflammatory factors, and improving IDD (87). In addition to the aforementioned
lncRNAs, numerous lncRNAs discovered in previous studies also have
important effects on the occurrence and development of IDD
(56), requiring further
verification experiments (Table I
and Fig. 3).
Senescence of NP cells plays an important role in
the development of IDD. During ageing, the regenerative capacity of
NP cells weakens, apoptosis and inflammation increase, and cell
metabolism becomes abnormal (65),
ultimately promoting the occurrence of IDD under the combined
action of various factors. In vivo and in vitro
experiments have shown that the expression levels of adipogenesis
regulatory factor (ADIRF)-AS1 and SERPINA1 are downregulated in
high-grade degenerated NP tissues and that both have binding sites
for miR-214-3p. Subsequent experiments revealed that ADIRF-AS1 can
bind to miR-214-3p, thereby increasing SERPINA1 expression and
ultimately delaying NP cell ageing and apoptosis (88). As aforementioned, HOTAIR can
promote the apoptosis and ECM degradation of NP cells, and
accelerate the ageing of NP cells, which can comprehensively affect
the progress of IDD in these numerous aspects (74). Furthermore, transient receptor
potential canonical 7 RNA 1-AS1 adsorbs miR-4769-5p through the
classical ceRNA mechanism, inhibiting HPN and regulating ageing,
vitality and ECM synthesis in NP cells (89). The methylation level of lncRNA
activated by DNA damage (NORAD) has been shown to be significantly
increased in ageing NP cells, and the expression of WTAP was
revealed to be increased in the degenerated NP, significantly
promoting the m6 modification of NORAD; the lack of NORAD can
promote cellular senescence by affecting the expression of
Pumilio-homology domain (90).
In general, research on the mechanisms of lncRNAs in
IDD has made great progress since the discovery of the first
lncRNAs, H19, and the mechanisms of numerous lncRNAs in IDD have
been clarified step-by-step. The same lncRNAs can control IDD
through different signalling channels. Different lncRNAs can also
function through the same signalling pathway. The impact of lncRNAs
on NP cells has also impacted a number of aspects, such as
apoptosis, ECM degradation and synthesis, inflammatory response,
autophagy and ageing. lncRNAs form a large and complex regulatory
network with miRNAs, circRNAs and proteins. To date, most studies
have focused on the role of lncRNAs in nucleus pulposus, and there
has been little research on CEP and AF. Future research should
endeavour to further the understanding of CEP and AF (Table I and Fig. 3).
There are numerous differences in lncRNA expression
between patients with AS and healthy individuals, thus warranting
studies into the specific mechanisms governing this. Fang et
al (98) found that
NONHSAT227927.1 (a type of lncRNA) and tumour necrosis factor
receptor-associated factor 2 (TRAF2) were significantly increased
in the peripheral blood cells of patients with AS, and were
positively associated with clinical inflammatory indicators.
Xinfeng capsules, a traditional Chinese medicine, reduced immune
inflammation in AS by inhibiting lncRNA NONHSAT227927.1/TRAF2.
Furthermore, NONHSAT227927.1 was shown to activate the nuclear
factor-κB-p65 pathway by promoting TRAF2 expression, thereby
affecting the inflammatory process of AS (98). Osteoblasts are important regulators
of bone formation; however, their specific mechanisms of action in
AS remain unclear. Liu et al (99) collected serum and mesenchymal stem
cells from patients with AS and healthy donors and found that MEG3
and TNFα-induced protein 3 (TNFAIP3) were downregulated, whereas
miR-125a-5p was upregulated. Overexpression and knockdown
experiments showed that MEG3 reduced osteogenic differentiation and
inhibited AS progression through the
miR-125a-5p/TNFAIP3/Wnt/β-catenin axis (99). As the first lncRNA discovered, H19
plays a regulatory role in diseases such as tumours and IDD, and
affects the progression of inflammatory diseases such as AS. A
total of three molecules, H19, VDR and TGF-β, can bind to miR22-5p
and miR675-5p, and experiments have shown that H19 can regulate AS
through the miR22-5p/miR675-5p/VDR-IL-17A/IL-23 signalling pathway
(100). As a chronic spinal
disease, AS causes severe pain, and understanding the role of
lncRNAs in AS regulation provides a potential therapeutic strategy
for AS (Table II).
SCI is a severe consequence of trauma to the spinal
cord. The spinal cord is affected by external forces or chronic
compression, and the corresponding sensory, motor and autonomic
nervous functions are impaired owing to factors such as
inflammation and apoptosis (101). Nerve cells have poor regenerative
ability and cannot be cured after being damaged; therefore,
treatment and rehabilitation after SCI are challenging (102). Differentially expressed lncRNAs
play a regulatory role in SCI, providing new ideas for its
treatment (103). Notably,
menstrual blood-derived stem cells (MenSCs) have important
therapeutic effects in degenerative and traumatic diseases, such as
premature ovarian failure in mice (104) and cutaneous wound (105). Compared with SCI rats, rats
treated with MenSCs exhibited significant upregulation of 89
lncRNAs and other RNAs, and significant downregulation of 65
lncRNAs and other RNAs. The lncRNA-miRNA-mRNA and
circRNA-miRNA-mRNA ceRNA networks of SCI indicated that the role of
lncRNAs in SCI may be mediated by a large interconnected regulatory
network (103). Liu et al
(106) examined changes in the
expression of lncRNAs in the proximal tissue of the T10 layer at
different time points during the right hemisection of T10
laminectomy. The expression of 68 lncRNAs first increased and then
remained high 3 days after injury. Meanwhile, the expression of 56
lncRNAs decreased initially and remained low 3 days after injury.
