The cytoskeleton is composed of three primary
components: Microtubules, actin filaments and intermediate
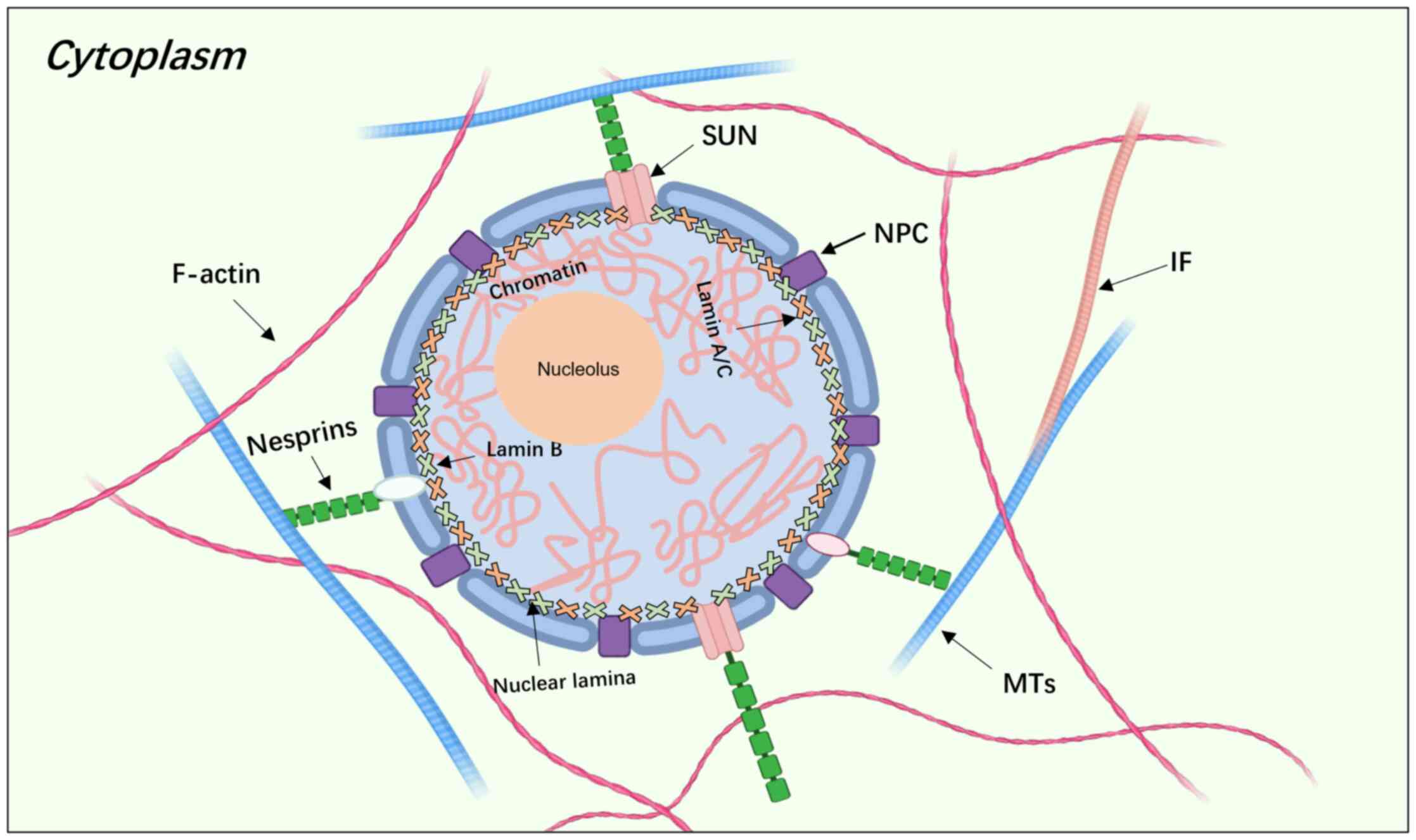
filaments (1). The nuclear lamina,
a thin and dense protein network, is situated beneath the nuclear
membrane, forming a highly organized mesh (2,3)
(Fig. 1). Lamins, which are
intermediate filament proteins located in the nuclear lamina, are
categorized into A-type and B-type proteins (4,5). The
nuclear lamina performs multiple cellular functions, including
providing mechanical stability, facilitating protein nuclear
localization, enabling cell migration, regulating chromatin
organization, and participating in DNA replication and repair
(6–10). Research has indicated that the
nuclear lamina is also implicated in laminopathies, such as
Emery-Dreifuss muscular dystrophy (EDMD) and dilated cardiomyopathy
(DCM), and premature aging syndromes, such as Hutchinson-Gilford
progeria syndrome (HGPS) and Werner syndrome (11,12).
In human cells, A-type lamins, comprising lamin A
and lamin C, are encoded by the LMNA gene. Lamin C is a splice
variant of lamin A, and, due to their structural similarity, they
are often collectively referred to as lamin A/C for research
purposes (13–15). Impairment of lamin functions can
lead to the development of a wide range of disorders (11,16).
The function and expression of lamin A/C vary across tissue types
and cancer sub-types, resulting in no specific pattern of lamin A/C
expression in cancer (17–19). Lamin A and lamin C expression
levels are typically measured together; however, lamin A and lamin
C expression may not always be equally altered in certain diseases,
further complicating the understanding of the effect of lamin A/C
on disease progression (17–19).
An increasing number of diseases have been reported to be
associated with mutations or altered expression levels of genes
controlling nuclear lamina, such as intestinal polyps and cancer,
which adds to the complexity of studying these proteins (17–19).
The present review aimed to examine the molecular structure of the
lamin family, explore the relationship between the LMNA gene and
various diseases, and discuss the association of nuclear lamina
with tumors and signaling pathways that may be perturbed by lamin.
The objective of the study was to provide valuable insights for
basic and clinical translational research into laminopathies and
related cancer.
The nuclear fibrillar layer, located at the
underside of the nuclear membrane, is composed of lamina and its
associated proteins. These proteins connect nuclear proteins to
heterochromatin and have been identified as intermediate filament
proteins with a V-shaped structure. Based on differences in gene
structure and nucleotide sequences, lamin is classified into six
isoforms: Types I and II, which include acidic and basic keratins;
type III, which includes vimentin, desmin, peripherin and glial
fibrillary acidic protein; type IV intermediate filament group,
which includes neurofilament proteins and a fourth neurofilament
subunit, α-internexin protein; type V, which consists of lamins
located in the nucleus; and group VI, which consists of
lens-specific intermediate filaments, CP49/crystallin and filensin.
All lamins have good elasticity and are usually related to cell
morphology (20). Intermediate
filament proteins, positioned between actin microfilaments and
microtubules, have an average diameter of 10–12 nm (21). While the majority of lamins are
found in the nuclear lamina, a small proportion are present in the
nucleoplasm (4,22–24).
The functionality of lamin proteins varies across different tissues
due to its interactions with membrane proteins and heterochromatin,
facilitated by diverse binding chaperones (23–25).
