Tetraspanins, also termed TSPANs or the
transmembrane 4 super-family proteins, are members of a protein
family widely conserved across different species. These proteins
are ubiquitously distributed within various organisms (1). Tetraspanins participate in a broad
spectrum of biological processes, such as regulating cell
morphology, motility, invasion, fusion and signaling. Specifically,
tetraspanins serve crucial roles in tumor metastasis, membrane
dynamics, infection, synapse formation, platelet aggregation,
fertilization and the immune response (1). Although tetraspanins have a small
molecular size of 200–350 amino acids and only extend 3–5 nm above
the cell membrane (2,3), their function is significant. Studies
have emphasized their essential biological and physiological
functions in promoting cell movement and growth by serving as
coordinators of complex structures within cell membranes (1,4–7).
Tetraspanins, form a protein group consisting of 33
identified members in humans (8).
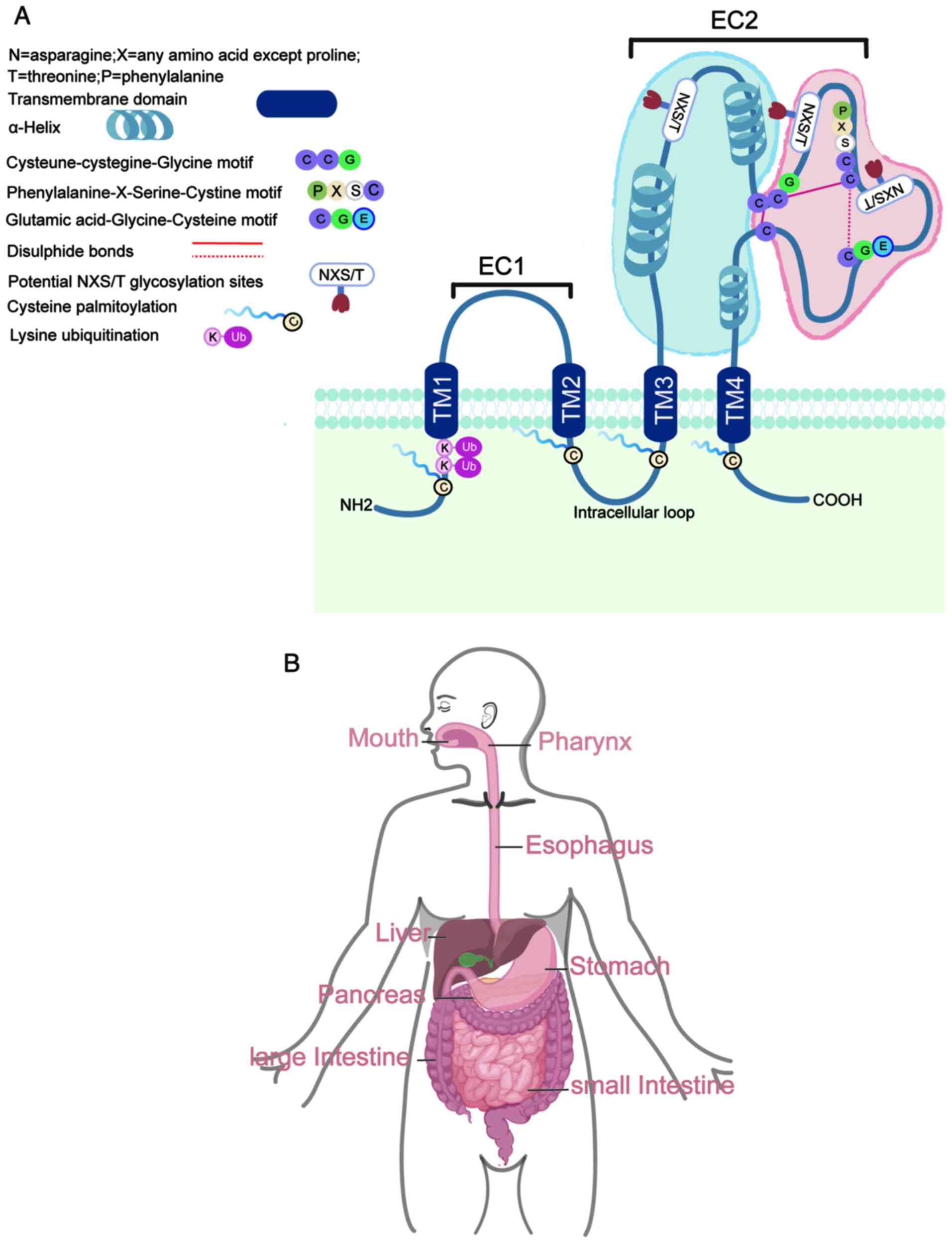
Members of the tetraspanin family possess four transmembrane (TM)
domains, conserved amino acids, a small extracellular loop, a
larger extracellular loop (EC2 loop), a cytoplasmic loop and short
N- and C-terminal cytoplasmic tails. The EC2 loop is divided into a
constant region, with three α-helices, and a variable region, which
houses most tetraspanin protein-protein interaction sites (Fig. 1A). Tetraspanins typically undergo
post-translational palmitoylation at cysteine residues proximal to
the membrane, which is key for their interaction with other TM
proteins, cholesterol and gangliosides (1,4,6,8).
This modification is necessary to create ‘tetraspanin-enriched
microdomains’ (TEMs), which are distinct from lipid rafts and can
be disrupted by Triton X-100 (9).
Furthermore, tetraspanins undergo additional post-translational
modifications at several highly conserved cysteine and lysine
residues, including N-glycosylation (common in the EC2
structural domain) and ubiquitination (in the N-terminal structural
domain) (10). The general
structure of a typical tetraspanin protein is shown in Fig. 1A.
Globally, cancer (excluding nonmelanoma skin
cancer), estimated to cause 250 million (95% UI, 235–264 million)
in 2019, is the second most common cause of mortality, with
digestive system-related cancer being the most widespread form of
malignancy (11). In the context
of digestive system cancer (Fig.
1B), tetraspanins have emerged as significant contributors to
tumorigenesis and disease progression. Therefore, understanding the
expression patterns and functional implications of tetraspanins
within the intricate network of digestive system malignancies is
imperative for developing targeted therapeutic interventions and
diagnostic approaches. Over the last 10 years, studies have linked
tetraspanins to different cancer-related traits, such as tissue
specialization, cell growth, migration, attachment, mobility,
apoptosis, angiogenesis, cellular pluripotency and metastasis
(1,12–18).
For instance, tetraspanins have been implicated in the regulation
of tumor cell metastasis, migration and altered adhesion through
integrin signaling (1,19). Tetraspanins also bind to
peptidases, a disintegrin, ADAM metalloproteinases (particularly
ADAM10) (20), matrix
metalloproteinases (21) and the
urokinase-type plasminogen activator receptor (22) to regulate cell invasiveness.
Although tetraspanins share a highly conserved structure, they can
either promote or inhibit tumor development. In the context of
digestive system tumors, the precise function of tetraspanins
warrants further research and exploration. The present review
comprehensively explored the current understanding of the
expression profiles and diverse functions of tetraspanins in
different types of digestive system-related cancer (Table I)(23–67),
shedding light on their potential as biomarkers and therapeutic
targets for combating these challenging malignancies.
In terms of worldwide prevalence, GC ranks fourth
for men and fifth for women, and >1 million new cases are
attributed to GC annually globally (68). Nevertheless, individuals diagnosed
with late-stage GC generally face a poor outlook, experiencing a
decreased survival rate; the 5-year survival rate is <10%
(69). In recent years, a number
of experiments have been conducted to explore the pathogenesis of
GC, revealing a close association with tetraspanins. These diverse
proteins serve critical roles in various biological functions
within GC cells (70). In this
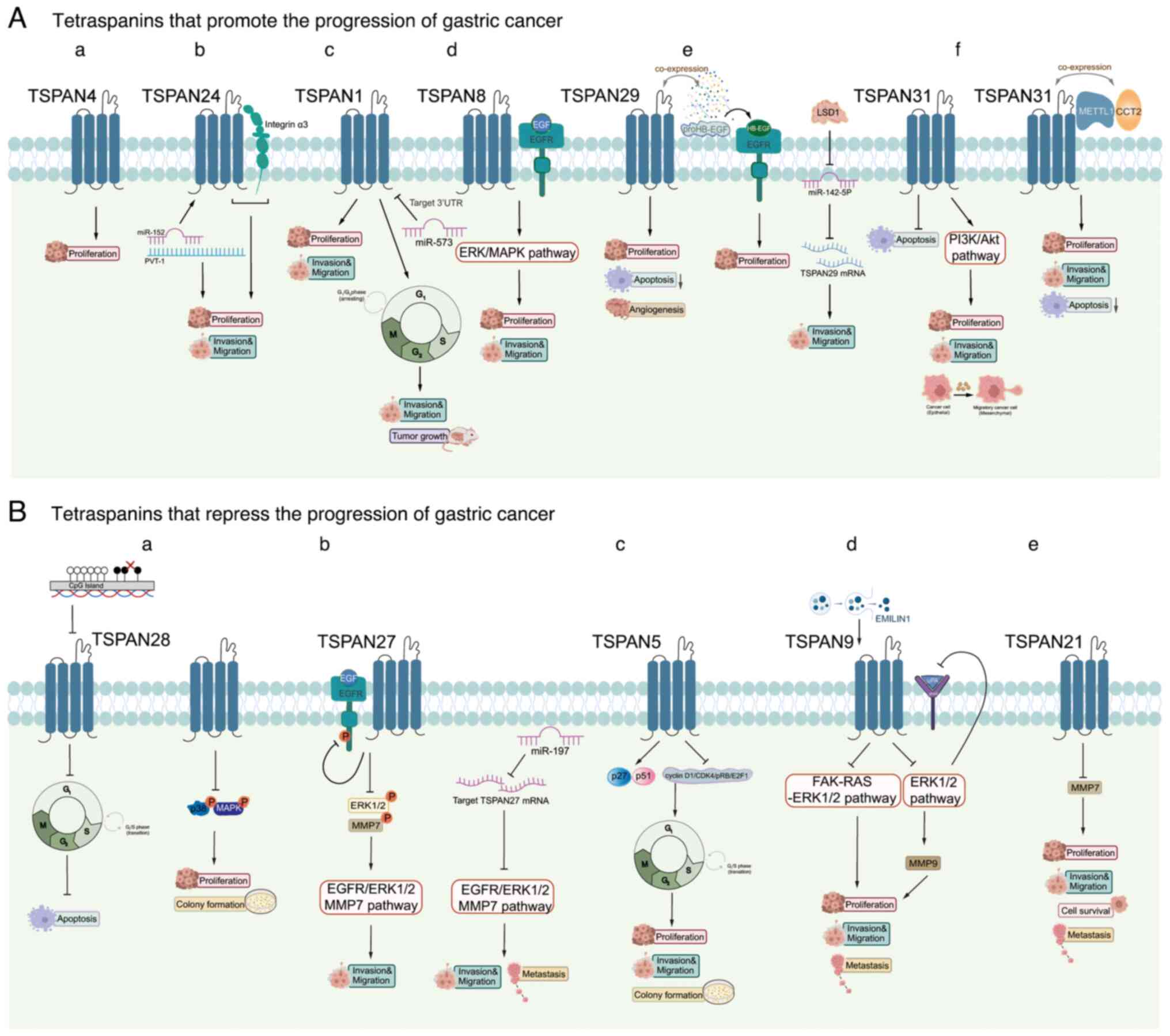
section, the effect of specific tetraspanins on GC cell
proliferation and invasion are summarized. Notably, TSPAN8,
TSPAN24/CD151, TSPAN4, TSPAN1, TSPAN29/CD9 and TSPAN31 enhance
these processes, while TSPAN28/CD81, TSPAN27/CD82, TSPAN5, TSPAN9
and TSPAN21 suppress the growth of GC cells (Fig. 2).
TSPAN8 is associated with numerous types of cancer,
contributing to increased metastasis, proliferation, angiogenesis
and thrombosis (71–75). Upregulated TSPAN8 is also
recognized as a standalone predictor of outcomes for patients with
GC. Knockdown of TSPAN8 has been shown to reduce the invasive
ability of GC cells, but TSPAN8-containing exosomes restores this
ability (76). Research has
demonstrated that TSPAN8 functions as an oncogene in GC,
contributing to the growth and metastasis of GC cells by activating
the EGFR (33) and ERK/MAPK
(34) pathways. In conclusion,
TSPAN8 is involved in promoting the progression of GC.
TSPAN24/CD151 creates TEMs by interacting with
different TM proteins, which control integrin-dependent cell
adhesion, shape and movement. TSPAN24 serves a crucial role as a
binding partner to laminin-binding integrins (77,78).
A study suggests that the complex interaction of TSPAN24 with
integrin α3 in GC cells is correlated with increased invasiveness
(45). Additionally, non-coding
RNA plasmacytoma variant translocation (PVT)-1 enhances TSPAN24
expression by interacting with microRNA (miR)-152, promoting GC and
highlighting PVT-1 as a potential therapeutic target (79).
TSPAN1, also referred to as neuroepithelial
transforming gene 1, functions as a guanine nucleotide exchange
factor (80). TSPAN1 can enhance
tumor growth across various types of cancer, contributing to
metastases (81–84). Lu et al (23) observed a significant upregulation
of TSPAN1 expression in GC tissues, identifying TSPAN1 as a
potential oncogene that facilitates GC cell proliferation and
invasion. Additionally, it was demonstrated that miR-573 hinders
the growth and invasion of GC by directly targeting the
3′-untranslated region (UTR) of TSPAN1. This regulated axis offers
a fresh insight into the mechanisms underlying GC (23).
TSPAN29/CD9 expression is markedly elevated in
primary GC tissues and becomes more prominent in advanced GC
(90). Additionally, TSPAN29
expression is positively correlated with tumor status, including
the extent of infiltration and metastasis of the lymph nodes
(27). There is evidence for the
co-expression of proHB-epidermal growth factor (EGF; the
membrane-anchored form of HB-EGF) and TSPAN29 in GC, and one study
has clearly demonstrated the presence of HB-EGF mRNA and
EGF-receptor mRNA in GC tissues. These findings suggest that the
direct interaction could play a role in GC development and tumor
growth through paracrine, autocrine, and/or juxtacrine mechanisms
(58). Nakamoto et al
(59) successfully suppressed
tumor progression by administering an anti-TSPAN29 antibody to mice
bearing a human GC cell xenograft. Notably, TSPAN29 protein
expression is low in metastatic gastrointestinal stromal tumors
(GISTs), while low TSPAN29 expression in primary GIST is associated
with poorer disease-free survival (DFS). However, TSPAN29
expression in intestinal stromal tumors is not significantly
associated with the DFS of patients, suggesting that its role in
regulating GIST metastasis may be related to organ type (91). Additionally, a study by Zhao et
al (92) reveals that histone
lysine-specific demethylase 1 promotes GC migration by decreasing
miR-142-5p levels and increasing TSPAN29 expression.