In a similar study, circulating exosomes were extracted from the
blood of rats with SCI and control rats, and ncRNAs in the exosomes
were identified, ultimately showing that the expression of
ENSRN0T00000067908, XR_590093, XR_591455, XR_360081 and XR_346933
was upregulated, whereas the expression of XR_351404, XR_591426,
XR_353833, XR_590076 and XR_590719 was downregulated. These 10
lncRNAs were at the centre of the constructed lncRNA-miRNA-mRNA
co-expression network (107).
These studies indicated that lncRNAs form important regulatory
networks in SCI.
Growth-associated protein 43 (GAP43) is important
for axonal growth and elongation. Hu et al (108) reported the protective role of
vof16 (ischemia-related factor Vof-16) in SCI through the
miR-185-5p-GAP43 regulatory network. Tanshinone IIA (TSIIA), a
traditional Chinese medicine ingredient, can protect against SCI
and tectonic family member 2 (TCTN2) expression has been shown to
be low in an LPS-induced cell injury model. By contrast, TSIIA
reduced cell damage and increased the expression of TCTN2;
follow-up experiments demonstrated that TCTN2 may act as a
regulator of dual-specificity phosphatase 1 (DUSP1) expression
through miR-125a-5p; that is, TSIIA may regulate SCI through the
TCTN2/miR-125a-5p/DUSP1 axis (109). In another previous study
(110), researchers injected
small extracellular vesicles (sEVs) derived from human umbilical
cord mesenchymal stem cells induced by micro-electric field
stimulation (EF-sEVs) and normally conditioned human umbilical cord
mesenchymal stem cells-derived sEVs. Within the lesions of rats
with SCI, MALAT1 expression was elevated in tissues treated with
EF-sEVs. In addition, a luciferase reporter gene assay showed that
MALAT1 can competitively bind to miR-22-3p and reduce its
inhibitory effect on SIRT1, thereby improving SCI (110). A study has indicated that
photobiomodulation (PBM) reduces inflammation by inhibiting bone
marrow-derived macrophages. Transcriptome sequencing and
bioinformatics analyses indicated that taurine upregulated gene 1
(TUG1) may be a target of PBM, and that TUG1 could competitively
bind to miR-1192 and reduce miR-1192-induced inhibition of TLR3,
leading to increased TLR3 expression and promotion of nerve
survival and motor recovery (111) (Table II).
The role of lncRNAs in treating spinal diseases is
mostly exerted by reducing or increasing the effective expression
of the corresponding lncRNAs. Several methods can reduce the
expression of lncRNAs. Firstly, silencing lncRNAs, such as siPART1,
can enhance the growth of NP cells and increase the synthesis of
the ECM, which can be used to treat IDD (67). Secondly, in a previous study,
changing the chemical modification of lncRNAs increased WTAP
expression in degenerated NP cells and significantly promoted the
m6 modification of NORAD, promoting cell senescence. Therefore,
therapeutic purposes can be achieved by reducing the m6
modification of NORAD (90).
Finally, the formation of a complex with lncRNAs can interfere with
its function; for example, some small molecule inhibitors can hide
the functional sites of lncRNAs and interfere with their functions.
Notably, BRD4 can bind to the HOTAIR promoter, affect HOTAIR
function and control glioblastoma (112).
Methods to increase lncRNA expression include lncRNA
mimics transfection. A previous study reported that KLF3-AS1
overexpression may improve IDD through the miR-10a-3p/ZBTB20 axis
(70). Another method includes
exogenously supplementing lncRNAs. Li et al (87) revealed that BMSC-Exo delivered
exogenous CAHM can regulate macrophage polarisation and improve IDD
(87). LncRNAs have therapeutic
potential in spinal diseases, such as IDD, but some problems remain
to be solved. When regulating the expression of lncRNAs, their
impact cannot be accurately controlled, and their effect is not
permanent and may decrease over time. Therefore, treating spinal
diseases using lncRNAs requires further research to promote their
progress.
Numerous lncRNAs have complex functions. The present
article explains the classification and functions of lncRNAs and
their roles in IDD, AS and SCI. In spinal diseases such as IDD,
lncRNAs affect different aspects of disease progression, such as
the ECM, cell death, inflammation and ageing. Taking IDD as an
example, thousands of lncRNAs show differential expression in IDD
compared with normal tissues. However, only a handful of lncRNAs
have been studied. Increased research is required to fully
understand the role of lncRNAs in spinal diseases, and experiments
are needed to explore, supplement and improve ceRNA regulatory
networks. Application of lncRNA in treating spinal diseases, such
as IDD, is still in the preclinical experimental stage. However,
lncRNAs have been used in the treatment of some malignant diseases
(113) and have shown good
efficacy; therefore, lncRNAs show great potential for clinical
application in treating spinal diseases such as IDD.