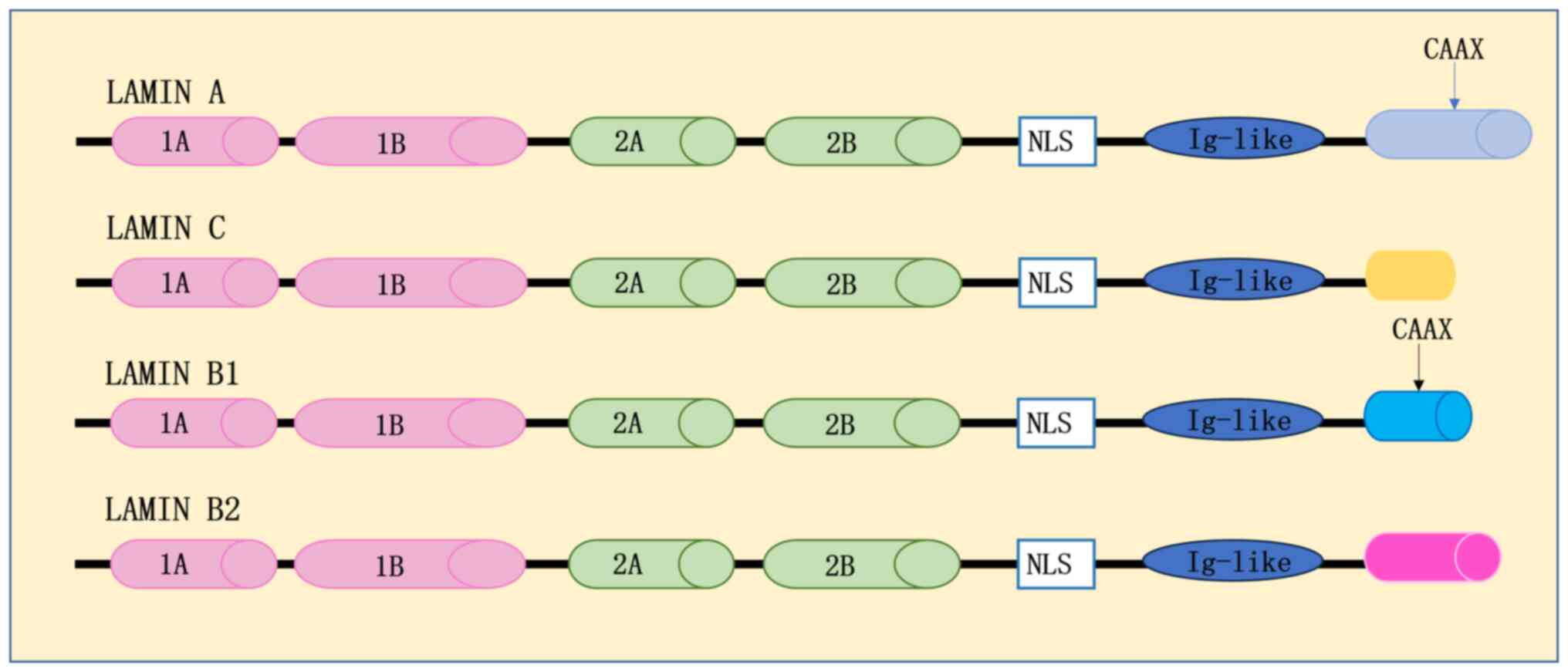
Structurally, lamins comprise three primary components: A central
α-helical structural domain; a bulbous amino-terminal in the head
region; and a carboxy-terminal in the tail domain (4). The central α-helical structural
domain is further subdivided into four α-helical fragments,
designated as d1A, 1B, 2A and 2B. These fragments are
interconnected by short sub-structural domains termed L1, L12 and
L2. The amino-terminal head structural domain exhibits variable
size, while the carboxy-terminal tail structural domain encompasses
nuclear localization signals, Ig structural domains and the CAAX
box (4) (Fig. 2).
The LMNA gene encodes lamin A/C proteins, which are
classified as lamin A-type proteins based on their structural
configuration (26–28). In mammals, seven main types of
lamin have been identified. The lamin A proteins, including the
major isoforms lamins A and C and the minor isoforms lamins A∆10
and C2, are all encoded by the LMNA gene (29). Conversely, the lamin B proteins
comprise three isoforms: Lamin B1, encoded by the LMNB1 gene, and
lamins B2 and B3, which are encoded by the LMNB2 gene (29). Lamins A, C, B1 and B2 are the
predominant lamin proteins in human cells, serving a crucial role
in maintaining nuclear integrity. While the amino-terminal head and
central rod-like structural domains of lamin A closely resemble
those of lamin B, lamin A is distinguished by a unique 90 amino
acid fragment at the carboxyl-terminal tail, forming an extended
structural domain (4) (Fig. 2). It has been suggested that lamin
B predates lamin A in evolutionary terms. Notably, the intron
position of human LMNB1 is conserved in LMNA and LMNB2; however,
LMNB2 contains an additional intron between the regions coding for
helix 1A and helix 1B that is absent in other intermediate filament
proteins, including LMNA, thus suggesting that LMNB2 and LMNA may
have evolved from LMNB1 (29,30).
Lamin A-type proteins are expressed in most differentiated somatic
cells, but their production is minimal in embryonic stem cells and
the sub-epidermis, which suggests a potential association between
lamin A and the differentiation process of organelles (31). By contrast, lamin B-type proteins
are expressed regardless of the degree of cellular differentiation
and are essential for normal organ development. Furthermore, lamin
B is expressed in almost all cells (31,32).
The LMNA gene is translated and spliced to form the lamin A
precursor, pre-lamin A, which undergoes farnesylation of the CAAX
box to produce mature lamin A (4,22,23,33).
Conversely, lamin C precursors directly form mature lamin C due to
the absence of the CAAX box (33).
The LMNB1 and LMNB2 genes are located on human chromosomes
5q23.2-q31.3 and 19p13.3, respectively. Lamin B-type proteins
contain the CAAX box but do not undergo farnesylation, forming
mature lamin B-type proteins directly after translation (34). In addition to farnesylation, lamin
proteins undergo phosphorylation and ubiquitination. Various
protein kinases can phosphorylate lamins, with CDC2, protein kinase
(PK)C and PKA being the three known kinases that
post-translationally modify lamins (4). CDC2 induces lamin cleavage, PKC
phosphorylation regulates lamin uptake into the nucleus, and PKA
phosphorylation inhibits lamin polymerization (35–37).
Lamin A/C proteins possess ubiquitination-like modification sites
within their rod and tail structural domains; mutation of the site
within the rod structural domain results in reduced levels of
SUMOylation of intracellular lamins and altered sub-nuclear
localization (38).
In somatic cells, lamin A-type and B-type proteins
form spatially independent reticular structures with overlapping
regions, fulfilling distinct cellular roles (39–45).
Transcriptionally silent genomic regions, including mitoplasts,
telomeres and inactivated X chromosomes, are predominantly
localized within the nuclear lamina. Lamin B proteins has been
identified as a global chromatin regulator (46,47).
At least two chromatin-binding regions have been identified in
lamins: One situated between the rod structural domain terminus and
the IG structural domains in the tail region, and another within
the rod structural domains (48,49).
Whole genome sequencing techniques have revealed genomic regions
preferentially associating with Lamin proteins, termed
lamina-associated domains (LADs) (50). These results suggest that lamin may
have an important role in maintaining cell homeostasis. In-depth
investigation of lamin proteins is essential for analyzing nuclear
homeostasis at the molecular level and exploring its association
with disease pathogenesis.
The LMNA gene encodes the multifunctional
intermediate filament proteins lamin A/C. Mutations in LMNA,
dysregulation of expression levels and improper protein processing
can result in various diseases collectively referred to as
laminopathies (13). Lamin
proteins are absent in a number of human diseases, and mutations
affect the course of some human diseases (Table I). Laminopathies are classified as
primary or secondary based on their etiology. Primary laminopathies
are caused by LMNA gene mutations and include striated muscle
disorders, lipodystrophy syndromes and peripheral neuropathy
(11,16,34,51).