TSPAN28/CD81 was first identified as the binding
site for an anti-growth antibody (95). Thus far, TSPAN28 has been shown to
affect cell shape, adhesion and growth and the differentiation of B
cells and T cells (96). In
addition, Yoo et al (56)
suggest that a reduction in TSPAN28 mRNA levels in GC tissues and
GC cell lines is a result of abnormal CpG hypermethylation in its
promoter region rather than genetic mutations. This decrease in
TSPAN8 expression encourages the progression from the G1
to the S phase of the cell cycle and reduces the cellular responses
to different types of stress that can lead to cell death, including
towards 5-fluorouracil (5-FU), etoposide, γ-irradiation,
doxorubicin and hypoxia. In addition, TSPAN28 expression decreases
the ability of GC cells to form colonies and hinders the activation
of p38 MAPK phosphorylation (56).
TSPAN27, also known as CD82 and KAI1, has been
recognized as a gene that suppresses metastasis in various types of
cancer (50,97). Furthermore, it has a strong
connection to the invasion of GC cells. Xu et al (48) reveal that TSPAN27 hinders the
infiltration of GC by reducing the phosphorylated (p-)EGFR,
p-ERK1/2 and MMP7 levels. Meanwhile, the nuclear endonuclease RNase
III enzyme, Drosha, facilitates the production of miR-197, and
elevated miR-197 inhibits TSPAN27, leading to activation of the
EGFR-ERK1/2-MMP7 pathway and ultimately enhancing GC cell invasion
and metastasis (48).
In summary, certain tetraspanins can either promote
or inhibit GC. Hence, investigating the internal molecular pathways
involved in the progression of GC can lead to the discovery of
improved treatment options and provide optimism for patients.
Hepatocellular carcinoma (HCC) is responsible for
almost 80% of liver cancer cases, making it the sixth most
prevalent cancer globally. HCC is known for its high occurrence,
disease burden and mortality rate (101). At present, there is no effective
remedy for advanced HCC (102).
Tetraspanins have been linked to hepatitis B virus (HBV) infection,
hepatitis C virus infection and cirrhosis and may therefore have a
significant impact on the progression of pre-cancerous conditions
resulting in HCC (103). For
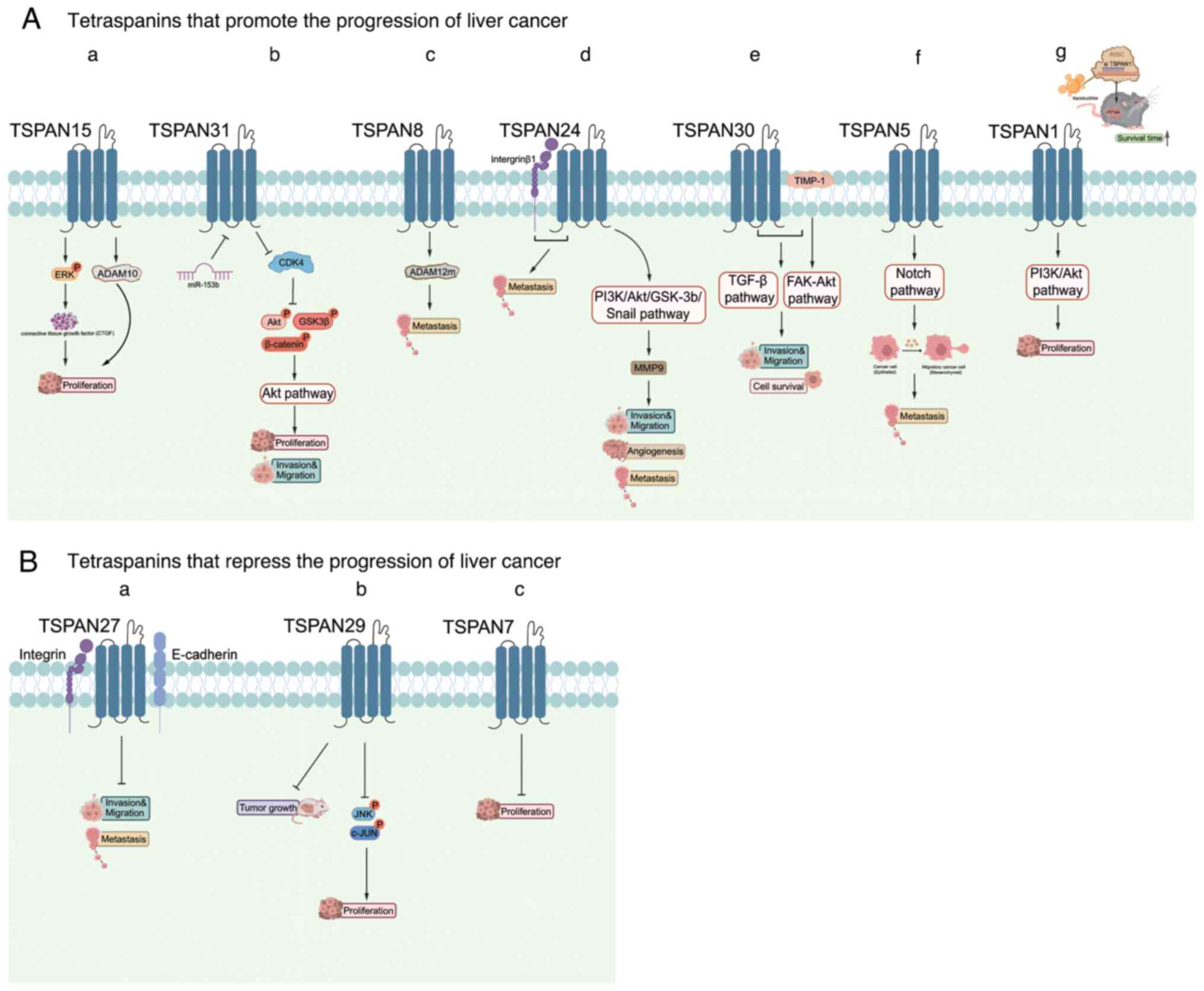
instance, TSPAN1 and TSPAN15 promote the proliferation of HCC,
while TSPAN5, TSPAN8, TSPAN24/CD151, TSPAN28/CD81, TSPAN30/CD63 and
TSPAN31 promote the metastasis of HCC. Conversely, TSPAN7/CD231,
TSPAN27/CD82 and TSPAN29/CD9 inhibit the progression of HCC
(Fig. 3).
Heterogeneous TSPAN15 expression patterns have been
observed in HCC patients with hepatitis C virus or HBV positive
using bioinformatic analysis (42). In HCC (TSPAN15-high tumors),
elevated TSPAN15 levels are positively correlated with cancer cell
stemness and recurrence. TSPAN15 enhances the phosphorylation of
ERK, which controls the expression and secretion of connective
tissue growth factor, ultimately stimulating the proliferation of
HCC cells. This pro-growth effect can also be associated with the
ability of TSPAN15 to increase the release of ADAM10 to a certain
degree, although the exact mechanism of this remains to be
elucidated (42).
TSPAN8, which is correlated with tumor metastasis,
is upregulated in a variety of tumor types (107). Fang et al (108) propose that TSPAN8 can potentially
enhance metastasis in HCC cells through the activation of ADAM12.
In the resting cells, the combination of TSPAN8 and integrin is
positioned at the rear of cancer cells (109). This process could serve a role in
the metastasis of HCC cells and establish a foundation for
determining if TSPAN8 is a viable prognostic indicator for HCC. In
addition, long non-coding RNA-TSPAN8 controls the TSPAN8 expression
level (57), which is also
influenced by astrocyte elevated gene-1 in HCC, a gene that is
upregulated in different types of cancer. Therefore, TSPAN8 has a
crucial role in promoting metastasis (35).
TSPAN24/CD151, the first tetraspanin linked to
metastasis, plays a metastasis-promoting role in a variety of types
of cancer (110,111). TSPAN24 may promote metastasis by
activating integrin β1 (112).
Additionally, TSPAN24 is a distinct pro-angiogenic factor that is
abundantly produced by HCC cells (113). TSPAN24 is able to initiate the
PI3K/Akt/GSK-3β signaling pathway, leading to the upregulation of
MMP9 and facilitating the breakdown of the extracellular matrix and
movement of cancer cells, both of which play a role in angiogenesis
and the invasion of HCC (46).
Therefore, TSPAN24 may be a sensitive biomarker for HCC metastasis.
Furthermore, the TSPAN24 expression level can be regulated by
miR-199a-3p, miR-124 and miR-128. However, the specific crosstalk
between TSPAN24 and its upstream effectors remains complicated
(114–116).
It is considered that TSPAN30/CD63 facilitates the
TGF-β signaling pathway by interacting with tissue inhibitors of
metalloproteinase-1 (TIMP-1), an important participant in the
communication between hepatic stellate cells and HCC cells mediated
by TGF-β. The FAK-Akt signaling pathway is triggered by TIMP-1,
promoting the migration and survival of these cells (65).
TSPAN5 has only been included in HCC research in the
last few years and, as with most family members, serves a
promotional role in HCC. Increased TSPAN5 levels have been
confirmed to enhance the metastasis of cancer, vascular
angiogenesis, clinical stage and overall survival in HCC (29). Additionally, validation of Notch
signaling activation by TSPAN5 in clinical HCC samples leads to
increased epithelial-mesenchymal transition (EMT) and actin
rearrangement, as shown in mechanistic studies (29). Deletion of TSPAN5 is also shown to
mediate the p16INK4a/pRb pathway, inducing oncogene-induced
senescence. Specifically, the inhibition of TSPAN5 results in
decreased actin polymerization, which consequently decreased the
formation of the myocardin-related transcription factor A
(MRTF-A)-filamentosamine A complex. This leads to a decrease in the
expression of MRTF/serum response factor-dependent target genes and
the initiation of senescence both in vitro and in
vivo (117).
It has been reported that TSPAN27/CD82 and other
members of the tetraspanin family can interact with each other,
integrins and E-cadherin (49).
Tetraspanins appear to facilitate cell-cell and cell-matrix
interactions, which could affect the ability of cancer cells to
invade and metastasize (49,50).
Additionally, a reduction in TSPAN27 mRNA levels in HCC cells
appears to affect their metastatic ability (118). HBx, a component of HBV, induces
methylation of the TSPAN27 promoter and thus hinders TSPAN27
expression during transcription. Nevertheless, examination of
clinical specimens did not reveal a significant disparity in
TSPAN27 levels between samples that were positive or negative for
HBV surface antigen (119).
PC is a highly aggressive tumor, with a 5-year
survival rate of only 5%. Furthermore, ~90% of all PC cases are
attributed to pancreatic ductal adenocarcinoma (PDAC) (123), which is known for its early
metastasis and limited responsiveness to radiotherapy. Despite
advances in diagnostic and surgical treatments aimed at prolonging
survival and alleviating symptoms, few approaches have proven
effective so far (124,125). Therefore, the early
identification of specific therapeutic targets in PC is of
paramount importance. This will not only allow for an earlier
diagnosis of PC but will also facilitate more personalized
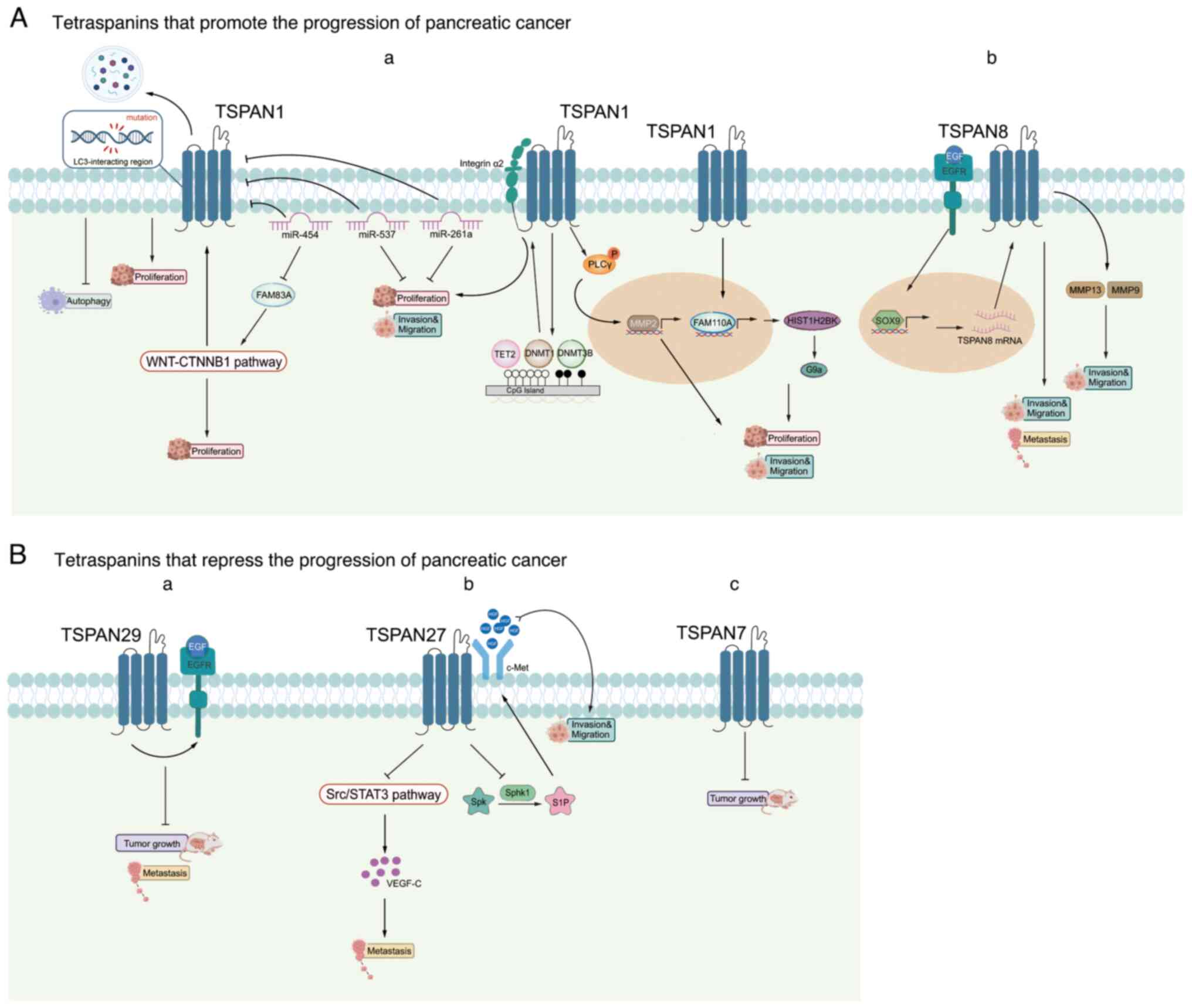
treatments. In this section, the roles of five tetraspanins are
summarized in the context of PC. TSPAN1 boosts the growth, movement
and infiltration of PC cells, while TSPAN8 facilitates PC
metastasis. Conversely, TSPAN7/CD231, TSPAN29/CD9 and TSPAN27/CD82
inhibit the progression of PC (Fig.