In summary, lncRNAs are important regulatory factors
affecting the occurrence and development of spinal diseases, such
as IDD. However, current research on lncRNAs in spinal diseases has
mainly focused on cells, rats and human degenerative tissues, and
studies on more advanced large animals such as monkeys are lacking.
Experimental and clinical treatments are meaningful directions for
future research.
Not applicable.
This work was supported by the CuiYing Science and Technology
Innovation plan project of Lanzhou University Second Hospital
(grant no. CY2021-MS-A03).
Not applicable.
ZM collected the literature and wrote the article;
ZL and JA revised the article; ZM and ZL designed the study; and
XL, XZ and SL prepared the figures and tables. ZM, XL, XZ, SL, JA
and ZL contributed to data analysis, drafted and critically revised
the paper, and agreed to be accountable for all aspects of the
work. Data authentication is not applicable. All authors read and
approved the final version of the manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Wong CK, Mak RY, Kwok TS, Tsang JS, Leung
MY, Funabashi M, Macedo LG, Dennett L and Wong AY: Prevalence,
incidence, and factors associated with non-specific chronic low
back pain in community-dwelling older adults aged 60 years and
older: A systematic review and meta-analysis. J Pain. 23:509–534.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Speed C: Low back pain. BMJ.
328:1119–1121. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
GBD 2017, . Disease and Injury Incidence
and Prevalence Collaborators: Global, regional, and national
incidence, prevalence, and years lived with disability for 354
diseases and injuries for 195 countries and territories, 1990–2017:
A systematic analysis for the global burden of disease study 2017.
Lancet. 392:1789–1858. 2018.PubMed/NCBI
|
|
4
|
Samartzis D, Karppinen J, Mok F, Fong DYT,
Luk KDK and Cheung KMC: A population-based study of juvenile disc
degeneration and its association with overweight and obesity, low
back pain, and diminished functional status. J Bone Joint Surg Am.
93:662–670. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen WK, Yu XH, Yang W, Wang C, He WS, Yan
YG, Zhang J and Wang WJ: lncRNAs: Novel players in intervertebral
disc degeneration and osteoarthritis. Cell Prolif. 50:e123132017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Song C, Hu P, Peng R, Li F, Fang Z and Xu
Y: Bioenergetic dysfunction in the pathogenesis of intervertebral
disc degeneration. Pharmacol Res. 202:1071192024. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ohnishi T, Sudo H, Tsujimoto T and Iwasaki
N: Age-related spontaneous lumbar intervertebral disc degeneration
in a mouse model. J Orthop Res. 36:224–232. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rinaldi S, Moroni E, Rozza R and
Magistrato A: Frontiers and challenges of computing ncRNAs
biogenesis, function and modulation. J Chem Theory Comput.
20:993–1018. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Parasramka MA, Maji S, Matsuda A, Yan IK
and Patel T: Long non-coding RNAs as novel targets for therapy in
hepatocellular carcinoma. Pharmacol Ther. 161:67–78. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jiang C, Chen Z, Wang X, Zhang Y, Guo X,
Xu Z, Yang H and Hao D: The potential mechanisms and application
prospects of non-coding RNAs in intervertebral disc degeneration.
Front Endocrinol (Lausanne). 13:10811852022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mehmandar-Oskuie A, Jahankhani K,
Rostamlou A, Mardafkan N, Karamali N, Razavi ZS and Mardi A:
Molecular mechanism of lncRNAs in pathogenesis and diagnosis of
auto-immune diseases, with a special focus on lncRNA-based
therapeutic approaches. Life Sci. 336:1223222024. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lei HT, Wang JH, Yang HJ, Wu HJ, Nian FH,
Jin FM, Yang J, Tian XM and Wang HD: LncRNA-mediated cell
autophagy: An emerging field in bone destruction in rheumatoid
arthritis. Biomed Pharmacother. 168:1157162023. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jafari-Raddani F, Davoodi-Moghaddam Z,
Yousefi AM, Ghaffari SH and Bashash D: An overview of long
noncoding RNAs: Biology, functions, therapeutics, analysis methods,
and bioinformatics tools. Cell Biochem Funct. 40:800–825. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Khan K, Irfan M, Sattar AA, Faiz MB,
Rahman AU, Athar H, Calina D, Sharifi-Rad J and Cho WC: LncRNA
SNHG6 role in clinicopathological parameters in cancers. Eur J Med
Res. 28:3632023. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Monteiro JP, Bennett M, Rodor J,
Caudrillier A, Ulitsky I and Baker AH: Endothelial function and
dysfunction in the cardiovascular system: The long non-coding road.
Cardiovasc Res. 115:1692–1704. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dai L, Liang W, Shi Z, Li X, Zhou S, Hu W,
Yang Z and Wang X: Systematic characterization and biological
functions of non-coding RNAs in glioblastoma. Cell Prolif.
56:e133752023. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jayasuriya R, Ganesan K, Xu B and Ramkumar
KM: Emerging role of long non-coding RNAs in endothelial
dysfunction and their molecular mechanisms. Biomed Pharmacother.