Secondary laminopathies primarily result from Zmpste24 defects,
where lamin A precursors fail to undergo Zmpste24-dependent
cleavage to form mature lamin A (11,16,51).
Additionally, secondary laminopathies can arise from mutations in
non-lamin genes that interact with lamin, leading to disorders such
as progeria and restrictive dermopathy (11,16,51).
LMNA gene mutations have been found to significantly impact cardiac
function, with dilated cardiomyopathy (DCM) being a major
contributor to heart failure. It has been reported that ~6% of DCM
cases worldwide can be attributed to LMNA gene mutations. Notably,
missense mutations in the α-helix-rod structural domain of LMNA
result in lamin C defects, ultimately leading to the onset of DCM
(11,52,53).
DCM is characterized by reduced left ventricular or biventricular
pumping function, potentially causing hypertension, valvular
disease or coronary artery disease. DCM caused by LMNA mutations
has been associated with a more severe clinical phenotype and
poorer prognosis compared with DCM of other etiologies (54). Treatment for cardiac dysfunction
caused by LMNA mutations remains marginally effective, with
interventions aimed at reversing molecular changes (55). Notably, EDMD was the first disease
identified as being associated with LMNA (12). EDMD clinical manifestations often
include muscle weakness and cardiac conduction abnormalities
(28). Wang et al (11) demonstrated that mutations in
specific structural domains of the LMNA gene, leading to LMNA
functional inactivation, can cause EDMD. Elucidating the
mechanistic role of LMNA in laminopathies is crucial for developing
novel diagnostic and therapeutic strategies for nuclear
laminopathies in clinical settings.
Lamin A/C expression has been observed to be
significantly upregulated in the adipose tissue of individuals
diagnosed with obesity and type 2 diabetes (41). Kim et al (56) revealed that obesity can induce an
increase in lamin C expression within adipose tissue macrophages
(ATMs). Furthermore, lamin C was found to contribute to
obesity-induced insulin resistance through activation of the NF-κB
signaling pathway, which may mediate ATM inflammation (56). Current therapeutic approaches for
type 2 diabetes primarily focus on lifestyle modifications,
including dietary changes and exercise regimens, coupled with the
administration of metformin and hypoglycemic agents (including
insulin); however, there remains a notable absence of targeted
treatments for this condition (57). Additionally, research has uncovered
that alterations in lamin C can lead to the development of
lipodystrophy syndromes. Notably, certain familial forms of
lipodystrophy have been predominantly attributed to a heterozygous
substitution occurring at amino acid position 482 within the
terminal structural domain of lamin C (58).
The expression level of LMNA gene is closely related
to the aging process. HGPS is a typical disease of aging, and ~90%
of HGPS cases worldwide are reported to be caused by mutations in
the LMNA gene (16). Specifically,
base substitutions within LMNA exon 11 (c.1824C>T) can activate
a cryptic splice site, resulting in the production of an
irreversible form of farnesylated mutation known as progerin
protein (59,60). Although progerins cannot be
incorporated into the nuclear fiber layer, their expression
interferes with normal cellular mitotic processes. This
interference induces genomic instability and premature senescence
(61–63). The expression of lamin A/C proteins
is intimately linked to nuclear stiffness, which gradually
decreases with age. In cardiomyocytes, a decrease in lamin A/C
expression has been observed with advancing age, corresponding to
changes in nuclear stiffness (64). The reduction in lamin A/C can also
lead to the downregulation of growth-associated transcription
factors and cytoskeletal regulators in cardiomyocytes, resulting in
cardiac dysfunction. Kirkland et al (64) demonstrated that maintaining lamin C
expression prevented an age-dependent decline in cardiac function.
Furthermore, lamin C may exert a significant influence on the aging
process by impairing PGC1α and inhibiting the NAMPT-NAD+
signaling pathway (65). Simon
et al (66) discovered that
genomic stability can be maintained by enhancing the interaction of
LMNA with mADPr, which contributes to human longevity. During cell
differentiation, increased expression of lamin A/C has been shown
to mediate cellular senescence by regulating the expression of
p16/INK4A through the lamin A/C-p53 network; this regulation is
crucial for selectively inducing cellular senescence (67). These findings collectively
suggested that LMNA may serve a decisive role in progeria and is
associated with the development of several myotonic dystrophy
syndromes. Mutations in the LMNA gene may ultimately lead to the
development of LMNA-associated disorders by affecting cellular
physiology. Consequently, LMNA may serve as a key target in the
treatment of these disorders.
Cancer development is characterized by dysregulated
gene expression, altered signaling pathways, increased overall
genomic instability and abnormal nuclear morphology (17,68).
The LMNA gene encodes lamin A/C proteins, which interact with
proteins such as emerin, retinoblastoma protein (pRb), c-Fos,
SREBP1 and MOK2, influencing cellular physiological functions.
Lamin A/C proteins serve a crucial role in various signaling
pathways, including p53, MAPK, ERK 1/2, Wnt, TGF-β, Notch and NF-κB
(17). The abnormal activation of
these pathways is frequently associated with tumor development.
Consequently, the expression level of the LMNA gene may be closely
linked to the process of tumor progression.
In certain tumor cells, the expression levels of the
LMNA gene are aberrantly altered, and this alteration may be used
as a molecular marker for the clinical diagnosis of specific tumors
(35). In developed countries,
where lung cancer exhibits a high mortality rate, Siegel et
al (69) and Broers et
al (70,71) observed that lamin A/C expression
was reduced in undifferentiated lung epithelial cells. The
expression of LMNA varies across different lung cancer cell lines.
Kaufmann et al (72) and
Broers et al (70,71) discovered that LMNA expression was
elevated in non-small cell lung cancer cell lines but was absent or
lowly expressed in small cell lung cancer cell lines. Furthermore,
Guinde and Frankel (73)
demonstrated that upregulation of microRNA (miRNA/miR)-9 in lung
cancer cells inhibited the expression of lamin A without affecting
lamin C expression; the ratio of lamin A to lamin C was observed to
be 1:8 in lung cancer cells, deviating from the typical 1:1 ratio
in other cell lines. This alteration in ratio may lead to increased
nuclear deformation, ultimately enhancing the migratory ability of
lung cancer cells (73,74). These findings may provide novel
targets for small molecule drug design to inhibit lung cancer
metastasis.
In breast cancer, it has been reported that Akt
targets the degradation of lamin A/C by altering its downstream
gene expression (75). LMNA is
involved in the developmental process of breast cancer and is
typically aberrantly expressed in breast cancer cells. Capo-chichi
et al (75) demonstrated
that knockdown of LMNA by short hairpin RNA resulted in the
appearance of cancer-like morphology and an increase in aneuploid
cells in primary mammary epithelial cells, which are typical
features of cancer (76–80). This finding suggested that LMNA may
have potential as an early clinical marker for breast cancer
diagnosis.
Ovarian cancer, the most lethal gynecological tumor
worldwide, presents complex treatment challenges and a generally
poor prognosis due to the frequent spread of cancer cells to organs
beyond the ovary at the time of diagnosis (81,82).
In ovarian cancer cells, downregulation of LMNA expression has been
shown to promote carcinogenesis, possibly due to caspase-6-mediated
downregulation of LMNA gene expression (83).
In leukemia and lymphoma, hypermethylation of the
CpG island promoter region can silence LMNA gene expression,
resulting in lamin A/C protein deficiency (84). Additionally, low expression of LMNA
has been detected in neuroblastoma and keratoacanthoma (85,86).