4).
A number of studies have established an intimate
connection between TSPAN1 and PC (126–131). Zhou et al (127) found that depletion of TSPAN1 not
only decreases PC cell proliferation in vitro and in
vivo, but its expression is also linked to a poor survival rate
among patients with PC. It was also discovered that TSPAN1
functions as an enhancer of macroautophagy/autophagy, with
mutations in the LC3-interacting region pattern of TSPAN1 leading
to the inability to trigger autophagy and support the growth of PC
cells. Additionally, it shows that family with sequence similarity
83 member A (FAM83A) increases the expression of TSPAN1 via the
Wnt-β-catenin signaling pathway and that miR-454 directly targets
both TSPAN1 and FAM83A. These results indicate that elements of the
miR454-FAM83A-TSPAN1 pathway can serve as important indicators for
predicting outcomes or as targets for treatment in PC (127). In addition to miR-454, Wang et
al (129) also found that
miR-573 is downregulated and TSPAN1 is upregulated in PC tissues
and cell lines. In addition, miR-573 suppresses the growth,
movement and infiltration of PC cells through the regulation of
TSPAN1. miR-216a is also found to be a controlling factor of
TSPAN1, interacting with the 3′-UTR of TSPAN1 and affecting the
levels of ten-eleven-translocation-2, methyltransferase 3B and DNA
methyltransferase 1 to influence the expression of integrin α2
through epigenetic mechanisms. Silencing integrin α2 prevents the
metastasis and growth of PC cells induced by elevated levels of
TSPAN1. In summary, the newly discovered miR-216a-TSPAN1-integrin
α2 pathway plays a role in controlling the advancement of PC and
offers a fresh approach for the potential treatment of PC (131). Prior to this, Zhang et al
(132) demonstrated that TSPAN1
siRNAs inhibits PC cell migration and infiltration and MMP2 mRNA
expression by preventing the translocation and phosphorylation of
phospholipase Cγ, which is crucial in human PC cell migration and
invasion.
A previous study demonstrates that TSPAN1 positively
transcriptionally regulates FAM110A, an oncogene that promotes
tumorigenesis in PC cells (25).
H2B clustered histone 12 (HIST1H2BK) is also recognized as a target
of FAM110A, with its tumor-promoting ability potentially related to
its regulation of G9a. The newly identified
TSPAN1-FAM110A-HIST1H2BK-G9a pathway serves a role in controlling
the advancement of PC and offers a fresh approach for predicting
and treating PC outcomes (25).
Mayado et al (133) found
that linking antibodies to mucin16, TSPAN1 and epithelial cell
adhesion molecule (EpCAM) can enhance the capturing efficiency of
circulating tumor cells in the bloodstream of patients with PDAC,
suggesting additional potential therapeutic implications of this
tetraspanin.
Prior research has established that TSPAN8 is a key
player in the advancement of PC (134,135). TSPAN8 may enhance the movement of
cancer cells via the recruitment and stimulation of the matrix
metalloproteinases, MMP9 and MMP13 (135). Tetraspanins are also considered
to be involved in exosome targeting. Tumor exosomes exhibiting
TSPAN8 activity facilitate stromal degradation, reprogramme stromal
and hematopoietic cells and induce a motile phenotype in a
non-metastatic rat PC lineage harboring a TSPAN8 knockdown
(136). A study revealed that in
PDAC, increased TSPAN8 promoted metastasis both in vivo and
in vitro (36). SRY-box
transcription factor 9 (SOX9) enhances the production of TSPAN8 in
response to EGF, which is associated with phenotypic changes
including decreased adhesion to the extracellular matrix and
heightened invasion capabilities. This research emphasizes the
connection between the EGF-SOX9-TSPAN8 signaling pathway and the
tumor stage, negative prognosis and decreased survival rate in
patients with PDAC (36).
In the context of PC treatment, TSPAN8 serves as a
potential target. One of the contributing factors to primary tumor
growth and metastasis is the presence of cancer stem cells. These
cells are characterized by a long life, self-renewal,
differentiation, slow cell cycle progression and resistance to
radiation and drugs (137,138).
As TSPAN8 is an indicator of PC stem cells (139), it is plausible that it may
facilitate cancer cell motility, penetration and homing through
various mechanisms such as signaling, gene regulation and
modulation of the tumor microenvironment (TME).
TSPAN7/CD231 is found to be downregulated in the
majority of tumors. An analysis that combined a Gene Expression
Omnibus dataset and the Human Cancer Metastasis Database identified
differentially expressed metastasis-related genes, among which
TSPAN7 was included. The results predict that TSPAN7 may be
downregulated in PDAC, identifying it as a tumor suppressor
(140). Using bioinformatics
methods involving differentially expressed genes, weighted gene
co-expression network analysis and miRNA target genes linked to
overall survival in patients with PDAC, Luo et al (141) also conclude that TSPAN7 could
serve as a PDAC diagnostic marker.
A notable inverse relationship is noted between the
expression of TSPAN29 and the metastatic capabilities and survival
rates of various types of tumors. In addition, TSPAN29 expression
is shown to be reduced in PC cells compared with pancreatic tissues
(142). Mechanistically, Tang
et al (61) found that
TSPAN29 inhibits the proliferation, migration and invasion of PC
cells. Furthermore, TSPAN29 may suppress tumor growth and
metastasis by moderating the endocytosis of EGFR.
CRC is a common type of cancer globally, ranked as
the second highest contributor to cancer-related mortality and
responsible for ~10% of all cancer fatalities (147,148). At present, the main treatment
modality for CRC is surgical resection, supplemented by
radiotherapy and chemotherapy, based on the stage and severity of
the cancer (149). Recent data
has indicated that the 5-year survival rate of CRC has risen by 15%
in recent years, largely due to improvements in early detection,
surgical methods and innovative treatments (149). In addition, the discovery of
tetraspanins and the numerous studies on their roles and mechanisms
in CRC have provided new directions for the treatment of this
disease. In this section, several tetraspanins associated with CRC
are summarized, including TSPAN1, TSPAN8, TSPAN9, TSPAN5 and
TSPAN12 that promote CRC progression, and TSPAN6, TSPAN29/CD9 and
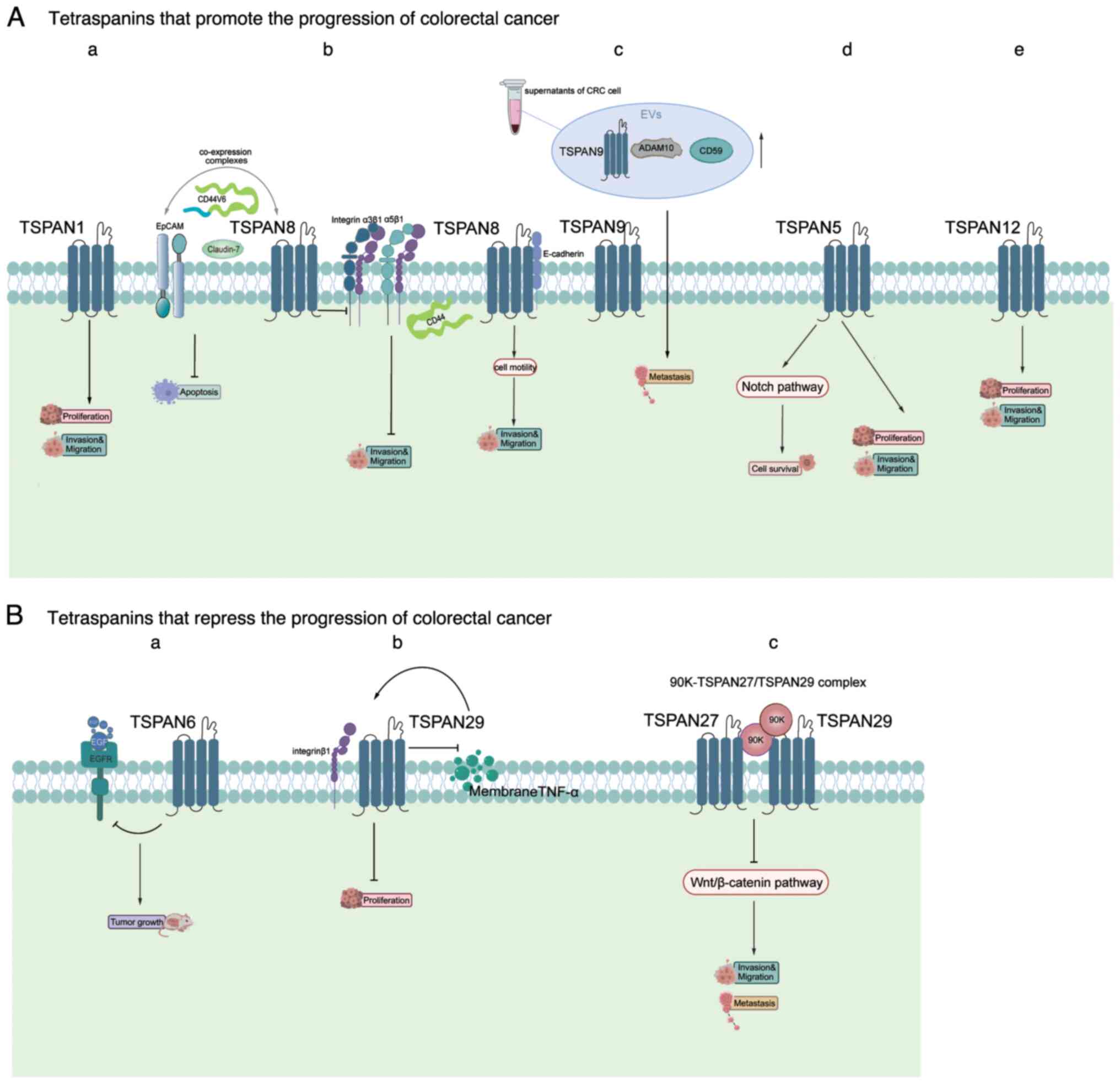
TSPAN27/CD82 that inhibit CRC progression (Fig. 5).
The role of TSPAN1 in CRC aligns with its function
in other tumors as a promoter. Previous research has demonstrated
that reducing TSPAN1 expression significantly suppresses the growth
and infiltration of CRC cells (26). Additionally, a study using small
extracellular vehicles (EVs) from CRC cell lines found that TSPAN1
is upregulated in EVs from CRC plasma compared with healthy
controls. Previous research has also demonstrated that reducing
TSPAN1 expression significantly suppresses the growth and
metastasis of CRC (150). Guo
et al (151) predicted and
scored the infiltration characteristics, mutations and prognostic
ability of CRC into immune cell infiltration (ICI) subtypes and ICI
scores using a series of bioinformatics methods. In this study,
TSPAN1 is identified as being closely associated with CRC.
Tetraspanins are important for organizing
multimolecular complexes in the plasma membrane. TSPAN8, found
exclusively in epithelial cells, is associated with cell-cell
junction proteins such as claudins and E-cadherin (152). It has been reported that TSPAN8,
EpCAM, claudin-7 and CD44v6 are co-expressed and can form complexes
in CRC liver metastases, as well as anaplastic colon and liver
tissues. Although the co-expression of these proteins is not
associated with tumor stage and grade in CRC, it is associated with
DFS and a higher degree of anti-apoptotic properties (153). Cancer-initiating cells (CICs),
which drive tumor progression, express markers that contribute to
exon binding and uptake, promoting signaling cascade activation,
transcription initiation and translational control (154). TSPAN8, EpCAM, claudin-7 and
CD44v6 are among these markers and are highly expressed in CICs of
the colon. These markers, along with other CIC markers, promote the
development of CRC (155).
Although co-expression in itself cannot be considered a prognostic
marker for CRC, it does provide valuable insights. Guo et al
(37) discovered that TSPAN8 acts
as a controller of adhesion between cells and between cells and the
extracellular matrix. Silencing TSPAN8 in CRC cells leads to
increased laminin integrin α3β1 and fibronectin integrin α5β1 on
the cell surface, and reduces CD44 levels. This alteration in
cell-matrix adhesion significantly reduces cell migration. A
further study demonstrating a direct interaction between E-cadherin
and TSPAN8 reveals that TSPAN8 functions as a regulator of cancer
cell motility. Therefore, targeting TSPAN8 may hinder tumor
progression (156).
A study of the ONCOMINE database demonstrated that
TSPAN9 expression is reduced in patients with CRC (32). A research study that used targeted
mass spectrometry to analyze EVs obtained from the supernatants of
four CRC cell lines and plasma EVs from patients with CRC observed
increased levels of TSPAN9, ADAM10 and CD59. Furthermore, elevated
levels of TSPAN9 in EVs is significantly correlated with lymph node
metastasis, distant metastasis upon diagnosis and advanced TNM
stage. This suggests that TSPAN9 could serve as a novel biomarker
for the detection of early CRC (40).
Examination of the ONCOMINE database reveals that
TSPAN5 is highly expressed in CRC (32), a result supported by the research
conducted by Roh et al (30). This study found that inhibition of
TSPAN5 expression suppresses the proliferation, migration and
invasion of CRC cells. It was also discovered that TSPAN5 may
promote CRC development by activating the Notch signaling pathway.
This discovery of the promotional effect of TSPAN5 provides new
insights for the treatment of CRC.
Relatively few studies have investigated the role
of TSPAN6 in tumors. However, a study showed that the low survival
rate of patients with CRC is due, at least in part, to the frequent
reduction or loss of TSPAN6 expression, identifying TSPAN6 as a
controller of CRC progression and a predictive biomarker for
EGFR-targeted treatment in CRC, in addition to RAS pathway
mutations (31). Additionally,
deletion of TSPAN6 results in an increased incidence of adenomas
and larger tumor sizes. This outcome is activated by an
EGF-dependent signaling pathway. Furthermore, TSPAN6 expression is
associated with an improved response to EGFR-targeted therapies in
patients with CRC, including cetuximab (31). However, there is little difference
in the TSPAN6 levels in plasma from patients with CRC and plasma
from healthy controls (158).