145:1124212022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang L, Xu X and Su X: Modifications of
noncoding RNAs in cancer and their therapeutic implications. Cell
Signal. 108:1107262023. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang JH, Chen JH, Guo B, Fang Y, Xu ZY,
Zhan L and Cao YX: Recent insights into noncoding RNAs in primary
ovarian insufficiency: Focus on mechanisms and treatments. J Clin
Endocrinol Metab. 108:1898–1908. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu X, Wang X, Li J, Hu S, Deng Y, Yin H,
Bao X, Zhang QC, Wang G, Wang B, et al: Identification of mecciRNAs
and their roles in the mitochondrial entry of proteins. Sci China
Life Sci. 63:1429–1449. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Statello L, Guo CJ, Chen LL and Huarte M:
Gene regulation by long non-coding RNAs and its biological
functions. Nat Rev Mol Cell Biol. 22:96–118. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Areeb Z, Stuart SF, West AJ, Gomez J,
Nguyen HPT, Paradiso L, Zulkifli A, Jones J, Kaye AH, Morokoff AP
and Luwor RB: Reduced EGFR and increased miR-221 is associated with
increased resistance to temozolomide and radiotherapy in
glioblastoma. Sci Rep. 10:177682020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang K, Shi ZM, Chang YN, Hu ZM, Qi HX
and Hong W: The ways of action of long non-coding RNAs in cytoplasm
and nucleus. Gene. 547:1–9. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lazorthes S, Vallot C, Briois S,
Aguirrebengoa M, Thuret JY, St Laurent G, Rougeulle C, Kapranov P,
Mann C, Trouche D and Nicolas E: A vlincRNA participates in
senescence maintenance by relieving H2AZ-mediated repression at the
INK4 locus. Nat Commun. 6:59712015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mattick JS: A new paradigm for
developmental biology. J Exp Biol. 210:1526–1547. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fatica A and Bozzoni I: Long non-coding
RNAs: New players in cell differentiation and development. Nat Rev
Genet. 15:7–21. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dueva R, Akopyan K, Pederiva C, Trevisan
D, Dhanjal S, Lindqvist A and Farnebo M: Neutralization of the
positive charges on histone tails by RNA promotes an open chromatin
structure. Cell Chem Biol. 26:1436–1449.e5. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li Y, Syed J and Sugiyama H: RNA-DNA
triplex formation by long noncoding RNAs. Cell Chem Biol.
23:1325–1333. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Niehrs C and Luke B: Regulatory R-loops as
facilitators of gene expression and genome stability. Nat Rev Mol
Cell Biol. 21:167–178. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhou X, Lv Y, Xie H, Li Y, Liu C, Zheng M,
Wu R, Zhou S, Gu X, Li J and Mi D: RNA sequencing of exosomes
secreted by fibroblast and Schwann cells elucidates mechanisms
underlying peripheral nerve regeneration. Neural Regen Res.
19:1812–1821. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang L, Hu L, Wang X, Geng Z, Wan M, Hao
J, Liu H, Fan Y, Xu T and Li Z: Long non-coding RNA LncCplx2
regulates glucose homeostasis and pancreatic β cell function. Mol
Metab. 80:1018782024. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hacisuleyman E, Goff LA, Trapnell C,
Williams A, Henao-Mejia J, Sun L, McClanahan P, Hendrickson DG,
Sauvageau M, Kelley DR, et al: Topological organization of
multichromosomal regions by the long intergenic noncoding RNA
Firre. Nat Struct Mol Biol. 21:198–206. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Naganuma T and Hirose T: Paraspeckle
formation during the biogenesis of long non-coding RNAs. RNA Biol.
10:456–461. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kopp F: Molecular functions and biological
roles of long non-coding RNAs in human physiology and disease. J
Gene Med. 21:e31042019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Núñez-Martínez HN and Recillas-Targa F:
Emerging functions of lncRNA loci beyond the transcript itself. Int
J Mol Sci. 23:62582022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang PS, Liu Z, Sweef O, Xie J, Chen J,
Zhu H, Zeidler-Erdely PC, Yang C and Wang Z: Long noncoding RNA
ABHD11-AS1 interacts with SART3 and regulates CD44 RNA alternative
splicing to promote lung carcinogenesis. Environ Int.
185:1084942024. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Rashid F, Shah A and Shan G: Long
non-coding RNAs in the cytoplasm. Genomics Proteomics
Bioinformatics. 14:73–80. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Huang X, Zhou X, Hu Q, Sun B, Deng M, Qi X
and Lü M: Advances in esophageal cancer: A new perspective on
pathogenesis associated with long non-coding RNAs. Cancer Lett.
413:94–101. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Han I, Ropper AE, Konya D, Kabatas S,
Toktas Z, Aljuboori Z, Zeng X, Chi JH, Zafonte R and Teng YD:
Biological approaches to treating intervertebral disk degeneration:
Devising stem cell therapies. Cell Transplant. 24:2197–2208. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang L, Hu S, Xiu C, Li M, Zheng Y, Zhang
R, Li B and Chen J: Intervertebral disc-intrinsic Hedgehog
signaling maintains disc cell phenotypes and prevents disc
degeneration through both cell autonomous and non-autonomous
mechanisms. Cell Mol Life Sci. 81:742024. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Boubriak OA, Watson N, Sivan SS, Stubbens
N and Urban JPG: Factors regulating viable cell density in the
intervertebral disc: Blood supply in relation to disc height. J
Anat. 222:341–348. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Woods BI, Vo N, Sowa G and Kang JD: Gene
therapy for intervertebral disk degeneration. Orthop Clin North Am.