While downregulation of LMNA expression is observed
in some types of cancer, upregulation occurs in others. Studies
have detected increased LMNA expression in prostate cancer and
human pre-metastatic colorectal adenocarcinoma, where it is
correlated with the enhanced metastatic ability of tumor cells
(87,88). In colorectal and prostate cancer
cells, LMNA may enable enhanced invasion of surrounding tissues and
growth by increasing cell motility (18,88).
The relationship between LMNA expression and cancer cell migration
is complex. Decreased LMNA expression in highly invasive and
proliferative breast cancer cells has been shown to lead to
increased nuclear deformability, enhancing cell migration in the
interstitial space (89).
Similarly, downregulation of LMNA expression resulted in a
significant increase in the migratory capacity of ovarian cancer
cells (90). However, this effect
may not apply universally. In lung cancer, distal metastasis of
cancer cells typically occurs through pulmonary capillaries, which
are less permeable compared with other metastatic pathways
(91,92). Roncato et al (93) demonstrated that knockdown of LMNA
in the melanoma metastatic cell line B16F10 reduced lung metastasis
and impaired cell survival in the lung. LMNA also promoted
circulating tumor cell migration by protecting the nucleus from
mechanical stress (94,95). During distal metastasis, tumor
cells must withstand mechanical stresses encountered in the
bloodstream. Lamin A/C proteins provide mechanical stability to the
nucleus. In addition, knockdown of LMNA in breast cancer cell lines
resulted in increased apoptosis due to fluid shear stress, thus
inhibiting the migratory ability of tumor cells (94). Conversely, when LMNA is
overexpressed in cancer cells, it can enhance their ability to
withstand circulating fluid shear stress, promoting distal
metastasis (15,94,96).
Kaspi et al (97) revealed
that LMNA deficiency in lung cancer cells may be associated with
the loss of epithelial membrane antigen during
epithelial-to-mesenchymal transition (EMT), and that LMNA
deficiency in these cells could increase cancer cell motility and
migration (97). In circulating
tumor cell cells, reduced LMNA expression has been reported to
result in nuclei that are more easily deformed, allowing migration
through narrower spaces compared with in cells with high LMNA
expression (94). Wang et
al (90) revealed that LMNA
overexpression in ovarian cancer cells resulted the nuclei were so
rigid that molecules could not pass through nuclear pores.
Conversely, when LMNA expression was low, the nucleus became highly
susceptible to damage, leading to increased genomic instability and
cell death after passing through pores. This suggests that both
high and low levels of LMNA expression negatively regulate the
migratory ability of cells (90).
Based on the aforementioned research, the clinical
significance of lamin A/C expression abundance in tumors has been
explored. In colon cancer, a correlation has been observed between
low LMNA expression and increased recurrence in patients with stage
II and III colon cancer; therefore, low LMNA expression may serve
as a biomarker for risk prediction of colorectal carcinogenesis
(98). In non-muscle-invasive
bladder cancer (NMIBC), LMNA dysregulation was not associated with
its prognosis, but it could be used as a diagnostic biomarker to
distinguish patients with NMIBC from healthy subjects (99). In breast cancer, a lower expression
of LMNA appears to be associated with worse clinical outcomes
(100).
LMNA serves a crucial role in maintaining cell
morphology, and severe nuclear aberrations in cancer cells can
accelerate cancer progression and lead to poor prognosis (101,102). In breast cells, nuclear
aberrations similar to those in breast cancer cells were observed
in mammary epithelial cells with low LMNA expression (75), and Bell et al (89) found that altered nuclear regulation
in invasive breast cancer cells was affected by abnormal LMNA
expression levels. Alhudiri et al (101) further corroborated that low LMNA
expression in breast cancer was associated with a worse prognosis
compared with high LMNA expression. The expression of LMNA differs
among various tumor cells (Table
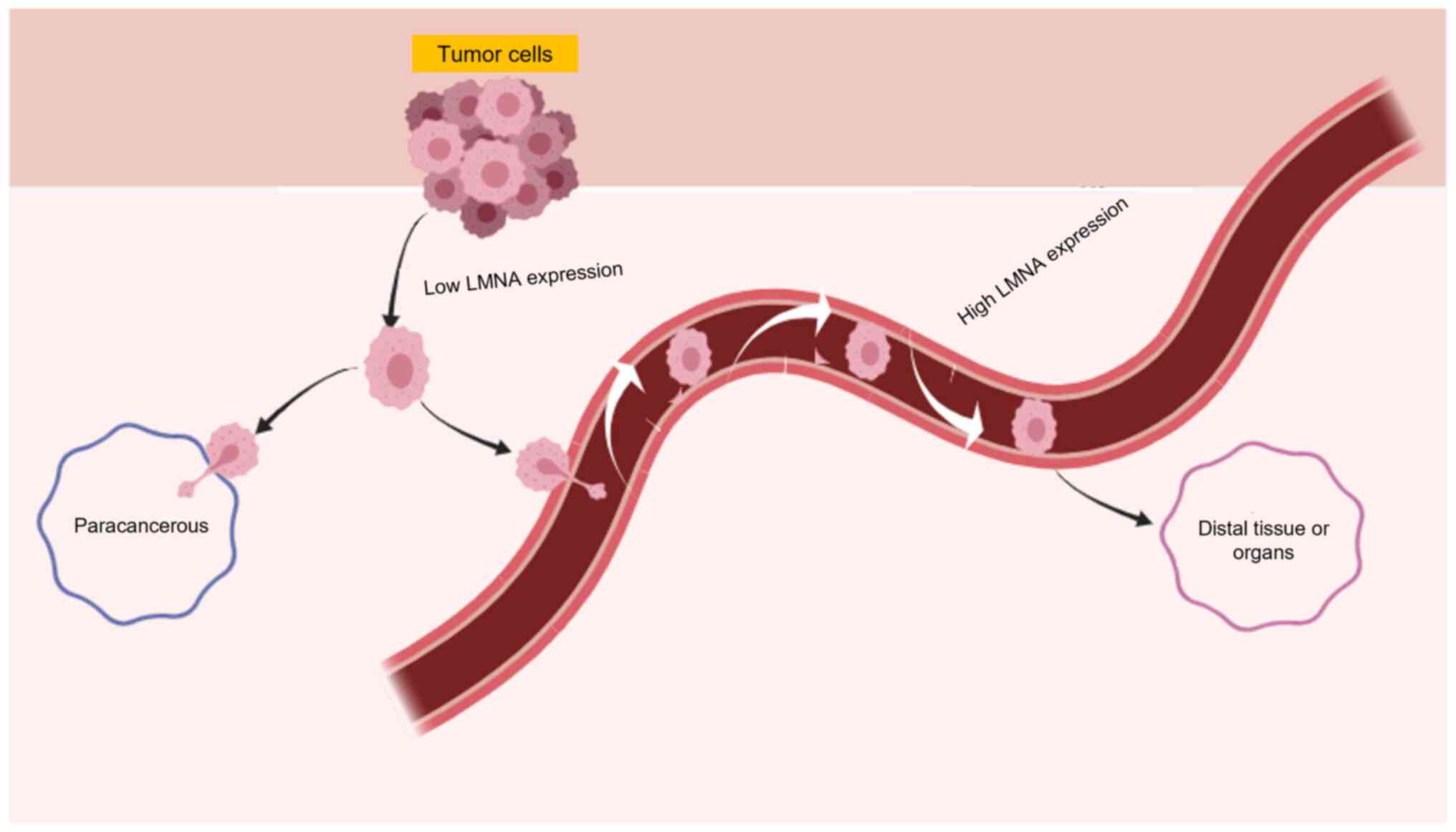
II). In tumor cells with low LMNA expression, the nucleus is
more likely to deform, which in turn makes it easier to cross the
interstitial space and enhance the ability of tumor cells to invade
adjacent tissues. However, these cells are less prone to distal
metastasis due to their susceptibility to mechanical damage and
death during the metastatic process. Conversely, tumor cells with
high LMNA expression may be more likely to undergo distal
metastasis (Fig. 3). Consequently,
the high or low expression of LMNA may be associated with the
metastatic ability of tumor cells. In-depth studies of the
mechanism of action of LMNA in tumors are required, which may
provide improved drug targets for metastatic tumors.