TSPAN27/CD82 is recognized as a biomarker for
predicting metastatic potential due to its unique role as a
well-defined suppressor of metastasis in various tumors that does
not affect the growth of the original tumor (145). There is a wealth of research
regarding TSPAN27 in CRC. In a study by Wu et al (161), the findings indicate that TSPAN27
gene expression is higher in CRC tissues than in normal colon
epithelial tissues and lymphatic metastatic tissues. Notably,
elevated expression of TSPAN27 is observed in early CRC, which
decreases in the advanced stages and may be lost in metastasis.
Certain research studies have demonstrated that TSPAN27 hinders the
migration and penetration of CRC cells (52,53).
Lee et al (162) found
that a 90K-TSPAN27/TSPAN29 complex, which inhibits Wnt/β-catenin
signaling in CRC, provides a useful treatment for inhibiting the
progression of CRC. Downregulation of this complex may be a sign of
poor prognosis in CRC.
Compared with gastric, liver, pancreatic and
colorectal cancer, the role of tetraspanins in esophageal and oral
cancer has been less extensively studied. In this section, the
latest findings on tetraspanins in these types of cancer are
briefly summarized.
Globally, esophageal cancer ranks as the eighth
most common type of cancer, contributing to 3% of all cancer
diagnoses (163). The prognosis
of esophageal cancer is poor, with one in every 18 cancer
mortalities in 2020 (148).
Furthermore, the occurrence of esophageal cancer is strongly
associated with dietary and lifestyle factors (164). Several tetraspanins have been
implicated in esophageal cancer pathogenesis. For instance, TSPAN1,
TSPAN8 and TSPAN24/CD151 are upregulated in esophageal
adenocarcinoma, whereas TSPAN29/CD9 is downregulated (165). Additionally, TSPAN4 expression is
elevated in esophageal squamous cell carcinoma (ESCC), where it
potentially enhances resistance to chemotherapy and suppresses
apoptosis (166). Upregulation of
TSPAN8 is associated with ESCC metastasis and drives invasion and
migration in vitro (38).
Upregulation of TSPAN15 in ESCC promotes cellular migration,
infiltration and metastasis (43).
TSPAN24/CD151 expression is correlated with proliferation,
invasiveness and the survival of patients with esophageal cancer
(47,167). Conversely, TSPAN27 expression has
an inverse relationship with lymph node metastasis, distant
metastasis and advanced stage in ESCC, and hinders the
proliferation and migration of ESCC cells through the TGF-β1/Smad
pathway (54,55).
With oral cancer, most cases comprise oral squamous
cell carcinoma (OSCC), with 75% of cases linked to lifestyle
factors (168–170). Increased expression of TSPAN15 in
OSCC can promote the advancement and metastasis of tumors by
activating ADAM10 (171).
Additionally, while TSPAN25/CD53 and TSPAN29/CD9 are expressed in
OSCC, TSPAN26/CD37, TSPAN28/CD81 and TSPAN30/CD63 are not (63,172). Furthermore, TSPAN29/CD9
expression is strongly associated with OSCC metastasis (63,64).
As with esophageal cancer, TSPAN27/CD82 may have a role in oral
cancer, but it may not be a reliable indicator of prognosis in OSCC
due to the non-significant relationship between patient survival
and expression of TSPAN27/CD82 by tumors; further studies still
need in future to make this clear. (63,173,174).
In summary, several tetraspanins appear to act as
oncogenes promoting proliferation, metastasis and chemotherapy
resistance in esophageal and oral cancer, while TSPAN27/CD82 may
act as a tumor suppressor gene. However, further research is
warranted to clarify the pathogenic and prognostic utility of
tetraspanins in these malignancies.
It is essential to understand the biological and
molecular processes involved in cancer advancement, specifically in
metastasis, to develop improved treatment approaches. Tetraspanins
are involved in numerous physiological and pathological processes,
and both experimental and clinical studies in the field of cancer
have reported a relationship between tetraspanin expression levels
and metastasis. Each tetraspanin specifically binds to one or
several other membrane proteins to form primary complexes,
underscoring the role of tetraspanins as organizers of
multimolecular complexes. TEMs, which are rich in tetraspanins,
create a signaling platform that serves a role in a number of
important cellular functions and cancer-related activities.
Therefore, tetraspanins play a significant role in multiple stages
of tumor formation and metastasis (175). As such, targeting tetraspanins
for cancer treatment holds promising therapeutic potential. In this
section, the potential clinical significance of tetraspanins in
digestive system tumors is summarized (Table II).
Potential molecules used for targeted cancer
therapies may include miRNA fragments (176,177), drugs (small molecule inhibitors),
siRNA fragments, toxins, soluble proteins and monoclonal antibodies
(mAb)(178). It is noteworthy
that several antibodies targeting TSPAN8 have been indicated as
potential candidates for the treatment of CRC and ovarian cancer.
Notably, Kim et al (179)
identify the extracellular large loop of TSPAN8 (TSPAN8-LEL) as a
key domain for its antitumor effect. A new antibody directed at
TSPAN8-LEL was created through the use of phage display technology,
potentially serving as an effective means to block the invasion of
metastatic CRC. Additionally, antibodies targeting TSPAN8-LEL
hinder the metastasis of ovarian cancer cells in vivo and
ex vivo (180). TSPAN8
stands out as a potential target for therapy due to its reliable
involvement and increased expression in HCC. Furthermore, according
to prior research that demonstrated an interaction between
TSPAN24/CD151 and integrin β1, Ke et al (181) produced a monoclonal antibody that
identifies the integrin β1 binding region of TSPAN24/CD151. This
monoclonal antibody (mAb) has broad potential for detecting CD151
antigen expression and localization in HCCs and could serve as an
effective strategy to inhibit HCC progression.
Several antibodies targeting TSPAN26/CD37 have
achieved significant results in treating malignant tumors. For
instance, in 2011, it was announced that TRU-016 (otlertuzumab), a
humanized anti-TSPAN26 fusion protein developed from SMIP-16 for
B-cell malignancies treatment, had been approved by the U.S. Food
and Drug Administration for the treatment of patients with chronic
lymphocytic leukemia (182).
Given that TSPAN26/CD37 expression is elevated in tumors of the
gastrointestinal system, it is considered to be a valuable
therapeutic target in these malignancies.
Inhibition of TSPAN1 expression through RNA
interference (RNAi)-mediated or miRNA-targeted methods has been
shown to suppress the growth and invasion of colon cancer and PC
cells (26,129), suggesting a potential therapeutic
strategy for these diseases. At present, approaches involving siRNA
and antibodies have been utilized to target TSPAN29/CD9 in tumors
(183,184). The connection between TSPAN29 and
the motility and metastasis of digestive system-related cancer is
significant, as it mainly inhibits the growth and tumorigenicity of
CRC (62). As such, the prospect
of utilizing TSPAN29 as a target to treat human cancer appears
promising.
Furthermore, the importance of tumor immunotherapy
in the treatment of cancer has grown significantly in the past few
years. Immune cells penetrate the TME to control the advancement of
cancer. In tumor immunology, the main focus is on studying T cells
and antigen presentation is the most crucial step for T cell
surveillance of cancer cells. Major histocompatibility complex
(MHC) II proteins stimulate CD4+ T lymphocytes, whereas
MHC I proteins stimulate CD8+ T lymphocytes. Monocytes,
B cells and bone marrow-derived dendritic cells (BMDCs), which are
specialized antigen-presenting cells, activate T cells that are
specific to antigens by binding to MHC on T cell receptors (TCRs)
(185). It has been noted that
certain tetraspanins are key players in antigen presentation during
immune responses, thereby influencing the TME. For instance,
TSPAN29/CD9 is essential for facilitating the association of
heterologous MHC II to enhance TCR stimulation by DCs (186). Additionally, knockout of
TSPAN29/CD9 in mice has been shown to increase TNF-α levels and
macrophage infiltration in lung cancer following lipopolysaccharide
treatment (187). TSPAN29/CD9 can
also induce the retention of MHC II by enhancing MHC II trafficking
and decreasing MHC II recycling, expecting to enhance T cell
receptor stimulation by DCs in tumor immunotherapy (188). TSPAN27/CD82 expression is
enhanced following the activation of monocyte-derived DCs and
BMDCs, promoting stable interactions between MHC II and T cells,
making TSPAN27/CD82 a modulator to enhance Ag presentation
machinery in tumor immunotherapy (189). Knocking out TSPAN26/CD37 also
inhibits DC migration, potentially affecting immunity (190). Furthermore, TSPAN25/CD53 antibody
ligation enhances natural killer (NK) cell adhesion and reduces NK
cell degranulation in response to tumor target cells (191).
However, certain tetraspanins, such as TSPAN30/CD63
and TSPAN1/CD151, inhibit the activation of T cells. Specifically,
knockdown of TSPAN30/CD6 in B lymphocytes activates CD4+
T cells (192) and DCs lacking
TSPAN1/CD15 stimulate T cells by co-stimulation (TCR activation and
Ag presentation) (193). A recent
study indicated that the clustering of MHC I by TSPAN5 is essential
for effective CD8+ T cell stimulation, potentially
affecting TCR and CD8 clustering (194). Another study highlights TSPAN8 as
a potential candidate for chimeric antigen receptor T (CAR-T) cells
in combating PC. While TSPAN8-specific CAR-T cells notably decrease
tumor formation in subcutaneous xenotransplantation models, the
potential mechanisms still require further exploration (195). Despite a lack of direct evidence
regarding their function in tumor immunology, particularly in
digestive system-related cancer, immunotherapy based on
tetraspanins still represents a promising strategy.
Although the tetraspanin family appears to have a
notable effect on the treatment of cancer of the digestive system,
there are still some obstacles to overcome in terms of
chemoresistance and expression heterogeneity. First, tetraspanins,
such as TSPAN29/CD9, TSPAN8, TSPAN4, TSPAN30/CD63, TSPAN12,
TSPAN27/CD82, TSPAN28/CD81 and TSPAN1, are involved in cancer
chemoresistance, which could be a challenge in cancer of the
digestive system therapy. mAbs directed at TSPAN29/CD9 induce
apoptosis in small cell lung cancer (SCLC) cells that are resistant
to cisplatin (CDDP) and etoposide (196). Furthermore, TSPAN29 plays a
crucial role in resistance to doxorubicin and 5-FU in breast cancer
(197). However, TSPAN8
inhibition can suppress drug resistance in glioma and breast cancer
(198,199). Additionally, ribophorin II
knockdown reduces TSPAN30/CD63 glycosylation, leading to decreased
chemoresistance and aggressiveness in breast cancer (200). TSPAN12 enhances chemoresistance
in small cell lung cancer cells through miR-495 regulation
(201). Enhanced expression of
TSPAN27/CD82 leads to increased resistance to chemotherapy in acute
myeloid leukemia through the activation of p38 MAPK via the PKCα
and β1 integrin signaling pathways (202). Reduced TSPAN28/CD81 levels make
acute lymphoblastic leukemia cells more responsive to chemotherapy
and interferes with Bruton tyrosine kinase phosphorylation
(203). In vitro,
inhibition of TSPAN1 in head and neck squamous cell carcinoma cells
resistant to cisplatin results in the suppression of resistance to
chemotherapy, EMT, autophagy and proliferation, as well as the
induction of apoptosis (84).
Prior research in the field of gastrointestinal cancer has shown
that TSPAN8 increases drug resistance and decreases cell mortality
in stomach cancer cells by triggering the Wnt/β-catenin pathway
through Notch2 interaction (204). High levels of TSPAN4 are observed
in ESCC, leading to resistance to chemotherapy and suppression of
apoptosis (166).
Overall, while targeting tetraspanins represents a
promising strategy in cancer therapy, chemoresistance should be
considered carefully during future research directions. When
considering a specific type of cancer, it is important to examine
the expression pattern of certain tetraspanins, as they may be
related to chemoresistance. In such cases, combination therapy may
be necessary to enhance the effectiveness of the treatment.
Tetraspanins exhibit extensive expression
variations across various tissues and even expression heterogeneity
between patients. For instance, the variable expression of TSPAN15
in HCC highlights the importance of evaluating TSPAN15 expression
in these patients prior to treatment initiation (68). This variety could potentially
affect the precision of targeted therapies. Therefore, researchers
should gain a deeper understanding of the expression patterns of
tetraspanins in diverse tissues, develop tissue-specific drug
delivery systems (205) or focus
on the functional characteristics of tetraspanins, which are often
exerted through interactions with other proteins, to design more
precise and specific drugs and minimize the effects on non-target
tissues. A potential strategy to address this challenge is the
development of more precise treatment methods, such as personalized
therapies, based on individual suitability. By designing more
specific RNAi sequences or small molecule inhibitors, it may become
possible to selectively modulate the expression and functionality
of particular tetraspanins, minimizing interference with normal
cells and enhancing treatment specificity.
Although the proteins of the tetraspanin family
have similar structures, the significant functional differences are
observed in different digestive system-related types of cancer.
Since stetraspanins can interact with numerous proteins, including
those on the cell membrane, identifying proteins that interact with
tetraspanins may help explain the mechanisms behind
tetraspanin-induced resistance, expression differences and
individual variability. Additionally, increasing evidence has
confirmed that tetraspanin family proteins are involved in the
formation of a new type of organelle termed migrasomes (206). Migrasomes are newly discovered
organelles formed by migrating cells, first identified in 2015
(85), which have roles in cell
communication, disposal of damaged organelles, the lateral transfer
of mRNA and proteins and mitochondrial quality control (88,207,208) and have been reported to be
associated with certain diseases such as stoke and podocyte injury
(86,209). However, there is still limited
research on the relationship between migrasomes and cancer, despite
the presence of migrasomes in cancer cells (210). A previous report linked the
development and prognosis of glioma to a key migrasome regulator
TSPAN4, though the specific mechanisms remain unclear (211). Another study reported the role of
TSPAN1/CD151 in migrasome formation in the invasiveness and
angiogenesis of HCC (212). A
pan-cancer analysis identified migrasome-related genes as a
potential immunotherapeutic target among 22 tumors, including CRC
(213). There have been a number
of studies regarding tetraspanins, exosomes and types of cancer;
therefore, it is worth investigating the impact of tetraspanins on
cancer through migrasomes. Similarly, discovering the interactions
between tetraspanins and other proteins or organelles may reveal
new roles in cancer, potentially providing new strategies for
treating digestive system-related and other types of cancer.