42563–574. (ix)2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Pooni JS, Hukins DW, Harris PF, Hilton RC
and Davies KE: Comparison of the structure of human intervertebral
discs in the cervical, thoracic and lumbar regions of the spine.
Surg Radiol Anat. 8:175–182. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Shapiro IM, Vresilovic EJ and Risbud MV:
Is the spinal motion segment a diarthrodial polyaxial joint: What a
nice nucleus like you doing in a joint like this? Bone. 50:771–776.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wang S, Sun J, Yang H, Zou W, Zheng B,
Chen Y, Guo Y and Shi J: Profiling and bioinformatics analysis of
differentially expressed circular RNAs in human intervertebral disc
degeneration. Acta Biochim Biophys Sin (Shanghai). 51:571–579.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ohnishi T, Iwasaki N and Sudo H: Causes of
and molecular targets for the treatment of intervertebral disc
degeneration: A review. Cells. 11:3942022. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Battié MC and Videman T: Lumbar disc
degeneration: epidemiology and genetics. J Bone Joint Surg Am. 88
(Suppl 2):S3–S9. 2006. View Article : Google Scholar
|
|
49
|
Roberts S, Evans H, Trivedi J and Menage
J: Histology and pathology of the human intervertebral disc. J Bone
Joint Surg Am. 88 (Suppl 2):S10–S14. 2006. View Article : Google Scholar
|
|
50
|
Ye F, Lyu FJ, Wang H and Zheng Z: The
involvement of immune system in intervertebral disc herniation and
degeneration. JOR Spine. 5:e11962022. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Takahashi J, Shono Y, Hirabayashi H,
Kamimura M, Nakagawa H, Ebara S and Kato H: Usefulness of white
blood cell differential for early diagnosis of surgical wound
infection following spinal instrumentation surgery. Spine (Phila Pa
1976). 31:1020–1025. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Tsujimoto T, Sudo H, Todoh M, Yamada K,
Iwasaki K, Ohnishi T, Hirohama N, Nonoyama T, Ukeba D, Ura K, et
al: An acellular bioresorbable ultra-purified alginate gel promotes
intervertebral disc repair: A preclinical proof-of-concept study.
EBioMedicine. 37:521–534. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Kuo CC, Soliman MAR, Baig RA, Aguirre AO,
Ruggiero N, Donnelly BM, Siddiqi M, Khan A, Quiceno E, Mullin JP
and Pollina J: Vertebral bone quality score as a predictor of
adjacent segment disease after lumbar interbody fusion.
Neurosurgery. Feb 9–2024.(Epub ahead of print). View Article : Google Scholar
|
|
54
|
Wan ZY, Song F, Sun Z, Chen YF, Zhang WL,
Samartzis D, Ma CJ, Che L, Liu X, Ali MA, et al: Aberrantly
expressed long noncoding RNAs in human intervertebral disc
degeneration: A microarray related study. Arthritis Res Ther.
16:4652014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Chen Y, Ni H, Zhao Y, Chen K, Li M, Li C,
Zhu X and Fu Q: Potential role of lncRNAs in contributing to
pathogenesis of intervertebral disc degeneration based on
microarray data. Med Sci Monit. 21:3449–3458. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wu Y, Li S, Shen J, Wang Z and Liu H:
Nucleus pulposus related lncRNA and mRNA expression profiles in
intervertebral disc degeneration. Genomics. 115:1105702023.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Shi Y, Guo R, Zeng Y, Fang Q, Wang X, Liu
W, Huang G and Wu W: SNHG5/miR-299-5p/ATF2 axis as a biomarker in
immune microenvironment of intervertebral disc degeneration.
Mediators Inflamm. 2022:25582752022. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhang Y, Zhang J, Sun Z, Wang H, Ning R,
Xu L, Zhao Y, Yang K, Xi X and Tian J: MAPK8 and CAPN1 as potential
biomarkers of intervertebral disc degeneration overlapping immune
infiltration, autophagy, and ceRNA. Front Immunol. 14:11887742023.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Iwashina T, Mochida J, Sakai D, Yamamoto
Y, Miyazaki T, Ando K and Hotta T: Feasibility of using a human
nucleus pulposus cell line as a cell source in cell transplantation
therapy for intervertebral disc degeneration. Spine (Phila Pa
1976). 31:1177–1186. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Chen X, Zhu L, Wu G, Liang Z, Yang L and
Du Z: A comparison between nucleus pulposus-derived stem cell
transplantation and nucleus pulposus cell transplantation for the
treatment of intervertebral disc degeneration in a rabbit model.
Int J Surg. 28:77–82. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Li Z, Li X, Chen C, Li S, Shen J, Tse G,
Chan MTV and Wu WKK: Long non-coding RNAs in nucleus pulposus cell
function and intervertebral disc degeneration. Cell Prolif.