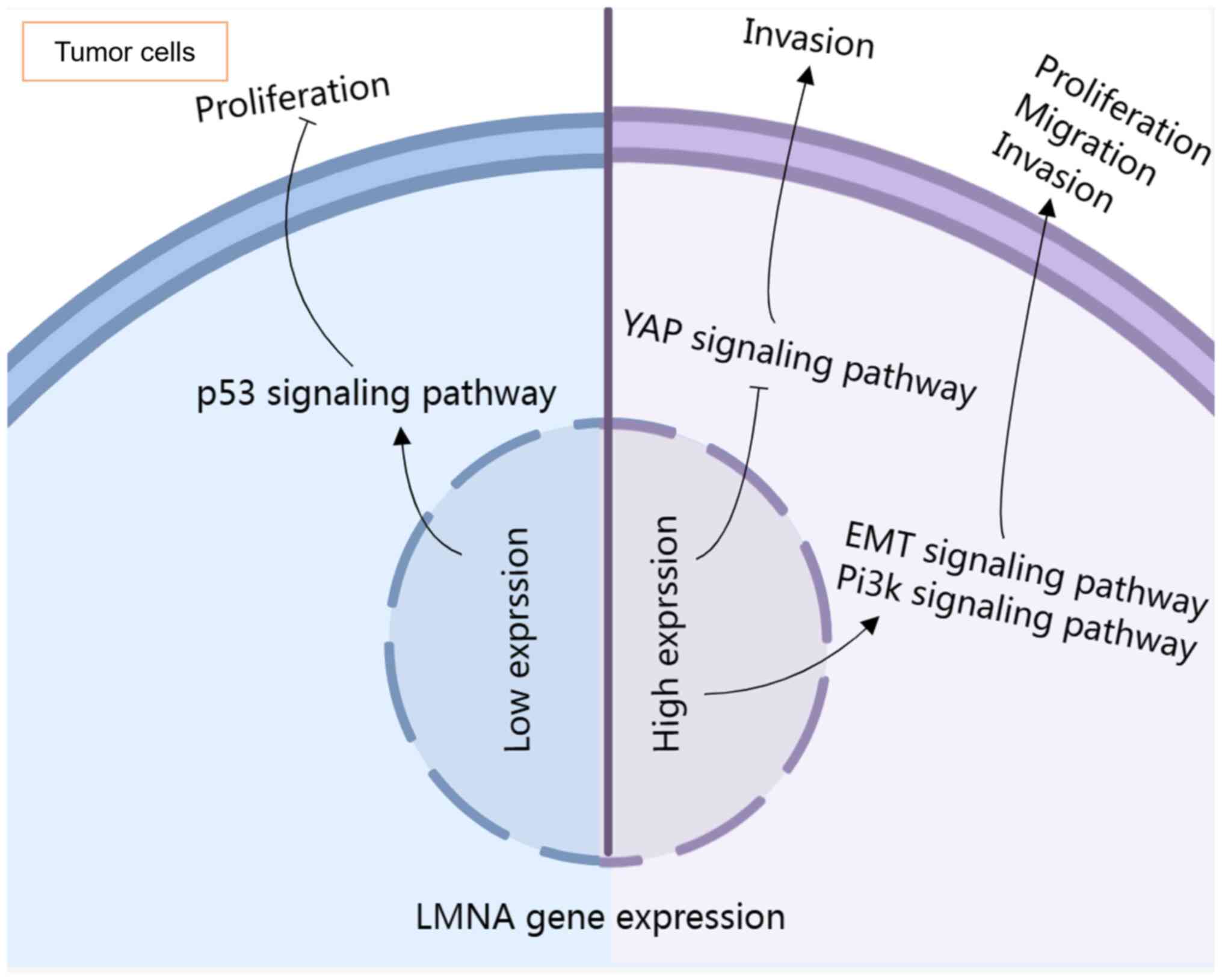
LMNA performs essential functions in cells, and
previous studies have established an association between LMNA and
tumor development. Notably, tumor-related signaling pathways may be
affected by abnormally expressed LMNA (Fig. 4). Elucidating the perturbation of
signaling pathways by LMNA in tumors may aid in the establishment
of a solid foundation for its in-depth study in oncology.
LMNA is closely associated with cancer development
and progression. In colorectal cancer cells, the motility of cells
expressing GFP-lamin A has been reported to be increased, and this
effect may be attributed to the activation of the EMT pathway by
LMNA gene expression, which promotes tumor cell metastasis
(103,104). Prostate cancer, one of the most
common types of cancer in men, is associated with an increasing
incidence with age, and has a poor prognosis and a high metastatic
potential. The identification of novel biomarkers, such as LMNA,
may provide improved treatment options for prostate cancer
(105–107). Kong et al (88) observed heterogeneous LMNA
expression in prostate cancer, with higher expression in
paracancerous tissues infiltrated with pre-existing tumor cells
compared with the tumor center. This finding suggested that LMNA
may be associated with the metastasis and motility of prostate
cancer cells, and could serve as a biomarker for differentiating
between tumor grades (88). This
previous study indicated that LMNA was highly expressed in
tissue-invasive prostate cancer, and may promote prostate cancer
cell proliferation, migration and invasion through the
phosphatidylinositol-3-kinase/AKT/PTEN axis (88). The interaction between lamin A/C
and emerin may also enhance cancer cell metastasis, possibly due to
increased nuclear stability resulting from high LMNA expression in
breast cancer cells (108).
Therefore, LMNA may not only serve as a diagnostic marker but also
as a potential target for inhibiting prostate cancer
metastasis.
In breast cancer cells, reduced LMNA expression has
been reported to lead to the formation of tetraploid and aneuploid
mammary epithelial cells. Aneuploidy and polyploidy can
subsequently induce growth arrest through the p53/p21 pathway
(75,104). The clinical treatment of highly
invasive triple-negative breast cancer (TNBC) faces significant
challenges, and miRNAs present a potential target for the
diagnosis, treatment and prognosis of TNBC (109). Chiarini et al (109) discovered that upregulation of
miR-129 could reduce LMNA expression in breast cancer cells. Given
the close relationship between LMNA and breast cancer development,
the miR-LMNA axis may represent a novel therapeutic pathway for
breast cancer. In invasive bone tumors, Chiarini et al
(109) revealed that high
expression of LMNA reduced the nuclear recruitment of
Yes-associated protein/TAZ and decreased the invasive ability of
tumor cells.