In conclusion, while the tetraspanin family holds
notable potential in cancer therapy, overcoming challenges related
to their widespread expression and achieving specificity in
treatment remains critical. Future research efforts should focus on
delving into the functionalities and interrelationships with
proteins and organelles, chemoresistance, expression patterns and
the structure of tetraspanin family members across different
tissues and patients. Simultaneously, advances in personalized
therapies and innovative treatment technologies, such as
immunotherapy, are essential for providing more effective treatment
options for patients with cancer.
Cancer presents a significant public health
challenge due to treatment resistance and fatal metastases.
Understanding the molecular mechanisms of cancer progression is
crucial for developing effective therapies. Tetraspanins, integral
membrane proteins, have a key role in cancer metastasis,
particularly in digestive system-related types of cancer. Research
on tetraspanins is therefore vital for cancer treatment and
molecular biology advancement, as they regulate cancer initiation,
progression and metastasis. Additionally, specific tetraspanins
have both tumor-promoting and tumor-suppressive roles by
influencing oncogenic pathways, cell motility and the TME.
Identifying tetraspanin expression patterns as prognostic
biomarkers in different digestive system-related types of cancer is
a future focus for research. Furthermore, targeting dysregulated
tetraspanins could halt tumor growth and progression. Advanced
techniques such as super-resolution microscopy and single-cell
sequencing may aid in understanding the complex roles of
tetraspanins. Despite challenges in targeting tetraspanins, further
research is needed before they are used as therapeutic targets and
biomarkers for metastatic cancer. In conclusion, tetraspanins are
crucial regulators of metastatic competence in different digestive
system-related types of cancer, emphasizing the need for innovative
diagnostic and therapeutic approaches based on their molecular
functions to enhance clinical outcomes.
Not applicable.
The present study was supported by the National Natural Science
Foundation of China (grant no. 82030007); National Natural Science
Foundation of China (grant no. U23A20398); The Central Government
Guides Local Science and Technology Development Project (grant no.
2022ZYD0057); Natural Science Foundation of Sichuan Province (grant
no. 2024NSFSC0580); the Applied Basic Research Program of Sichuan
Province (grant nos. 2023NSFSC1840 and 2021YJ0200); the Sichuan
Science and Technology Program (grant nos. 2022YFS0630,
2022YFS0627, 2022YFS0578 and 2022YFS0614); the Science and
Technology Project of Luzhou Government (grant nos. 2022YFS0630-B1,
2022YFS0627-A1 and 2022YFS0614-A1); and the Southwest Medical
University (grant no. 2021ZKMS005).
Not applicable.
KC wrote the original draft and edited the
manuscript. QL was responsible for conceptualization, writing the
original draft and reviewing and editing the manuscript. YL wrote
the original draft and edited the manuscript. DJ was responsible
for writing the original draft and reviewing and editing the
manuscript LC and JJ reviewed and supervised the manuscript. SL
wrote the original draft reviewed and supervised the manuscript. CZ
was responsible for conceptualization, writing the original draft
and reviewing and editing the manuscript. Data authentication is
not applicable. All authors read and approved the final
manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Hemler ME: Tetraspanin proteins mediate
cellular penetration, invasion, and fusion events and define a
novel type of membrane microdomain. Annu Rev Cell Dev Biol.
19:397–422. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Min G, Wang H, Sun TT and Kong XP:
Structural basis for tetraspanin functions as revealed by the
cryo-EM structure of uroplakin complexes at 6-A resolution. J Cell
Biol. 173:975–983. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kitadokoro K, Bordo D, Galli G, Petracca
R, Falugi F, Abrignani S, Grandi G and Bolognesi M: CD81
extracellular domain 3D structure: Insight into the tetraspanin
superfamily structural motifs. EMBO J. 20:12–18. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Maecker HT, Todd SC and Levy S: The
tetraspanin superfamily: molecular facilitators. FASEB J.
11:428–442. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Boucheix C and Rubinstein E: Tetraspanins.
Cell Mol Life Sci. 58:1189–1205. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Boucheix C, Duc GH, Jasmin C and
Rubinstein E: Tetraspanins and malignancy. Expert Rev Mol Med.
2001:1–17. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tarrant JM, Robb L, van Spriel AB and
Wright MD: Tetraspanins: Molecular organisers of the leukocyte
surface. Trends Immunol. 24:610–617. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Berditchevski F: Complexes of tetraspanins
with integrins: More than meets the eye. J Cell Sci. 114:4143–4151.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Claas C, Stipp CS and Hemler ME:
Evaluation of prototype transmembrane 4 superfamily protein
complexes and their relation to lipid rafts. J Biol Chem.
276:7974–7984. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Garcia-Mayea Y, Mir C, Carballo L,
Sánchez-García A, Bataller M and LLeonart ME: TSPAN1, a novel
tetraspanin member highly involved in carcinogenesis and
chemoresistance. Biochim Biophys Acta Rev Cancer. 1877:1886742022.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Global Burden of Disease 2019 Cancer
Collaboration, . Kocarnik JM, Compton K, Dean FE, Fu W, Gaw BL,
Harvey JD, Henrikson HJ, Lu D, Pennini A, et al: Cancer incidence,
mortality, years of life lost, years lived with disability, and
disability-adjusted life years for 29 cancer groups from 2010 to
2019: A systematic analysis for the global burden of disease study
2019. JAMA Oncol. 8:420–444. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Berditchevski F, Gilbert E, Griffiths MR,
Fitter S, Ashman L and Jenner SJ: Analysis of the CD151-alpha3beta1
integrin and CD151-tetraspanin interactions by mutagenesis. J Biol
Chem. 276:41165–41174. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gesierich S, Paret C, Hildebrand D, Weitz
J, Zgraggen K, Schmitz-Winnenthal FH, Horejsi V, Yoshie O, Herlyn
D, Ashman LK and Zöller M: Colocalization of the, tetraspanins
CO-029 and CD151, with integrins in human pancreatic
adenocarcinoma: Impact on cell motility. Clin Cancer Res.
11:2840–2852. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lazo PA: Functional implications of
tetraspanin proteins in cancer biology. Cancer Sci. 98:1666–1677.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li J, Xu J, Li L, Ianni A, Kumari P, Liu
S, Sun P, Braun T, Tan X, Xiang R and Yue S: MGAT3-mediated
glycosylation of tetraspanin CD82 at asparagine 157 suppresses
ovarian cancer metastasis by inhibiting the integrin signaling
pathway. Theranostics. 10:6467–6482. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tiwari-Woodruff SK, Buznikov AG, Vu TQ,
Micevych PE, Chen K, Kornblum HI and Bronstein JM: OSP/claudin-11
forms a complex with a novel member of the tetraspanin super family
and beta1 integrin and regulates proliferation and migration of
oligodendrocytes. J Cell Biol. 153:295–305. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Otsubo C, Otomo R, Miyazaki M,
Matsushima-Hibiya Y, Kohno T, Iwakawa R, Takeshita F, Okayama H,
Ichikawa H, Saya H, et al: TSPAN2 is involved in cell invasion and
motility during lung cancer progression. Cell Rep. 7:527–538. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tardif MR and Tremblay MJ: Tetraspanin
CD81 provides a costimulatory signal resulting in increased human
immunodeficiency virus type 1 gene expression in primary CD4+ T
lymphocytes through NF-kappaB, NFAT, and AP-1 transduction
pathways. J Virol. 79:4316–4328. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Levy S and Shoham T: The tetraspanin web
modulates immune-signalling complexes. Nat Rev Immunol. 5:136–148.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Arduise C, Abache T, Li L, Billard M,
Chabanon A, Ludwig A, Mauduit P, Boucheix C, Rubinstein E and Le
Naour F: Tetraspanins regulate ADAM10-mediated cleavage of
TNF-alpha and epidermal growth factor. J Immunol. 181:7002–7013.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yañez-Mó M, Barreiro O, Gonzalo P, Batista
A, Megías D, Genís L, Sachs N, Sala-Valdés M, Alonso MA, Montoya
MC, et al: MT1-MMP collagenolytic activity is regulated through
association with tetraspanin CD151 in primary endothelial cells.
Blood. 112:3217–3226. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bass R, Werner F, Odintsova E, Sugiura T,
Berditchevski F and Ellis V: Regulation of urokinase receptor
proteolytic function by the tetraspanin CD82. J Biol Chem.
280:14811–14818. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lu Z, Luo T, Nie M, Pang T, Zhang X, Shen
X, Ma L, Bi J, Wei G, Fang G and Xue X: TSPAN1 functions as an
oncogene in gastric cancer and is downregulated by miR-573. FEBS
Lett. 589:1988–1994. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sun X, Wang M, Zhang F and Kong X:
Inhibition of NET-1 suppresses proliferation and promotes apoptosis
of hepatocellular carcinoma cells by activating the PI3K/AKT
signaling pathway. Exp Ther Med. 17:2334–2340. 2019.PubMed/NCBI
|
|
25
|
Huang H, Li H, Zhao T, Khan AA, Pan R,
Wang S, Wang S and Liu X: TSPAN1-elevated FAM110A promotes
pancreatic cancer progression by transcriptionally regulating
HIST1H2BK. J Cancer. 13:906–917. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen L, Yuan D, Zhao R, Li H and Zhu J:
Suppression of TSPAN1 by RNA interference inhibits proliferation
and invasion of colon cancer cells in vitro. Tumori. 96:744–750.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen Z, Gu S, Trojanowicz B, Liu N, Zhu G,
Dralle H and Hoang-Vu C: Down-regulation of TM4SF is associated
with the metastatic potential of gastric carcinoma TM4SF members in
gastric carcinoma. World J Surg Oncol. 9:432011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
He P, Wang S, Zhang X, Gao Y, Niu W, Dong
N, Shi X, Geng Y, Ma Q, Li M, et al: Tspan5 is an independent
favourable prognostic factor and suppresses tumour growth in
gastric cancer. Oncotarget. 7:40160–40173. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xie Q, Guo H, He P, Deng H, Gao Y, Dong N,
Niu W, Liu T, Li M, Wang S, et al: Tspan5 promotes
epithelial-mesenchymal transition and tumour metastasis of
hepatocellular carcinoma by activating Notch signalling. Mol Oncol.
15:3184–3202. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Roh S, Kim S, Hong I, Lee M, Kim HJ, Ahn
TS, Kang DH, Baek MJ, Kwak HJ, Kim CJ and Jeong D: High expression
of tetraspanin 5 as a prognostic marker of colorectal cancer. Int J
Mol Sci. 24:64762023. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Andrijes R, Hejmadi RK, Pugh M, Rajesh S,
Novitskaya V, Ibrahim M, Overduin M, Tselepis C, Middleton GW,
Győrffy B, et al: Tetraspanin 6 is a regulator of carcinogenesis in
colorectal cancer. Proc Natl Acad Sci USA. 118:e20114111182021.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Qi Y, Li H, Lv J, Qi W, Shen L, Liu S,
Ding A, Wang G, Sun L and Qiu W: Expression and function of
transmembrane 4 superfamily proteins in digestive system cancers.
Cancer Cell Int. 20:3142020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhu H, Wu Y, Zheng W and Lu S: CO-029 is
overexpressed in gastric cancer and mediates the effects of EGF on
gastric cancer cell proliferation and invasion. Int J Mol Med.
35:798–802. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wei L, Li Y and Suo Z: TSPAN8 promotes
gastric cancer growth and metastasis via ERK MAPK pathway. Int J
Clin Exp Med. 8:8599–8607. 2015.PubMed/NCBI
|
|
35
|
Akiel MA, Santhekadur PK, Mendoza RG,
Siddiq A, Fisher PB and Sarkar D: Tetraspanin 8 mediates
AEG-1-induced invasion and metastasis in hepatocellular carcinoma
cells. FEBS Lett. 590:2700–2708. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li J, Chen X, Zhu L, Lao Z, Zhou T, Zang
L, Ge W, Jiang M, Xu J, Cao Y, et al: SOX9 is a critical regulator
of TSPAN8-mediated metastasis in pancreatic cancer. Oncogene.
40:4884–4893. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Guo Q, Xia B, Zhang F, Richardson MM, Li
M, Zhang JS, Chen F and Zhang XA: Tetraspanin CO-029 inhibits
colorectal cancer cell movement by deregulating cell-matrix and
cell-cell adhesions. PLoS One. 7:e384642012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhou Z, Ran YL, Hu H, Pan J, Li ZF, Chen
LZ, Sun LC, Peng L, Zhao XL, Yu L, et al: TM4SF3 promotes
esophageal carcinoma metastasis via upregulating ADAM12m
expression. Clin Exp Metastasis. 25:537–548. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Qi Y, Lv J, Liu S, Sun L, Wang Y, Li H, Qi
W and Qiu W: TSPAN9 and EMILIN1 synergistically inhibit the
migration and invasion of gastric cancer cells by increasing TSPAN9
expression. BMC Cancer. 19:6302019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Dash S, Wu CC, Wu CC, Chiang SF, Lu YT,
Yeh CY, You JF, Chu LJ, Yeh TS and Yu JS: Extracellular vesicle
membrane protein profiling and targeted mass spectrometry unveil
CD59 and tetraspanin 9 as novel plasma biomarkers for detection of
colorectal cancer. Cancers (Basel). 15:1772022. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu J, Chen C, Li G, Chen D and Zhou Q:
Upregulation of TSPAN12 is associated with the colorectal cancer
growth and metastasis. Am J Transl Res. 9:812–822. 2017.PubMed/NCBI
|
|
42
|
Sidahmed-Adrar N, Ottavi JF, Benzoubir N,
Ait Saadi T, Bou Saleh M, Mauduit P, Guettier C, Desterke C and Le
Naour F: Tspan15 is a new stemness-related marker in hepatocellular
carcinoma. Proteomics. 19:e19000252019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang B, Zhang Z, Li L, Qin YR, Liu H,
Jiang C, Zeng TT, Li MQ, Xie D, Li Y, et al: TSPAN15 interacts with
BTRC to promote oesophageal squamous cell carcinoma metastasis via
activating NF-κB signaling. Nat Commun. 9:14232018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zheng Y, Wang DD, Wang W, Pan K, Huang CY,
Li YF, Wang QJ, Yuan SQ, Jiang SS, Qiu HB, et al: Reduced
expression of uroplakin 1A is associated with the poor prognosis of
gastric adenocarcinoma patients. PLoS One. 9:e930732014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yang YM, Zhang ZW, Liu QM, Sun YF, Yu JR
and Xu WX: Overexpression of CD151 predicts prognosis in patients
with resected gastric cancer. PLoS One. 8:e589902013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ke AW, Shi GM, Zhou J, Wu FZ, Ding ZB, Hu
MY, Xu Y, Song ZJ, Wang ZJ, Wu JC, et al: Role of overexpression of
CD151 and/or c-Met in predicting prognosis of hepatocellular
carcinoma. Hepatology. 49:491–503. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Suzuki S, Miyazaki T, Tanaka N, Sakai M,
Sano A, Inose T, Sohda M, Nakajima M, Kato H and Kuwano H:
Prognostic significance of CD151 expression in esophageal squamous
cell carcinoma with aggressive cell proliferation and invasiveness.