51:e124832018. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Jiang C, Liu Y, Zhao W, Yang Y, Ren Z,
Wang X, Hao D, Du H and Yin S: microRNA-365 attenuated
intervertebral disc degeneration through modulating nucleus
pulposus cell apoptosis and extracellular matrix degradation by
targeting EFNA3. J Cell Mol Med. 28:e180542024. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Lin Y, Jiao Y, Yuan Y, Zhou Z, Zheng Y,
Xiao J, Li C, Chen Z and Cao P: Propionibacterium acnes
induces intervertebral disc degeneration by promoting nucleus
pulposus cell apoptosis via the TLR2/JNK/mitochondrial-mediated
pathway. Emerg Microbes Infect. 7:12018. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zhao K, An R, Xiang Q, Li G, Wang K, Song
Y, Liao Z, Li S, Hua W, Feng X, et al: Acid-sensing ion channels
regulate nucleus pulposus cell inflammation and pyroptosis via the
NLRP3 inflammasome in intervertebral disc degeneration. Cell
Prolif. 54:e129412021. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Du J, Xu M, Kong F, Zhu P, Mao Y, Liu Y,
Zhou H, Dong Z, Yu Z, Du T, et al: CB2R attenuates intervertebral
disc degeneration by delaying nucleus pulposus cell senescence
through AMPK/GSK3β pathway. Aging Dis. 13:552–567. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Jiang W, Zhao P and Zhang X: Apelin
promotes ECM synthesis by enhancing autophagy flux via TFEB in
human degenerative NP cells under oxidative stress. Biomed Res Int.
2020:48971702020.PubMed/NCBI
|
|
67
|
Gao D, Hao L and Zhao Z: Long non-coding
RNA PART1 promotes intervertebral disc degeneration through
regulating the miR-93/MMP2 pathway in nucleus pulposus cells. Int J
Mol Med. 46:289–299. 2020.PubMed/NCBI
|
|
68
|
Yu X, Liu Q, Wang Y, Bao Y, Jiang Y, Li M,
Li Z, Wang B, Yu L, Wang S, et al: Depleted long noncoding RNA GAS5
relieves intervertebral disc degeneration via microRNA-17-3p/Ang-2.
Oxid Med Cell Longev. 2022:17924122022. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Hao Y, Zhu G, Yu L, Ren Z, Zhou W, Zhang P
and Lian X: FOXO3-activated HOTTIP sequesters miR-615-3p away from
COL2A1 to mitigate intervertebral disc degeneration. Am J Pathol.
194:280–295. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Chen S, Zhuang Q, Li P, Zeng J, Peng Y,
Ding Z, Cao H, Zheng R and Wang W: The long non-coding RNA
KLF3-AS1/miR-10a-3p/ZBTB20 axis improves the degenerative changes
in human nucleus pulposus cells. Cell Tissue Res. 393:97–109. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Shang L, Ma H, Zhang X, Mao R, Ma C and
Ruan Z: Docosahexaenoic acid alleviates the excessive degradation
of extracellular matrix in the nucleus pulposus by reducing the
content of lncRNA NEAT1 to prevent the progression of
intervertebral disc degeneration. Clin Exp Pharmacol Physiol.
50:403–414. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Wang Z, Liu B, Ma X, Wang Y, Han W and
Xiang L: lncRNA ZFAS1 promotes intervertebral disc degeneration by
upregulating AAK1. Open Med (Wars). 17:1973–1986. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Zhu M, Yan X, Zhao Y, Xue H, Wang Z, Wu B,
Li X and Shen Y: lncRNA LINC00284 promotes nucleus pulposus cell
proliferation and ECM synthesis via regulation of the
miR-205-3p/Wnt/β-catenin axis. Mol Med Rep. 25:1792022. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Zhan S, Wang K, Xiang Q, Song Y, Li S,
Liang H, Luo R, Wang B, Liao Z, Zhang Y and Yang C: lncRNA HOTAIR
upregulates autophagy to promote apoptosis and senescence of
nucleus pulposus cells. J Cell Physiol. 235:2195–2208. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Zhan S, Wang K, Song Y, Li S, Yin H, Luo
R, Liao Z, Wu X, Zhang Y and Yang C: Long non-coding RNA HOTAIR
modulates intervertebral disc degenerative changes via
Wnt/β-catenin pathway. Arthritis Res Ther. 21:2012019. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Zhou D, Mei Y, Song C, Cheng K, Cai W, Guo
D, Gao S, Lv J, Liu T, Zhou Y, et al: Exploration of the mode of
death and potential death mechanisms of nucleus pulposus cells. Eur
J Clin Invest. e142262024.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Gao ZX, Lin YC, Wu ZP, Zhang P, Cheng QH,
Ye LH, Wu FH, Chen YJ, Fu MH, Cheng CG and Gao YC: LncRNA SNHG6 can
regulate the proliferation and apoptosis of rat degenerate nucleus
pulposus cells via regulating the expression of miR-101-3p. Eur Rev
Med Pharmacol Sci. 24:8251–8262. 2020.PubMed/NCBI
|
|
78
|
Yu J and Li C: Role of lncRNA MAGI2-AS3 in
lipopolysaccharide-induced nucleus pulposus cells injury by
regulating miR-374b-5p/interleukin-10 axis. Immun Inflamm Dis.
11:e7722023. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Yang W, Huang XD, Zhang T, Zhou YB, Zou YC
and Zhang J: LncRNA MIR155HG functions as a ceRNA of miR-223-3p to
promote cell pyroptosis in human degenerative NP cells. Clin Exp
Immunol. 207:241–252. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Tao X, Xue F, Xu J and Wang W:
Platelet-rich plasma-derived extracellular vesicles inhibit
NF-κB/NLRP3 pathway-mediated pyroptosis in intervertebral disc
degeneration via the MALAT1/microRNA-217/SIRT1 axis. Cell Signal.