The influence of the LMNA gene on cancer progression
extends beyond its effects on tumor-associated pathways. In tumors,
the interactions of LMNA with various cellular proteins can
significantly impact disease progression. TPX2, a hallmark protein
in ovarian cancer that is associated with poor prognosis (110), has been shown to regulate lamin
A/C stability. Meng et al (111) and Sidera et al (112) demonstrated that TPX2 can modulate
lamin A/C phosphorylation levels in ovarian cancer cells, thereby
affecting their stability and inhibiting cellular processes. Hsp90,
a highly conserved chaperone protein, is crucial for tumor cell
invasion and DNA damage repair, making it a potential therapeutic
target. Wang et al (113)
observed that LMNA knockdown altered Hsp90 distribution in ovarian
cancer cells, increasing nuclear localization while decreasing
cytoplasmic presence. In hepatocellular carcinoma (HCC), a major
contributor to global cancer-related deaths (114–116), LMNA has been reported to interact
with sperm-associated antigen 4, a member of the SUN family, and
LINC complexes (117,118). This interaction can increase the
expression of SREBP1, a key regulator of adipogenesis (114,119,120), which can increase the expression
of enzymes related to lipid metabolism, thereby promoting HCC
development (121). Additionally,
post-translational modification of lamin A at the K265/270 site has
been shown to enhance HCC cell proliferation and prevent senescence
under hypoxic conditions (122).
In colon cancer, the interaction between LMNA and c-Fos, a
regulator of cell transformation (123,124), has been studied. Under specific
microenvironmental conditions, GDF15 was found to separate c-Fos
from lamin A/C, activating c-Fos, and promoting cancer cell
invasion and metastasis through EMT-related gene expression
(123). The antitumor activity of
polyphenols extracted from Artemisia annua L. (pKAL) in
colorectal cancer cells has been linked to p53-mediated
upregulation of the LMNA gene (104). Furthermore, LMNA has been shown
to protect pRb tumor suppressor proteins, key regulators of cell
proliferation and differentiation, from proteasomal degradation
(125). These findings highlight
the multifaceted role of LMNA in tumor progression through its
interactions with various proteins. LMNA participates in
post-translational modifications and protein interactions that
maintain stability or enhance activity of oncogenic proteins,
thereby influencing tumor physiology. Further investigation into
these interactions may provide comprehensive insights for
developing novel cancer treatment strategies.
The LMNA gene-encoded lamin A/C proteins, which are
crucial intermediate filament proteins for maintaining cellular
nuclear morphology, serve a pivotal role in various cellular
processes, including DNA damage repair, gene expression regulation
and cell differentiation. These proteins are intricately linked to
human health. While significant progress has been made in
elucidating the mechanisms of LMNA-induced diseases, substantial
challenges remain in developing specific clinical treatments.
Current experimental data have indicated that LMNA significantly
influences the treatment and prognosis of LMNA-associated diseases
and cancer in clinical settings, and studies have elucidated the
mechanisms underlying the role of LMNA in these conditions and
suggested its potential as a biomarker. However, there is a notable
lack of effective progress in the specific clinical treatment of
LMNA-induced diseases and tumors. For example, DCM caused by LMNA
mutations typically has a poorer prognosis compared with DCM
resulting from other etiologies; however, the underlying mechanisms
remain to be fully elucidated. Notably, the expression patterns of
LMNA in various types of cancer are inconsistent, thus adding
complexity to LMNA-related tumor studies. LMNA expression has been
reported to be downregulated in ovarian and breast cancer, whereas
it is upregulated in prostate and colorectal cancer. This
variability complicates the research on the role of LMNA in tumors.
Moreover, since the LMNA-encoded nuclear lamin A protein is an
integral component of the cell nucleus, targeting LMNA as a
therapeutic approach may potentially affect normal cells. The
present review delineated the tumor-associated pathways potentially
influenced by LMNA and the interacting proteins present in tumor
cells. This approach aims to identify associated pathways and
interacting proteins that could be targeted with enhanced
selectivity. Current research suggests that LMNA is associated with
the progression of highly invasive and metastatic tumors;
therefore, further in-depth investigation into the function and
molecular mechanisms of lamin A/C is warranted. In conclusion,
continued research into the role of LMNA holds positive theoretical
significance and clinical translational value for the prevention,
specific treatment and prognosis of LMNA-related diseases and
tumors.
Not applicable.
This work was supported by grants from the Scientific Research
Project of Education Department of Yunnan Province (grant nos.
2023Y0787 and 2024Y217).
Not applicable.
JZ and YT designed the review. JZ and HZ wrote the
first draft of this review, while CP, QH and KZ collected the
information needed for the review, and also helped design and write
review. YT revised the article. Data authentication is not
applicable. All authors read and approved the final version of the
manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Donnaloja F, Carnevali F, Jacchetti E and
Raimondi MT: Lamin A/C mechanotransduction in laminopathies. Cells.
9:13062020. View Article : Google Scholar
|
|
2
|
Burke B and Stewart CL: The nuclear
lamins: Flexibility in function. Nat Rev Mol Cell Biol. 14:13–24.
2013. View
Article : Google Scholar
|
|
3
|
Xie W and Burke B: Lamins. Curr Biol.
26:R348–R350. 2016. View Article : Google Scholar
|
|
4
|
Dittmer TA and Misteli T: The lamin
protein family. Genome Biol. 12:2222011. View Article : Google Scholar
|
|
5
|
Naetar N, Ferraioli S and Foisner R:
Lamins in the nuclear interior-life outside the lamina. J Cell Sci.
130:2087–2096. 2017. View Article : Google Scholar
|
|
6
|
Tajik A, Zhang Y, Wei F, Sun J, Jia Q,
Zhou W, Singh R, Khanna N, Belmont AS and Wang N: Transcription
upregulation via force-induced direct stretching of chromatin. Nat
Mater. 15:1287–1296. 2016. View
Article : Google Scholar
|
|
7
|
Ramdas NM and Shivashankar GV:
Cytoskeletal control of nuclear morphology and chromatin
organization. J Mol Biol. 427:695–706. 2015. View Article : Google Scholar
|
|
8
|
Davidson PM and Lammerding J: Broken
nuclei-lamins, nuclear mechanics, and disease. Trends Cell Biol.
24:247–256. 2014. View Article : Google Scholar
|
|
9
|
Lee JSH, Hale CM, Panorchan P, Khatau SB,
George JP, Tseng Y, Stewart CL, Hodzic D and Wirtz D: Nuclear lamin
A/C deficiency induces defects in cell mechanics, polarization, and
migration. Biophys J. 93:2542–2552. 2007. View Article : Google Scholar
|
|
10
|
Dahl KN, Kahn SM, Wilson KL and Discher
DE: The nuclear envelope lamina network has elasticity and a
compressibility limit suggestive of a molecular shock absorber. J
Cell Sci. 117:4779–4786. 2004. View Article : Google Scholar
|
|
11
|
Wang X, Zabell A, Koh W and Tang WH: Lamin
A/C cardiomyopathies: Current understanding and novel treatment
strategies. Curr Treat Options Cardiovasc Med. 19:212017.
View Article : Google Scholar
|
|
12
|
Chen SN, Sbaizero O, Taylor MRG and
Mestroni L: Lamin A/C cardiomyopathy: Implications for treatment.
Curr Cardiol Rep. 21:1602019. View Article : Google Scholar
|
|
13
|
Dubik N and Mai S: Lamin A/C: Function in
normal and tumor cells. Cancers (Basel). 12:36882020. View Article : Google Scholar
|
|
14
|
Lund E, Oldenburg AR, Delbarre E, Freberg
CT, Duband-Goulet I, Eskeland R, Buendia B and Collas P: Lamin
A/C-promoter interactions specify chromatin state-dependent
transcription outcomes. Genome Res. 23:1580–1589. 2013. View Article : Google Scholar
|
|
15
|
Nmezi B, Xu J, Fu R, Armiger TJ,
Rodriguez-Bey G, Powell JS, Ma H, Sullivan M, Tu Y, Chen NY, et al:
Concentric organization of A- and B-type lamins predicts their
distinct roles in the spatial organization and stability of the
nuclear lamina. Proc Natl Acad Sci USA. 116:4307–4315. 2019.