Ann Surg Oncol. 18:888–893. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Xu L, Hou Y, Tu G, Chen Y, Du YE, Zhang H,
Wen S, Tang X, Yin J, Lang L, et al: Nuclear Drosha enhances cell
invasion via an EGFR-ERK1/2-MMP7 signaling pathway induced by
dysregulated miRNA-622/197 and their targets LAMC2 and CD82 in
gastric cancer. Cell Death Dis. 8:e26422017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Hemler ME, Mannion BA and Berditchevski F:
Association of TM4SF proteins with integrins: Relevance to cancer.
Biochim Biophys Acta. 1287:67–71. 1996.PubMed/NCBI
|
|
50
|
Dong JT, Lamb PW, Rinker-Schaeffer CW,
Vukanovic J, Ichikawa T, Isaacs JT and Barrett JC: KAI1, a
metastasis suppressor gene for prostate cancer on human chromosome
11p11.2. Science. 268:884–886. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Liu X, Guo X, Li H, Chen J and Qi X:
Src/STAT3 signaling pathways are involved in KAI1-induced
downregulation of VEGF-C expression in pancreatic cancer. Mol Med
Rep. 13:4774–4778. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Huang X, Li Y, He X, Chen Y, Wei W, Yang X
and Ma K: Gangliosides and CD82 inhibit the motility of colon
cancer by downregulating the phosphorylation of EGFR at different
tyrosine sites and signaling pathways. Mol Med Rep. 22:3994–4002.
2020.PubMed/NCBI
|
|
53
|
Takaoka A, Hinoda Y, Satoh S, Adachi Y,
Itoh F, Adachi M and Imai K: Suppression of invasive properties of
colon cancer cells by a metastasis suppressor KAI1 gene. Oncogene.
16:1443–1453. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Miyazaki T, Kato H, Shitara Y, Yoshikawa
M, Tajima K, Masuda N, Shouji H, Tsukada K, Nakajima T and Kuwano
H: Mutation and expression of the metastasis suppressor gene KAI1
in esophageal squamous cell carcinoma. Cancer. 89:955–962. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zeng TD, Zheng B, Zheng W and Chen C:
CD82/KAI1 inhibits invasion and metastasis of esophageal squamous
cell carcinoma via TGF-β1. Eur Rev Med Pharmacol Sci. 22:5928–5937.
2018.PubMed/NCBI
|
|
56
|
Yoo TH, Ryu BK, Lee MG and Chi SG: CD81 is
a candidate tumor suppressor gene in human gastric cancer. Cell
Oncol (Dordr). 36:141–153. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Fang TT, Sun XJ, Chen J, Zhao Y, Sun RX,
Ren N and Liu BB: Long non-coding RNAs are differentially expressed
in hepatocellular carcinoma cell lines with differing metastatic
potential. Asian Pac J Cancer Prev. 15:10513–10524. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Murayama Y, Miyagawa J, Shinomura Y,
Kanayama S, Isozaki K, Yamamori K, Mizuno H, Ishiguro S, Kiyohara
T, Miyazaki Y, et al: Significance of the association between
heparin-binding epidermal growth factor-like growth factor and CD9
in human gastric cancer. Int J Cancer. 98:505–513. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Nakamoto T, Murayama Y, Oritani K,
Boucheix C, Rubinstein E, Nishida M, Katsube F, Watabe K, Kiso S,
Tsutsui S, et al: A novel therapeutic strategy with anti-CD9
antibody in gastric cancers. J Gastroenterol. 44:889–896. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Li Y, Yu S, Li L, Chen J, Quan M, Li Q and
Gao Y: KLF4-mediated upregulation of CD9 and CD81 suppresses
hepatocellular carcinoma development via JNK signaling. Cell Death
Dis. 11:2992020. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Tang M, Yin G, Wang F, Liu H, Zhou S, Ni
J, Chen C, Zhou Y and Zhao Y: Downregulation of CD9 promotes
pancreatic cancer growth and metastasis through upregulation of
epidermal growth factor on the cell surface. Oncol Rep. 34:350–358.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Ovalle S, Gutiérrez-López MD, Olmo N,
Turnay J, Lizarbe MA, Majano P, Molina-Jiménez F, López-Cabrera M,
Yáñez-Mó M, Sánchez-Madrid F and Cabañas C: The tetraspanin CD9
inhibits the proliferation and tumorigenicity of human colon
carcinoma cells. Int J Cancer. 121:2140–2152. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Buim ME, Lourenço SV, Carvalho KC, Cardim
R, Pereira C, Carvalho AL, Fregnani JH and Soares FA:
Downregulation of CD9 protein expression is associated with
aggressive behavior of oral squamous cell carcinoma. Oral Oncol.
46:166–171. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Kusukawa J, Ryu F, Kameyama T and Mekada
E: Reduced expression of CD9 in oral squamous cell carcinoma: CD9
expression inversely related to high prevalence of lymph node
metastasis. J Oral Pathol Med. 30:73–79. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Park SA, Kim MJ, Park SY, Kim JS, Lim W,
Nam JS and Yhong Sheen Y: TIMP-1 mediates TGF-β-dependent crosstalk
between hepatic stellate and cancer cells via FAK signaling. Sci
Rep. 5:164922015. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Takashima Y, Komatsu S, Ohashi T, Kiuchi
J, Kamiya H, Shimizu H, Arita T, Konishi H, Shiozaki A, Kubota T,
et al: Overexpression of Tetraspanin31 contributes to malignant
potential and poor outcomes in gastric cancer. Cancer Sci.
113:1984–1998. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Wang J, Zhou Y, Li D, Sun X, Deng Y and
Zhao Q: TSPAN31 is a critical regulator on transduction of survival
and apoptotic signals in hepatocellular carcinoma cells. FEBS Lett.
591:2905–2918. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Smyth EC, Nilsson M, Grabsch HI, van
Grieken NC and Lordick F: Gastric cancer. Lancet. 396:635–648.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Yang L, Ying X, Liu S, Lyu G, Xu Z, Zhang
X, Li H, Li Q, Wang N and Ji J: Gastric cancer: Epidemiology, risk
factors and prevention strategies. Chin J Cancer Res. 32:695–704.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Deng Y, Cai S, Shen J and Peng H:
Tetraspanins: Novel molecular regulators of gastric cancer. Front
Oncol. 11:7025102021. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Hemler ME: Tetraspanin proteins promote
multiple cancer stages. Nat Rev Cancer. 14:49–60. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Zöller M: Tetraspanins: Push and pull in
suppressing and promoting metastasis. Nat Rev Cancer. 9:40–55.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Bonnet M, Maisonial-Besset A, Zhu Y,
Witkowski T, Roche G, Boucheix C, Greco C and Degoul F: Targeting
the tetraspanins with monoclonal antibodies in oncology: Focus on
Tspan8/Co-029. Cancers (Basel). 11:1792019. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Claas C, Seiter S, Claas A, Savelyeva L,
Schwab M and Zöller M: Association between the rat homologue of
CO-029, a metastasis-associated tetraspanin molecule and
consumption coagulopathy. J Cell Biol. 141:267–280. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Gesierich S, Berezovskiy I, Ryschich E and
Zöller M: Systemic induction of the angiogenesis switch by the
tetraspanin D6.1A/CO-029. Cancer Res. 66:7083–7094. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Anami K, Oue N, Noguchi T, Sakamoto N,
Sentani K, Hayashi T, Naito Y, Oo HZ and Yasui W: TSPAN8,
identified by Escherichia coli ampicillin secretion trap, is
associated with cell growth and invasion in gastric cancer. Gastric
Cancer. 19:370–380. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Hemler ME: Tetraspanin functions and
associated microdomains. Nat Rev Mol Cell Biol. 16:801–811. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Wang HX, Li Q, Sharma C, Knoblich K and
Hemler ME: Tetraspanin protein contributions to cancer. Biochem Soc
Trans. 39:547–552. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Li T, Meng XL and Yang WQ: Long noncoding
RNA PVT1 acts as a ‘sponge’ to inhibit microRNA-152 in gastric
cancer cells. Dig Dis Sci. 62:3021–3028. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Murray D, Horgan G, Macmathuna P and Doran
P: NET1-mediated RhoA activation facilitates lysophosphatidic
acid-induced cell migration and invasion in gastric cancer. Br J
Cancer. 99:1322–1329. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Wang GL, Chen L, Wei YZ, Zhou JM, Wu YY,
Zhang YX, Qin J and Zhu YY: The effect of NET-1 on the
proliferation, migration and endocytosis of the SMMC-7721 HCC cell
line. Oncol Rep. 27:1944–1952. 2012.PubMed/NCBI
|
|
82
|
Shang H, Wu B, Liang X, Sun Y, Han X,
Zhang L, Wang Q and Cheng W: Evaluation of therapeutic effect of
targeting nanobubbles conjugated with NET-1 siRNA by shear wave
elastography: an in vivo study of hepatocellular carcinoma bearing
mice model. Drug Deliv. 26:944–951. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Li T, Xue Y, Wang G, Gu T, Li Y, Zhu YY
and Chen L: Multi-target siRNA: Therapeutic strategy for
hepatocellular carcinoma. J Cancer. 7:1317–1327. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Garcia-Mayea Y, Mir C, Carballo L,
Castellvi J, Temprana-Salvador J, Lorente J, Benavente S,
García-Pedrero JM, Allonca E, Rodrigo JP and LLeonart ME: TSPAN1: A
novel protein involved in head and neck squamous cell carcinoma
chemoresistance. Cancers (Basel). 12:32692020. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Ma L, Li Y, Peng J, Wu D, Zhao X, Cui Y,
Chen L, Yan X, Du Y and Yu L: Discovery of the migrasome, an
organelle mediating release of cytoplasmic contents during cell
migration. Cell Res. 25:24–38. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Wu L, Yang S, Li H, Zhang Y, Feng L, Zhang
C, Wei J, Gu X, Xu G, Wang Z and Wang F: TSPAN4-positive migrasome
derived from retinal pigmented epithelium cells contributes to the
development of proliferative vitreoretinopathy. J
Nanobiotechnology. 20:5192022. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Zhang C, Li T, Yin S, Gao M, He H, Li Y,
Jiang D, Shi M, Wang J and Yu L: Monocytes deposit migrasomes to
promote embryonic angiogenesis. Nat Cell Biol. 24:1726–1738. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Zhang Y, Zhang M, Xie Z, Ding Y, Huang J,
Yao J, Lv Y and Zuo J: Research progress and direction of novel
organelle-migrasomes. Cancers (Basel). 15:1342022. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Qi W, Sun L, Liu N, Zhao S, Lv J and Qiu
W: Tetraspanin family identified as the central genes detected in
gastric cancer using bioinformatics analysis. Mol Med Rep.
18:3599–3610. 2018.PubMed/NCBI
|
|
90
|
Hori H, Yano S, Koufuji K, Takeda J and
Shirouzu K: CD9 expression in gastric cancer and its significance.
J Surg Res. 117:208–215. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Setoguchi T, Kikuchi H, Yamamoto M, Baba
M, Ohta M, Kamiya K, Tanaka T, Baba S, Goto-Inoue N, Setou M, et
al: Microarray analysis identifies versican and CD9 as potent
prognostic markers in gastric gastrointestinal stromal tumors.
Cancer Sci. 102:883–889. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Zhao LJ, Fan QQ, Li YY, Ren HM, Zhang T,
Liu S, Maa M, Zheng YC and Liu HM: LSD1 deletion represses gastric
cancer migration by upregulating a novel miR-142-5p target protein
CD9. Pharmacol Res. 159:1049912020. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Wunder JS, Eppert K, Burrow SR, Gokgoz N,
Bell RS and Andrulis IL: Co-amplification and overexpression of
CDK4, SAS and MDM2 occurs frequently in human parosteal
osteosarcomas. Oncogene. 18:783–788. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Ma X, Qiu S, Tang X, Song Q, Wang P, Wang
J, Xia Q, Wang Z, Zhao Q and Lu M: TSPAN31 regulates the
proliferation, migration, and apoptosis of gastric cancer cells
through the METTL1/CCT2 pathway. Transl Oncol. 20:1014232022.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Oren R, Takahashi S, Doss C, Levy R and
Levy S: TAPA-1, the target of an antiproliferative antibody,
defines a new family of transmembrane proteins. Mol Cell Biol.
10:4007–4015. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Levy S, Todd SC and Maecker HT: CD81
(TAPA-1): A molecule involved in signal transduction and cell
adhesion in the immune system. Annu Rev Immunol. 16:89–109. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Dong JT, Suzuki H, Pin SS, Bova GS,
Schalken JA, Isaacs WB, Barrett JC and Isaacs JT: Down-regulation
of the KAI1 metastasis suppressor gene during the progression of
human prostatic cancer infrequently involves gene mutation or
allelic loss. Cancer Res. 56:4387–4390. 1996.PubMed/NCBI
|
|
98
|
García-Frigola C, Burgaya F, Calbet M, de
Lecea L and Soriano E: Mouse Tspan-5, a member of the tetraspanin
superfamily, is highly expressed in brain cortical structures.
Neuroreport. 11:3181–3185. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Li PY, Lv J, Qi WW, Zhao SF, Sun LB, Liu
N, Sheng J and Qiu WS: Tspan9 inhibits the proliferation, migration
and invasion of human gastric cancer SGC7901 cells via the ERK1/2
pathway. Oncol Rep. 36:448–454. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Qi Y, Qi W, Liu S, Sun L, Ding A, Yu G, Li
H, Wang Y, Qiu W and Lv J: TSPAN9 suppresses the chemosensitivity
of gastric cancer to 5-fluorouracil by promoting autophagy. Cancer
Cell Int. 20:42020. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Sayiner M, Golabi P and Younossi ZM:
Disease burden of hepatocellular carcinoma: A global perspective.