117:1111062024. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Zhang L, Zhou Y, Huang T, Cheng ASL, Yu J,
Kang W and To KF: The interplay of LncRNA-H19 and its binding
partners in physiological process and gastric carcinogenesis. Int J
Mol Sci. 18:4502017. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Sun Z, Tang X, Wang H, Sun H, Chu P, Sun L
and Tian J: LncRNA H19 aggravates intervertebral disc degeneration
by promoting the autophagy and apoptosis of nucleus pulposus cells
through the miR-139/CXCR4/NF-κB axis. Stem Cells Dev. 30:736–748.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Guo C, Liu Y, Zhao Z, Wu Y, Kong Q and
Wang Y: Regulating inflammation and apoptosis: A smart microgel
gene delivery system for repairing degenerative nucleus pulposus. J
Control Release. 365:1004–1018. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Jiang X and Chen D: LncRNA FAM83H-AS1
maintains intervertebral disc tissue homeostasis and attenuates
inflammation-related pain via promoting nucleus pulposus cell
growth through miR-22-3p inhibition. Ann Transl Med. 8:15182020.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Luo Y, He Y, Wang Y, Xu Y and Yang L:
LncRNA HCG18 promotes inflammation and apoptosis in intervertebral
disc degeneration via the miR-495-3p/FSTL1 axis. Mol Cell Biochem.
479:171–181. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Zhang C, Qiu Y and Yuan F: The long
non-coding RNA maternally expressed 3-micorRNA-15a-5p axis is
modulated by melatonin and prevents nucleus pulposus cell
inflammation and apoptosis. Basic Clin Pharmacol Toxicol.
133:603–619. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Li W, Xu Y and Chen W: Bone mesenchymal
stem cells deliver exogenous lncRNA CAHM via exosomes to regulate
macrophage polarization and ameliorate intervertebral disc
degeneration. Exp Cell Res. 421:1134082022. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Zhong H, Zhou Z, Guo L, Liu FS, Wang X, Li
J, Lv GH and Zou MX: SERPINA1 is a hub gene associated with
intervertebral disc degeneration grade and affects the nucleus
pulposus cell phenotype through the ADIRF-AS1/miR-214-3p axis.
Transl Res. 245:99–116. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Wang X, Li D, Wu H, Liu F, Liu F, Zhang Q
and Li J: LncRNA TRPC7-AS1 regulates nucleus pulposus cellular
senescence and ECM synthesis via competing with HPN for miR-4769-5p
binding. Mech Ageing Dev. 190:1112932020. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Li G, Ma L, He S, Luo R, Wang B, Zhang W,
Song Y, Liao Z, Ke W, Xiang Q, et al: WTAP-mediated m6A
modification of lncRNA NORAD promotes intervertebral disc
degeneration. Nat Commun. 13:14692022. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Mauro D, Thomas R, Guggino G, Lories R,
Brown MA and Ciccia F: Ankylosing spondylitis: An autoimmune or
autoinflammatory disease? Nat Rev Rheumatol. 17:387–404. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Kenar G, Yarkan-Tuğsal H, Çetin-Özmen P,
Solmaz D, Can G and Önen F: A lower frequency of inflammatory back
pain in male patients with ankylosing spondylitis compared with
female patients. Rheumatol Int. 44:477–482. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Braun J and Sieper J: Ankylosing
spondylitis. Lancet. 369:1379–1390. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Ward MM, Deodhar A, Akl EA, Lui A, Ermann
J, Gensler LS, Smith JA, Borenstein D, Hiratzka J, Weiss PF, et al:
American college of rheumatology/spondylitis association of
america/spondyloarthritis research and treatment network 2015
recommendations for the treatment of ankylosing spondylitis and
nonradiographic axial spondyloarthritis. Arthritis Rheumatol.
68:282–298. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Cen S, Cai M, Wang Y, Lu X, Chen Z, Chen
H, Fang Y, Wu C, Qiu S and Liu Z: Aberrant lncRNA-mRNA expression
profile and function networks during the adipogenesis of
mesenchymal stem cells from patients with ankylosing spondylitis.
Front Genet. 13:9918752022. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Huang D, Liu J, Wan L, Fang Y, Long Y,
Zhang Y and Bao B: Identification of lncRNAs associated with the
pathogenesis of ankylosing spondylitis. BMC Musculoskelet Disord.
22:2722021. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Wang JX, Jing FY, Xu YC, Zong HX, Chu YR,
Wang C, Chen KM, Tong WQ, Wang XL and Xu SQ: The potential
regulatory mechanism of lncRNA 122K13.12 and lncRNA 326C3.7 in
ankylosing spondylitis. Front Mol Biosci. 8:7454412021. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Fang Y, Liu J, Xin L, Jiang H, Wen J, Li
X, Wang F, He M and Han Q: Xinfeng capsule inhibits lncRNA
NONHSAT227927.1/TRAF2 to alleviate NF-κB-p65-induced
immuno-inflammation in ankylosing spondylitis. J Ethnopharmacol.
323:1176772024. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Liu C, Liang T, Zhang Z, Chen J, Xue J,
Zhan X and Ren L: MEG3 alleviates ankylosing spondylitis by
suppressing osteogenic differentiation of mesenchymal stem cells
through regulating microRNA-125a-5p-mediated TNFAIP3. Apoptosis.