View Article : Google Scholar
|
|
16
|
Kang SM, Yoon MH and Park BJ:
Laminopathies; mutations on single gene and various human genetic
diseases. BMB Rep. 51:327–337. 2018. View Article : Google Scholar
|
|
17
|
Sakthivel KM and Sehgal P: A novel role of
lamins from genetic disease to cancer biomarkers. Oncol Rev.
10:3092016.
|
|
18
|
Foster CR, Przyborski SA, Wilson RG and
Hutchison CJ: Lamins as cancer biomarkers. Biochem Soc Trans.
38:297–300. 2010. View Article : Google Scholar
|
|
19
|
Wang AS, Kozlov SV, Stewart CL and Horn
HF: Tissue specific loss of A-type lamins in the gastrointestinal
epithelium can enhance polyp size. Differentiation. 89:11–21. 2015.
View Article : Google Scholar
|
|
20
|
Fisher DZ, Chaudhary N and Blobel G: cDNA
sequencing of nuclear lamins A and C reveals primary and secondary
structural homology to intermediate filament proteins. Proc Natl
Acad Sci USA. 83:6450–6454. 1986. View Article : Google Scholar
|
|
21
|
Ishikawa H, Bischoff R and Holtzer H:
Mitosis and intermediate-sized filaments in developing skeletal
muscle. J Cell Biol. 38:538–555. 1968. View Article : Google Scholar
|
|
22
|
Lin F and Worman HJ: Structural
organization of the human gene encoding nuclear lamin A and nuclear
lamin C. J Biol Chem. 268:16321–16326. 1993. View Article : Google Scholar
|
|
23
|
Zwerger M and Medalia O: From lamins to
lamina: A structural perspective. Histochem Cell Biol. 140:3–12.
2013. View Article : Google Scholar
|
|
24
|
de Leeuw R, Gruenbaum Y and Medalia O:
Nuclear lamins: Thin filaments with major functions. Trends Cell
Biol. 28:34–45. 2018. View Article : Google Scholar
|
|
25
|
Gruenbaum Y and Medalia O: Lamins: The
structure and protein complexes. Curr Opin Cell Biol. 32:7–12.
2015. View Article : Google Scholar
|
|
26
|
Turgay Y, Eibauer M, Goldman AE, Shimi T,
Khayat M, Ben-Harush K, Dubrovsky-Gaupp A, Sapra KT, Goldman RD and
Medalia O: The molecular architecture of lamins in somatic cells.
Nature. 543:261–264. 2017. View Article : Google Scholar
|
|
27
|
Zwerger M, Roschitzki-Voser H, Zbinden R,
Denais C, Herrmann H, Lammerding J, Grütter MG and Medalia O:
Altering lamina assembly reveals lamina-dependent and -independent
functions for A-type lamins. J Cell Sci. 128:3607–3620. 2015.
|
|
28
|
Broers JLV, Peeters EAG, Kuijpers HJH,
Endert J, Bouten CVC, Oomens CWJ, Baaijens FPT and Ramaekers FCS:
Decreased mechanical stiffness in LMNA-/- cells is caused by
defective nucleo-cytoskeletal integrity: Implications for the
development of laminopathies. Hum Mol Genet. 13:2567–2580. 2004.
View Article : Google Scholar
|
|
29
|
Chiarini F, Evangelisti C, Cenni V, Fazio
A, Paganelli F, Martelli AM and Lattanzi G: The cutting edge: The
role of mTOR signaling in laminopathies. Int J Mol Sci. 20:8472019.
View Article : Google Scholar
|
|
30
|
Stick R: The gene structure of Xenopus
nuclear lamin A: A model for the evolution of A-type from B-type
lamins by exon shuffling. Chromosoma. 101:566–574. 1992. View Article : Google Scholar
|
|
31
|
Hanif M, Rosengardten Y, Sagelius H,
Rozell B and Eriksson M: Differential expression of A-type and
B-type lamins during hair cycling. PLoS One. 4:e41142009.
View Article : Google Scholar
|
|
32
|
Kim Y, Sharov AA, McDole K, Cheng M, Hao
H, Fan CM, Gaiano N, Ko MS and Zheng Y: Mouse B-type lamins are
required for proper organogenesis but not by embryonic stem cells.
Science. 334:1706–1710. 2011. View Article : Google Scholar
|
|
33
|
Al-Saaidi R and Bross P: Do lamin A and
lamin C have unique roles? Chromosoma. 124:1–12. 2015. View Article : Google Scholar
|
|
34
|
Gruenbaum Y and Foisner R: Lamins: Nuclear
intermediate filament proteins with fundamental functions in
nuclear mechanics and genome regulation. Annu Rev Biochem.
84:131–164. 2015. View Article : Google Scholar
|
|
35
|
Peter M, Nakagawa J, Dorée M, Labbé JC and
Nigg EA: In vitro disassembly of the nuclear lamina and M
phase-specific phosphorylation of lamins by cdc2 kinase. Cell.
61:591–602. 1990. View Article : Google Scholar
|
|
36
|
Collas P, Thompson L, Fields AP, Poccia DL
and Courvalin JC: Protein kinase C-mediated interphase lamin B
phosphorylation and solubilization. J Biol Chem. 272:21274–21280.
1997. View Article : Google Scholar
|
|
37
|
Molloy S and Little M: p34cdc2
kinase-mediated release of lamins from nuclear ghosts is inhibited
by cAMP-dependent protein kinase. Exp Cell Res. 201:494–499. 1992.
View Article : Google Scholar
|
|
38
|
Zhang YQ and Sarge KD: Sumoylation
regulates lamin A function and is lost in lamin A mutants
associated with familial cardiomyopathies. J Cell Biol. 182:35–39.
2008. View Article : Google Scholar
|
|
39
|
Shimi T, Kittisopikul M, Tran J, Goldman
AE, Adam SA, Zheng Y, Jaqaman K and Goldman RD: Structural
organization of nuclear lamins A, C, B1, and B2 revealed by
superresolution microscopy. Mol Biol Cell. 26:4075–4086. 2015.
View Article : Google Scholar
|
|
40
|
Grossman E, Dahan I, Stick R, Goldberg MW,
Gruenbaum Y and Medalia O: Filaments assembly of ectopically
expressed Caenorhabditis elegans lamin within Xenopus oocytes. J
Struct Biol. 177:113–118. 2012. View Article : Google Scholar
|
|
41
|
Kapinos LE, Schumacher J, Mücke N,
Machaidze G, Burkhard P, Aebi U, Strelkov SV and Herrmann H:
Characterization of the head-to-tail overlap complexes formed by
human lamin A, B1 and B2 ‘half-minilamin’ dimers. J Mol Biol.
396:719–731. 2010. View Article : Google Scholar
|
|
42
|
Goldberg MW, Huttenlauch I, Hutchison CJ
and Stick R: Filaments made from A- and B-type lamins differ in
structure and organization. J Cell Sci. 121:215–225. 2008.
View Article : Google Scholar
|
|
43
|
Schirmer EC and Gerace L: The stability of
the nuclear lamina polymer changes with the composition of lamin
subtypes according to their individual binding strengths. J Biol
Chem. 279:42811–42817. 2004. View Article : Google Scholar
|
|
44
|
Schirmer EC, Guan T and Gerace L:
Involvement of the lamin rod domain in heterotypic lamin
interactions important for nuclear organization. J Cell Biol.