Dig Dis Sci. 64:910–917. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Zhang K, Lai X, Song J, He L, Wang L, Ou
G, Tian X, Wang L, Deng J, Zhang J, et al: A novel cell culture
model reveals the viral interference during hepatitis B and C virus
coinfection. Antiviral Res. 189:1050612021. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Chen L, Wang Z, Zhan X, Li DC, Zhu YY and
Zhu J: Association of NET-1 gene expression with human
hepatocellular carcinoma. Int J Surg Pathol. 15:346–353. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Wu B, Shang H, Liang X, Sun Y, Jing H, Han
X and Cheng W: Preparation of novel targeting nanobubbles
conjugated with small interfering RNA for concurrent molecular
imaging and gene therapy in vivo. FASEB J. 33:14129–14136. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Wu B, Qiao Q, Han X, Jing H, Zhang H,
Liang H and Cheng W: Targeted nanobubbles in low-frequency
ultrasound-mediated gene transfection and growth inhibition of
hepatocellular carcinoma cells. Tumour Biol. 37:12113–12121. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Kanetaka K, Sakamoto M, Yamamoto Y,
Yamasaki S, Lanza F, Kanematsu T and Hirohashi S: Overexpression of
tetraspanin CO-029 in hepatocellular carcinoma. J Hepatol.
35:637–642. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Fang T, Lin J, Wang Y, Chen G, Huang J,
Chen J, Zhao Y, Sun R, Liang C and Liu B: Tetraspanin-8 promotes
hepatocellular carcinoma metastasis by increasing ADAM12m
expression. Oncotarget. 7:40630–40643. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Herlevsen M, Schmidt DS, Miyazaki K and
Zöller M: The association of the tetraspanin D6.1A with the
alpha6beta4 integrin supports cell motility and liver metastasis
formation. J Cell Sci. 116:4373–4390. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Sanjmyatav J, Steiner T, Wunderlich H,
Diegmann J, Gajda M and Junker K: A specific gene expression
signature characterizes metastatic potential in clear cell renal
cell carcinoma. J Urol. 186:289–294. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Tokuhara T, Hasegawa H, Hattori N, Ishida
H, Taki T, Tachibana S, Sasaki S and Miyake M: Clinical
significance of CD151 gene expression in non-small cell lung
cancer. Clin Cancer Res. 7:4109–4114. 2001.PubMed/NCBI
|
|
112
|
Devbhandari RP, Shi GM, Ke AW, Wu FZ,
Huang XY, Wang XY, Shi YH, Ding ZB, Xu Y, Dai Z, et al: Profiling
of the tetraspanin CD151 web and conspiracy of CD151/integrin β1
complex in the progression of hepatocellular carcinoma. PLoS One.
6:e249012011. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Shi GM, Ke AW, Zhou J, Wang XY, Xu Y, Ding
ZB, Devbhandari RP, Huang XY, Qiu SJ, Shi YH, et al: CD151
modulates expression of matrix metalloproteinase 9 and promotes
neoangiogenesis and progression of hepatocellular carcinoma.
Hepatology. 52:183–196. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Zhang PF, Wang F, Wu J, Wu Y, Huang W, Liu
D, Huang XY, Zhang XM and Ke AW: LncRNA SNHG3 induces EMT and
sorafenib resistance by modulating the miR-128/CD151 pathway in
hepatocellular carcinoma. J Cell Physiol. 234:2788–2794. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Liu T, Zu CH, Wang SS, Song HL, Wang ZL,
Xu XN, Liu HS, Wang YL and Shen ZY: PIK3C2A mRNA functions as a
miR-124 sponge to facilitate CD151 expression and enhance
malignancy of hepatocellular carcinoma cells. Oncotarget.
7:43376–43389. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Kim JH, Badawi M, Park JK, Jiang J, Mo X,
Roberts LR and Schmittgen TD: Anti-invasion and anti-migration
effects of miR-199a-3p in hepatocellular carcinoma are due in part
to targeting CD151. Int J Oncol. 49:2037–2045. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Schreyer L, Mittermeier C, Franz MJ, Meier
MA, Martin DE, Maier KC, Huebner K, Schneider-Stock R, Singer S,
Holzer K, et al: Tetraspanin 5 (TSPAN5), a novel gatekeeper of the
tumor suppressor DLC1 and myocardin-related transcription factors
(MRTFs), controls HCC growth and senescence. Cancers (Basel).
13:53732021. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Guo XZ, Friess H, Di Mola FF, Heinicke JM,
Abou-Shady M, Graber HU, Baer HU, Zimmermann A, Korc M and Büchler
MW: KAI1, a new metastasis suppressor gene, is reduced in
metastatic hepatocellular carcinoma. Hepatology. 28:1481–1488.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Yu G, Bing Y, Li W, Xia L and Liu Z:
Hepatitis B virus inhibits the expression of CD82 through
hypermethylation of its promoter in hepatoma cells. Mol Med Rep.
10:2580–2586. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Gilsanz A, Sánchez-Martín L,
Gutiérrez-López MD, Ovalle S, Machado-Pineda Y, Reyes R, Swart GW,
Figdor CG, Lafuente EM and Cabañas C: ALCAM/CD166 adhesive function
is regulated by the tetraspanin CD9. Cell Mol Life Sci. 70:475–493.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Wang J, Ma L, Weng W, Qiao Y, Zhang Y, He
J, Wang H, Xiao W, Li L, Chu Q, et al: Mutual interaction between
YAP and CREB promotes tumorigenesis in liver cancer. Hepatology.
58:1011–1120. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Ma L, Wang J, Lin J, Pan Q, Yu Y and Sun
F: Cluster of differentiation 166 (CD166) regulated by
phosphatidylinositide 3-Kinase (PI3K)/AKT signaling to exert its
anti-apoptotic role via yes-associated protein (YAP) in liver
cancer. J Biol Chem. 289:6921–6933. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Jentzsch V, Davis JAA and Djamgoz MBA:
Pancreatic cancer (PDAC): Introduction of evidence-based
complementary measures into integrative clinical management.
Cancers (Basel). 12:30962020. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Zeitouni D, Pylayeva-Gupta Y, Der CJ and
Bryant KL: KRAS mutant pancreatic cancer: No lone path to an
effective treatment. Cancers (Basel). 8:452016. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Ye H, Li T, Wang H, Wu J, Yi C, Shi J,
Wang P, Song C, Dai L, Jiang G, et al: TSPAN1, TMPRSS4, SDR16C5,
and CTSE as novel panel for pancreatic cancer: A bioinformatics
analysis and experiments validation. Front Immunol. 12:6495512021.
View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Zhou C, Liang Y, Zhou L, Yan Y, Liu N,
Zhang R, Huang Y, Wang M, Tang Y, Ali DW, et al: TSPAN1 promotes
autophagy flux and mediates cooperation between WNT-CTNNB1
signaling and autophagy via the MIR454-FAM83A-TSPAN1 axis in
pancreatic cancer. Autophagy. 17:3175–3195. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Liu S, Cai Y, Changyong E, Sheng J and
Zhang X: Screening and validation of independent predictors of poor
survival in pancreatic cancer. Pathol Oncol Res. 27:16098682021.
View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Wang L, Gao P, Yuan P, Zhou P, Fan H, Lin
X, Yuan X, Zhu M, Fan X, Lu Y and Wang Z: miR-573 suppresses
pancreatic cancer cell proliferation, migration, and invasion
through targeting TSPAN1. Strahlenther Onkol. 197:438–448. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Ma C, Cui Z, Wang Y, Zhang L, Wen J, Guo
H, Li N and Zhang W: Bioinformatics analysis reveals TSPAN1 as a
candidate biomarker of progression and prognosis in pancreatic
cancer. Bosn J Basic Med Sci. 21:47–60. 2021.PubMed/NCBI
|
|
131
|
Wang S, Liu X, Khan AA, Li H, Tahir M, Yan
X, Wang J and Huang H: miR-216a-mediated upregulation of TSPAN1
contributes to pancreatic cancer progression via transcriptional
regulation of ITGA2. Am J Cancer Res. 10:1115–1129. 2020.PubMed/NCBI
|
|
132
|
Zhang X, Shi G, Gao F, Liu P, Wang H and
Tan X: TSPAN1 upregulates MMP2 to promote pancreatic cancer cell
migration and invasion via PLCγ. Oncol Rep. 41:2117–2125.
2019.PubMed/NCBI
|
|
133
|
Mayado A, Orfao A, Mentink A, Gutierrez
ML, Muñoz-Bellvis L and Terstappen LWMM: Detection of circulating
tumor cells in blood of pancreatic ductal adenocarcinoma patients.
Cancer Drug Resist. 3:83–97. 2020.PubMed/NCBI
|
|
134
|
Wang H, Rana S, Giese N, Büchler MW and
Zöller M: Tspan8, CD44v6 and alpha6beta4 are biomarkers of
migrating pancreatic cancer-initiating cells. Int J Cancer.
133:416–426. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Yue S, Mu W and Zöller M: Tspan8 and CD151
promote metastasis by distinct mechanisms. Eur J Cancer.
49:2934–2948. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Yue S, Mu W, Erb U and Zöller M: The
tetraspanins CD151 and Tspan8 are essential exosome components for
the crosstalk between cancer initiating cells and their
surrounding. Oncotarget. 6:2366–2384. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Greenow K and Clarke AR: Controlling the
stem cell compartment and regeneration in vivo: The role of
pluripotency pathways. Physiol Rev. 92:75–99. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Sales KM, Winslet MC and Seifalian AM:
Stem cells and cancer: An overview. Stem Cell Rev. 3:249–255. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Heiler S, Wang Z and Zöller M: Pancreatic
cancer stem cell markers and exosomes-the incentive push. World J
Gastroenterol. 22:5971–6007. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Wu M, Li X, Liu R, Yuan H, Liu W and Liu
Z: Development and validation of a metastasis-related gene
signature for predicting the overall survival in patients with
pancreatic ductal adenocarcinoma. J Cancer. 11:6299–6318. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Luo L, Li Y, Huang C, Lin Y, Su Y, Cen H,
Chen Y, Peng S, Ren T, Xie R and Zeng L: A new 7-gene survival
score assay for pancreatic cancer patient prognosis prediction. Am
J Cancer Res. 11:495–512. 2021.PubMed/NCBI
|
|
142
|
Crnogorac-Jurcevic T, Efthimiou E, Capelli
P, Blaveri E, Baron A, Terris B, Jones M, Tyson K, Bassi C, Scarpa
A and Lemoine NR: Gene expression profiles of pancreatic cancer and
stromal desmoplasia. Oncogene. 20:7437–7446. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Feng J, Huang C, Wren JD, Wang DW, Yan J,
Zhang J, Sun Y, Han X and Zhang XA: Tetraspanin CD82: A suppressor
of solid tumors and a modulator of membrane heterogeneity. Cancer
Metastasis Rev. 34:619–633. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Liu WM and Zhang XA: KAI1/CD82, a tumor
metastasis suppressor. Cancer Lett. 240:183–194. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Yan W, Huang J, Zhang Q and Zhang J: Role
of metastasis suppressor KAI1/CD82 in different cancers. J Oncol.
2021:99244732021. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Liu X, Guo XZ, Zhang WW, Lu ZZ, Zhang QW,
Duan HF and Wang LS: KAI1 inhibits HGF-induced invasion of
pancreatic cancer by sphingosine kinase activity. Hepatobiliary
Pancreat Dis Int. 10:201–208. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Siegel RL, Miller KD, Wagle NS and Jemal
A: Cancer statistics, 2023. CA Cancer J Clin. 73:17–48. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Miller KD, Nogueira L, Devasia T, Mariotto
AB, Yabroff KR, Jemal A, Kramer J and Siegel RL: Cancer treatment
and survivorship statistics, 2022. CA Cancer J Clin. 72:409–436.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Lee CH, Im EJ, Moon PG and Baek MC:
Discovery of a diagnostic biomarker for colon cancer through
proteomic profiling of small extracellular vesicles. BMC Cancer.
18:10582018. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Guo JN, Chen D, Deng SH, Huang JR, Song
JX, Li XY, Cui BB and Liu YL: Identification and quantification of
immune infiltration landscape on therapy and prognosis in left- and
right-sided colon cancer. Cancer Immunol Immunother. 71:1313–1330.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Min J, Yang S, Cai Y, Vanderwall DR, Wu Z,
Li S, Liu S, Liu B, Wang J, Ding Y, et al: Tetraspanin Tspan8
restrains interferon signaling to stabilize intestinal epithelium
by directing endocytosis of interferon receptor. Cell Mol Life Sci.
80:1542023. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Kuhn S, Koch M, Nübel T, Ladwein M,
Antolovic D, Klingbeil P, Hildebrand D, Moldenhauer G, Langbein L,
Franke WW, et al: A complex of EpCAM, claudin-7, CD44 variant
isoforms, and tetraspanins promotes colorectal cancer progression.
Mol Cancer Res. 5:553–567. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Visvader JE: Cells of origin in cancer.
Nature. 469:314–322. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Wang Z and Zöller M: Exosomes, metastases,
and the miracle of cancer stem cell markers. Cancer Metastasis Rev.
38:259–295. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Greco C, Bralet MP, Ailane N,
Dubart-Kupperschmitt A, Rubinstein E, Le Naour F and Boucheix C:
E-cadherin/p120-catenin and tetraspanin Co-029 cooperate for cell
motility control in human colon carcinoma. Cancer Res.
70:7674–7583. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Knoblich K, Wang HX, Sharma C, Fletcher
AL, Turley SJ and Hemler ME: Tetraspanin TSPAN12 regulates tumor
growth and metastasis and inhibits β-catenin degradation. Cell Mol
Life Sci. 71:1305–1314. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Chiang SF, Kan CY, Hsiao YC, Tang R, Hsieh
LL, Chiang JM, Tsai WS, Yeh CY, Hsieh PS, Liang Y, et al: Bone
marrow stromal antigen 2 is a novel plasma biomarker and
prognosticator for colorectal carcinoma: A secretome-based
verification study. Dis Markers. 2015:8740542015. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Hashida H, Takabayashi A, Tokuhara T,
Hattori N, Taki T, Hasegawa H, Satoh S, Kobayashi N, Yamaoka Y and
Miyake M: Clinical significance of transmembrane 4 superfamily in
colon cancer. Br J Cancer. 89:158–167. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Mori M, Mimori K, Shiraishi T, Haraguchi
M, Ueo H, Barnard GF and Akiyoshi T: Motility related protein 1
(MRP1/CD9) expression in colon cancer. Clin Cancer Res.
4:1507–1510. 1998.PubMed/NCBI
|
|
161
|
Wu DH, Liu L, Chen LH and Ding YQ: KAI1
gene expression in colonic carcinoma and its clinical
significances. World J Gastroenterol. 10:2245–2249. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Lee JH, Bae JA, Lee JH, Seo YW, Kho DH,
Sun EG, Lee SE, Cho SH, Joo YE, Ahn KY, et al: Glycoprotein 90K,
downregulated in advanced colorectal cancer tissues, interacts with
CD9/CD82 and suppresses the Wnt/beta-catenin signal via ISGylation
of beta-catenin. Gut. 59:907–917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Morgan E, Soerjomataram I, Rumgay H,
Coleman HG, Thrift AP, Vignat J, Laversanne M, Ferlay J and Arnold
M: The Global landscape of esophageal squamous cell carcinoma and
esophageal adenocarcinoma incidence and mortality in 2020 and
projections to 2040: New estimates from GLOBOCAN 2020.