28:498–513. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Zhang X, Ji S, Cai G, Pan Z, Han R, Yuan
Y, Xu S, Yang J, Hu X, Chen M, et al: H19 increases IL-17A/IL-23
releases via regulating VDR by interacting with miR675-5p/miR22-5p
in ankylosing spondylitis. Mol Ther Nucleic Acids. 19:393–404.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Baskozos G, Dawes JM, Austin JS,
Antunes-Martins A, McDermott L, Clark AJ, Trendafilova T, Lees JG,
McMahon SB, Mogil JS, et al: Comprehensive analysis of long
noncoding RNA expression in dorsal root ganglion reveals cell-type
specificity and dysregulation after nerve injury. Pain.
160:463–485. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Yang Y, Fan R, Li H, Chen H, Gong H and
Guo G: Polysaccharides as a promising platform for the treatment of
spinal cord injury: A review. Carbohydr Polym. 327:1216722024.
View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Qi L, Jiang W, He W, Li X, Wu J, Chen S,
Liao Z, Yu S, Liu J, Sun Y, et al: Transcriptome profile analysis
in spinal cord injury rats with transplantation of menstrual
blood-derived stem cells. Front Mol Neurosci. 17:13354042024.
View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Wang Z, Wang Y, Yang T, Li J and Yang X:
Study of the reparative effects of menstrual-derived stem cells on
premature ovarian failure in mice. Stem Cell Res Ther. 8:112017.
View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Chen L, Qu J, Mei Q, Chen X, Fang Y, Chen
L, Li Y and Xiang C: Small extracellular vesicles from menstrual
blood-derived mesenchymal stem cells (MenSCs) as a novel
therapeutic impetus in regenerative medicine. Stem Cell Res Ther.
12:4332021. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Liu W, Tao JC, Zhu SZ, Dai CL, Wang YX, Yu
B, Yao C and Sun YY: Expression and regulatory network of long
noncoding RNA in rats after spinal cord hemisection injury. Neural
Regen Res. 17:2300–2304. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Li JA, Shi MP, Cong L, Gu MY, Chen YH,
Wang SY, Li ZH, Zan CF and Wei WF: Circulating exosomal lncRNA
contributes to the pathogenesis of spinal cord injury in rats.
Neural Regen Res. 18:889–894. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Hu Y, Sun YF, Yuan H, Liu J, Chen L, Liu
DH, Xu Y, Zhou XF, Ding L, Zhang ZT, et al: Vof16-miR-185-5p-GAP43
network improves the outcomes following spinal cord injury via
enhancing self-repair and promoting axonal growth. CNS Neurosci
Ther. 30:e145352024. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Yan Q, Xun Y, Lei D and Zhai H: Tanshinone
IIA protects motor neuron-like NSC-34 cells against
lipopolysaccharide-induced cell injury by the regulation of the
lncRNA TCTN2/miR-125a-5/DUSP1 axis. Regen Ther. 24:417–425. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Li K, Liu Z, Wu P, Chen S, Wang M, Liu W,
Zhang L, Guo S, Liu Y, Liu P, et al: Micro electrical fields
induced MSC-sEVs attenuate neuronal cell apoptosis by activating
autophagy via lncRNA MALAT1/miR-22-3p/SIRT1/AMPK axis in spinal
cord injury. J Nanobiotechnology. 21:4512023. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Ju C, Ma Y, Zuo X, Wang X, Song Z, Zhang
Z, Zhu Z, Li X, Liang Z, Ding T, et al: Photobiomodulation promotes
spinal cord injury repair by inhibiting macrophage polarization
through lncRNA TUG1-miR-1192/TLR3 axis. Cell Mol Biol Lett.
28:52023. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Pastori C, Kapranov P, Penas C, Peschansky
V, Volmar CH, Sarkaria JN, Bregy A, Komotar R, St Laurent G, Ayad
NG and Wahlestedt C: The bromodomain protein BRD4 controls HOTAIR,
a long noncoding RNA essential for glioblastoma proliferation. Proc
Natl Acad Sci USA. 112:8326–8331. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Li L, Gao Y, Yu B, Zhang J, Ma G and Jin
X: Role of LncRNA H19 in tumor progression and treatment. Mol Cell
Probes. 75:1019612024. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Cao S, Ma Y, Yang H, Luo G, Cheng H, Jin X
and Sun T: Long noncoding RNA HCG18 promotes extracellular matrix
degradation of nucleus pulposus cells in intervertebral disc
degeneration by regulating the miR-4306/EPAS1 axis. World
Neurosurg. 172:e52–e61. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Chen WK, Zhang HJ, Zou MX, Wang C, Yan YG,
Zhan XL, Li XL and Wang WJ: LncRNA HOTAIR influences cell
proliferation via miR-130b/PTEN/AKT axis in IDD. Cell Cycle.
21:323–339. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Chen W, Wang F, Wang J, Chen F and Chen T:
The molecular mechanism of long non-coding RNA MALAT1-mediated
regulation of chondrocyte pyroptosis in ankylosing spondylitis. Mol
Cells. 45:365–375. 2022. View Article : Google Scholar : PubMed/NCBI
|