Gastroenterology. 163:649–658.e2. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
van den Brandt PA: The impact of a healthy
lifestyle on the risk of esophageal and gastric cancer subtypes.
Eur J Epidemiol. 37:931–945. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Botelho NK, Schneiders FI, Lord SJ,
Freeman AK, Tyagi S, Nancarrow DJ, Hayward NK, Whiteman DC and Lord
RV: Gene expression alterations in formalin-fixed,
paraffin-embedded Barrett esophagus and esophageal adenocarcinoma
tissues. Cancer Biol Ther. 10:172–179. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Zhao WS, Yan WP, Chen DB, Dai L, Yang YB,
Kang XZ, Fu H, Chen P, Deng KJ, Wang XY, et al: Genome-scale CRISPR
activation screening identifies a role of ELAVL2-CDKN1A axis in
paclitaxel resistance in esophageal squamous cell carcinoma. Am J
Cancer Res. 9:1183–1200. 2019.PubMed/NCBI
|
|
167
|
Fisher OM, Levert-Mignon AJ, Lehane CW,
Botelho NK, Maag JL, Thomas ML, Edwards M, Lord SJ, Bobryshev YV,
Whiteman DC and Lord RV: CD151 Gene and protein expression provides
independent prognostic information for patients with adenocarcinoma
of the esophagus and gastroesophageal junction treated by
esophagectomy. Ann Surg Oncol. 23 (Suppl 5):S746–S754. 2016.
View Article : Google Scholar
|
|
168
|
Scully C and Porter S: ABC of oral health.
Oral cancer. BMJ. 321:97–100. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Scully C and Bedi R: Ethnicity and oral
cancer. Lancet Oncol. 1:37–42. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
170
|
D'souza S and Addepalli V: Preventive
measures in oral cancer: An overview. Biomed Pharmacother.
107:72–80. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Hiroshima K, Shiiba M, Oka N, Hayashi F,
Ishida S, Fukushima R, Koike K, Iyoda M, Nakashima D, Tanzawa H and
Uzawa K: Tspan15 plays a crucial role in metastasis in oral
squamous cell carcinoma. Exp Cell Res. 384:1116222019. View Article : Google Scholar : PubMed/NCBI
|
|
172
|
Nankivell P, Williams H, McConkey C,
Webster K, High A, MacLennan K, Senguven B, Rabbitts P and Mehanna
H: Tetraspanins CD9 and CD151, epidermal growth factor receptor and
cyclooxygenase-2 expression predict malignant progression in oral
epithelial dysplasia. Br J Cancer. 109:2864–2874. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Imai Y, Sasaki T, Shinagawa Y, Akimoto K
and Fujibayashi T: Expression of metastasis suppressor gene
(KAI1/CD82) in oral squamous cell carcinoma and its
clinico-pathological significance. Oral Oncol. 38:557–561. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
174
|
Farhadieh RD, Smee R, Ow K, Yang JL,
Russell PJ, Crouch R, Jackson P and Jacobson IV: Down-regulation of
KAI1/CD82 protein expression in oral cancer correlates with reduced
disease free survival and overall patient survival. Cancer Lett.
213:91–98. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
175
|
Matsumura N, Zembutsu H, Yamaguchi K,
Sasaki K, Tsuruma T, Nishidate T, Denno R and Hirata K:
Identification of novel molecular markers for detection of gastric
cancer cells in the peripheral blood circulation using genome-wide
microarray analysis. Exp Ther Med. 2:705–713. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
176
|
Lin H, Zhou AJ, Zhang JY, Liu SF and Gu
JX: MiR-324-5p reduces viability and induces apoptosis in gastric
cancer cells through modulating TSPAN8. J Pharm Pharmacol.
70:1513–1520. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
177
|
Zhai R, Kan X, Wang B, Du H, Long Y, Wu H,
Tao K, Wang G, Bao L, Li F and Zhang W: miR-152 suppresses gastric
cancer cell proliferation and motility by targeting CD151. Tumour
Biol. 35:11367–11373. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
178
|
Blanco E, Hsiao A, Ruiz-Esparza GU, Landry
MG, Meric-Bernstam F and Ferrari M: Molecular-targeted
nanotherapies in cancer: Enabling treatment specificity. Mol Oncol.
5:492–503. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
179
|
Kim TK, Park CS, Jeoung MH, Lee WR, Go NK,
Choi JR, Lee TS, Shim H and Lee S: Generation of a human antibody
that inhibits TSPAN8-mediated invasion of metastatic colorectal
cancer cells. Biochem Biophys Res Commun. 468:774–780. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
180
|
Park CS, Kim TK, Kim HG, Kim YJ, Jeoung
MH, Lee WR, Go NK, Heo K and Lee S: Therapeutic targeting of
tetraspanin8 in epithelial ovarian cancer invasion and metastasis.
Oncogene. 35:4540–4548. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
181
|
Ke AW, Zhang PF, Shen YH, Gao PT, Dong ZR,
Zhang C, Cai JB, Huang XY, Wu C, Zhang L, et al: Generation and
characterization of a tetraspanin CD151/integrin α6β1-binding
domain competitively binding monoclonal antibody for inhibition of
tumor progression in HCC. Oncotarget. 7:6314–6322. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
182
|
Jin L and Cambier JC: SMIP-016 in action:
CD37 as a death receptor. Cancer Cell. 21:597–598. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
183
|
Hwang JR, Jo K, Lee Y, Sung BJ, Park YW
and Lee JH: Upregulation of CD9 in ovarian cancer is related to the
induction of TNF-α gene expression and constitutive NF-κB
activation. Carcinogenesis. 33:77–83. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
184
|
Longo N, Yáñez-Mó M, Mittelbrunn M, de la
Rosa G, Muñoz ML, Sánchez-Madrid F and Sánchez-Mateos P: Regulatory
role of tetraspanin CD9 in tumor-endothelial cell interaction
during transendothelial invasion of melanoma cells. Blood.
98:3717–3726. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
185
|
Rock KL, Farfán-Arribas DJ, Colbert JD and
Goldberg AL: Re-examining class-I presentation and the DRiP
hypothesis. Trends Immunol. 35:144–152. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
186
|
Unternaehrer JJ, Chow A, Pypaert M, Inaba
K and Mellman I: The tetraspanin CD9 mediates lateral association
of MHC class II molecules on the dendritic cell surface. Proc Natl
Acad Sci USA. 104:234–239. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
187
|
Suzuki M, Tachibana I, Takeda Y, He P,
Minami S, Iwasaki T, Kida H, Goya S, Kijima T, Yoshida M, et al:
Tetraspanin CD9 negatively regulates lipopolysaccharide-induced
macrophage activation and lung inflammation. J Immunol.
182:6485–6493. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
188
|
Rocha-Perugini V, Martínez Del Hoyo G,
González-Granado JM, Ramírez-Huesca M, Zorita V, Rubinstein E,
Boucheix C and Sánchez-Madrid F: CD9 regulates major
histocompatibility complex class II trafficking in monocyte-derived
dendritic cells. Mol Cell Biol. 37:e00202–17. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
189
|
Jones EL, Wee JL, Demaria MC, Blakeley J,
Ho PK, Vega-Ramos J, Villadangos JA, van Spriel AB, Hickey MJ,
Hämmerling GJ and Wright MD: Dendritic cell migration and antigen
presentation are coordinated by the opposing functions of the
tetraspanins CD82 and CD37. J Immunol. 196:978–987. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
190
|
Gartlan KH, Wee JL, Demaria MC, Nastovska
R, Chang TM, Jones EL, Apostolopoulos V, Pietersz GA, Hickey MJ,
van Spriel AB and Wright MD: Tetraspanin CD37 contributes to the
initiation of cellular immunity by promoting dendritic cell
migration. Eur J Immunol. 43:1208–1219. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
191
|
Todros-Dawda I, Kveberg L, Vaage JT and
Inngjerdingen M: The tetraspanin CD53 modulates responses from
activating NK cell receptors, promoting LFA-1 activation and
dampening NK cell effector functions. PLoS One. 9:e978442014.
View Article : Google Scholar : PubMed/NCBI
|
|
192
|
Petersen SH, Odintsova E, Haigh TA,
Rickinson AB, Taylor GS and Berditchevski F: The role of
tetraspanin CD63 in antigen presentation via MHC class II. Eur J
Immunol. 41:2556–2561. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
193
|
Sheng KC, van Spriel AB, Gartlan KH, Sofi
M, Apostolopoulos V, Ashman L and Wright MD: Tetraspanins CD37 and
CD151 differentially regulate Ag presentation and T-cell
co-stimulation by DC. Eur J Immunol. 39:50–55. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
194
|
Colbert JD, Cruz FM, Baer CE and Rock KL:
Tetraspanin-5-mediated MHC class I clustering is required for
optimal CD8 T cell activation. Proc Natl Acad Sci USA.
119:e21221881192022. View Article : Google Scholar : PubMed/NCBI
|
|
195
|
Schäfer D, Tomiuk S, Küster LN, Rawashdeh
WA, Henze J, Tischler-Höhle G, Agorku DJ, Brauner J, Linnartz C,
Lock D, et al: Identification of CD318, TSPAN8 and CD66c as target
candidates for CAR T cell based immunotherapy of pancreatic
adenocarcinoma. Nat Commun. 12:14532021. View Article : Google Scholar : PubMed/NCBI
|
|
196
|
Kohmo S, Kijima T, Otani Y, Mori M, Minami
T, Takahashi R, Nagatomo I, Takeda Y, Kida H, Goya S, et al: Cell
surface tetraspanin CD9 mediates chemoresistance in small cell lung
cancer. Cancer Res. 70:8025–8035. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
197
|
Ullah M, Akbar A, Ng NN, Concepcion W and
Thakor AS: Mesenchymal stem cells confer chemoresistance in breast
cancer via a CD9 dependent mechanism. Oncotarget. 10:3435–3450.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
198
|
Pan SJ, Wu YB, Cai S, Pan YX, Liu W, Bian
LG, Sun B and Sun QF: Over-expression of tetraspanin 8 in malignant
glioma regulates tumor cell progression. Biochem Biophys Res
Commun. 458:476–482. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
199
|
Zhu R, Gires O, Zhu L, Liu J, Li J, Yang
H, Ju G, Huang J, Ge W, Chen Y, et al: TSPAN8 promotes cancer cell
stemness via activation of sonic Hedgehog signaling. Nat Commun.
10:28632019. View Article : Google Scholar : PubMed/NCBI
|
|
200
|
Tominaga N, Hagiwara K, Kosaka N, Honma K,
Nakagama H and Ochiya T: RPN2-mediated glycosylation of tetraspanin
CD63 regulates breast cancer cell malignancy. Mol Cancer.
13:1342014. View Article : Google Scholar : PubMed/NCBI
|
|
201
|
Ye M, Wei T, Wang Q, Sun Y, Tang R, Guo L
and Zhu W: TSPAN12 promotes chemoresistance and proliferation of
SCLC under the regulation of miR-495. Biochem Biophys Res Commun.
486:349–356. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
202
|
Floren M, Restrepo Cruz S, Termini CM,
Marjon KD, Lidke KA and Gillette JM: Tetraspanin CD82 drives acute
myeloid leukemia chemoresistance by modulating protein kinase C
alpha and β1 integrin activation. Oncogene. 39:3910–3925. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
203
|
Quagliano A, Gopalakrishnapillai A, Kolb
EA and Barwe SP: CD81 knockout promotes chemosensitivity and
disrupts in vivo homing and engraftment in acute lymphoblastic
leukemia. Blood Adv. 4:4393–4405. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
204
|
Li L, Yang D, Cui D, Li Y, Nie Z, Wang J
and Liang L: Quantitative proteomics analysis of the role of
tetraspanin-8 in the drug resistance of gastric cancer. Int J
Oncol. 52:473–484. 2018.PubMed/NCBI
|
|
205
|
Gao X, Ran N, Dong X, Zuo B, Yang R, Zhou
Q, Moulton HM, Seow Y and Yin H: Anchor peptide captures, targets,
and loads exosomes of diverse origins for diagnostics and therapy.
Sci Transl Med. 10:eaat01952018. View Article : Google Scholar : PubMed/NCBI
|
|
206
|
Huang Y, Zucker B, Zhang S, Elias S, Zhu
Y, Chen H, Ding T, Li Y, Sun Y, Lou J, et al: Migrasome formation
is mediated by assembly of micron-scale tetraspanin macrodomains.
Nat Cell Biol. 21:991–1002. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
207
|
Jiao H, Jiang D, Hu X, Du W, Ji L, Yang Y,
Li X, Sho T, Wang X, Li Y, et al: Mitocytosis, a migrasome-mediated
mitochondrial quality-control process. Cell. 184:2896–2910.e13.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
208
|
Yu S and Yu L: Migrasome biogenesis and
functions. FEBS J. 289:7246–7254. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
209
|
Schmidt-Pogoda A, Strecker JK, Liebmann M,
Massoth C, Beuker C, Hansen U, König S, Albrecht S, Bock S, Breuer
J, et al: Dietary salt promotes ischemic brain injury and is
associated with parenchymal migrasome formation. PLoS One.
13:e02098712018. View Article : Google Scholar : PubMed/NCBI
|
|
210
|
Chen L, Ma L and Yu L: WGA is a probe for
migrasomes. Cell Discov. 5:132019. View Article : Google Scholar : PubMed/NCBI
|
|
211
|
Zheng Y, Lang Y, Qi B, Wang Y, Gao W and
Li T: TSPAN4 is a prognostic and immune target in Glioblastoma
multiforme. Front Mol Biosci. 9:10300572023. View Article : Google Scholar : PubMed/NCBI
|
|
212
|
Zhang K, Zhu Z, Jia R, Wang NA, Shi M,
Wang Y, Xiang S, Zhang Q and Xu L: CD151-enriched migrasomes
mediate hepatocellular carcinoma invasion by conditioning cancer
cells and promoting angiogenesis. J Exp Clin Cancer Res.
43:1602024. View Article : Google Scholar : PubMed/NCBI
|
|
213
|
Qin Y, Yang J, Liang C, Liu J, Deng Z, Yan
B, Fu Y, Luo Y, Li X, Wei X and Li W: Pan-cancer analysis
identifies migrasome-related genes as a potential immunotherapeutic
target: A bulk omics research and single cell sequencing
validation. Front Immunol. 13:9948282022. View Article : Google Scholar : PubMed/NCBI
|