153:479–489. 2001. View Article : Google Scholar
|
|
45
|
Moir RD, Yoon M, Khuon S and Goldman RD:
Nuclear lamins A and B1: Different pathways of assembly during
nuclear envelope formation in living cells. J Cell Biol.
151:1155–1168. 2000. View Article : Google Scholar
|
|
46
|
Fawcett DW: On the occurrence of a fibrous
lamina on the inner aspect of the nuclear envelope in certain cells
of vertebrates. Am J Anat. 119:129–145. 1966. View Article : Google Scholar
|
|
47
|
Belmont AS, Zhai Y and Thilenius A: Lamin
B distribution and association with peripheral chromatin revealed
by optical sectioning and electron microscopy tomography. J Cell
Biol. 123:1671–1685. 1993. View Article : Google Scholar
|
|
48
|
Taniura H, Glass C and Gerace L: A
chromatin binding site in the tail domain of nuclear lamins that
interacts with core histones. J Cell Biol. 131:33–44. 1995.
View Article : Google Scholar
|
|
49
|
Bruston F, Delbarre E, Ostlund C, Worman
HJ, Buendia B and Duband-Goulet I: Loss of a DNA binding site
within the tail of prelamin A contributes to altered
heterochromatin anchorage by progerin. FEBS Lett. 584:2999–3004.
2010. View Article : Google Scholar
|
|
50
|
Guelen L, Pagie L, Brasset E, Meuleman W,
Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W and
van Steensel B: Domain organization of human chromosomes revealed
by mapping of nuclear lamina interactions. Nature. 453:948–951.
2008. View Article : Google Scholar
|
|
51
|
Stewart CL, Kozlov S, Fong LG and Young
SG: Mouse models of the laminopathies. Exp Cell Res. 313:2144–2156.
2007. View Article : Google Scholar
|
|
52
|
Tesson F, Saj M, Uvaize MM, Nicolas H,
Płoski R and Bilińska Z: Lamin A/C mutations in dilated
cardiomyopathy. Cardiol J. 21:331–342. 2014. View Article : Google Scholar
|
|
53
|
Fatkin D, MacRae C, Sasaki T, Wolff MR,
Porcu M, Frenneaux M, Atherton J, Vidaillet HJ Jr, Spudich S, De
Girolami U, et al: Missense mutations in the rod domain of the
lamin A/C gene as causes of dilated cardiomyopathy and
conduction-system disease. N Engl J Med. 341:1715–1724. 1999.
View Article : Google Scholar
|
|
54
|
van Tintelen JP, Tio RA,
Kerstjens-Frederikse WS, van Berlo JH, Boven LG, Suurmeijer AJ,
White SJ, den Dunnen JT, te Meerman GJ, Vos YJ, et al: Severe
myocardial fibrosis caused by a deletion of the 5′ end of the lamin
A/C gene. J Am Coll Cardiol. 49:2430–2439. 2007. View Article : Google Scholar
|
|
55
|
Tiwari V, Alam MJ, Bhatia M, Navya M and
Banerjee SK: The structure and function of lamin A/C: Special focus
on cardiomyopathy and therapeutic interventions. Life Sci.
341:1224892024. View Article : Google Scholar
|
|
56
|
Kim Y, Bayona PW, Kim M, Chang J, Hong S,
Park Y, Budiman A, Kim YJ, Choi CY, Kim WS, et al: Macrophage Lamin
A/C regulates inflammation and the development of obesity-induced
insulin resistance. Front Immunol. 9:6962018. View Article : Google Scholar
|
|
57
|
Vigouroux C, Guénantin AC, Vatier C, Capel
E, Le Dour C, Afonso P, Bidault G, Béréziat V, Lascols O, Capeau J,
et al: Lipodystrophic syndromes due to LMNA mutations: recent
developments on biomolecular aspects, pathophysiological hypotheses
and therapeutic perspectives. Nucleus. 9:235–248. 2018. View Article : Google Scholar
|
|
58
|
Cao H and Hegele RA: Nuclear lamin A/C
R482Q mutation in canadian kindreds with Dunnigan-type familial
partial lipodystrophy. Hum Mol Genet. 9:109–112. 2000. View Article : Google Scholar
|
|
59
|
De Sandre-Giovannoli A, Bernard R, Cau P,
Navarro C, Amiel J, Boccaccio I, Lyonnet S, Stewart CL, Munnich A,
Le Merrer M and Lévy N: Lamin a truncation in Hutchinson-Gilford
progeria. Science. 300:20552003. View Article : Google Scholar
|
|
60
|
Eriksson M, Brown WT, Gordon LB, Glynn MW,
Singer J, Scott L, Erdos MR, Robbins CM, Moses TY, Berglund P, et
al: Recurrent de novo point mutations in lamin A cause
Hutchinson-Gilford progeria syndrome. Nature. 423:293–298. 2003.
View Article : Google Scholar
|
|
61
|
Wong X, Melendez-Perez AJ and Reddy KL:
The nuclear lamina. Cold Spring Harb Perspect Biol. 14:a0401132022.
View Article : Google Scholar
|
|
62
|
Dechat T, Shimi T, Adam SA, Rusinol AE,
Andres DA, Spielmann HP, Sinensky MS and Goldman RD: Alterations in
mitosis and cell cycle progression caused by a mutant lamin A known
to accelerate human aging. Proc Natl Acad Sci USA. 104:4955–4960.
2007. View Article : Google Scholar
|
|
63
|
Ragnauth CD, Warren DT, Liu Y, McNair R,
Tajsic T, Figg N, Shroff R, Skepper J and Shanahan CM: Prelamin A
acts to accelerate smooth muscle cell senescence and is a novel
biomarker of human vascular aging. Circulation. 121:2200–2210.
2010. View Article : Google Scholar
|
|
64
|
Kirkland NJ, Skalak SH, Whitehead AJ,
Hocker JD, Beri P, Vogler G, Hum B, Wang M, Lakatta EG, Ren B, et
al: Age-dependent Lamin changes induce cardiac dysfunction via
dysregulation of cardiac transcriptional programs. Nat Aging.
3:17–33. 2023. View Article : Google Scholar
|
|
65
|
Maynard S, Hall A, Galanos P, Rizza S,
Yamamoto T, Gram HH, Munk SHN, Shoaib M, Sørensen CS, Bohr VA, et
al: Lamin A/C impairments cause mitochondrial dysfunction by
attenuating PGC1α and the NAMPT-NAD+ pathway. Nucleic Acids Res.
50:9948–9965. 2022. View Article : Google Scholar
|
|
66
|
Simon M, Yang J, Gigas J, Earley EJ,
Hillpot E, Zhang L, Zagorulya M, Tombline G, Gilbert M, Yuen SL, et
al: A rare human centenarian variant of SIRT6 enhances genome
stability and interaction with Lamin A. EMBO J. 41:e1103932022.
View Article : Google Scholar
|
|
67
|
Yoon MH, Kang SM, Lee SJ, Woo TG, Oh AY,
Park S, Ha NC and Park BJ: p53 induces senescence through Lamin A/C
stabilization-mediated nuclear deformation. Cell Death Dis.
10:1072019. View Article : Google Scholar
|
|
68
|
Lochs SJA, Kefalopoulou S and Kind J:
Lamina associated domains and gene regulation in development and
cancer. Cells. 8:2712019. View Article : Google Scholar
|
|
69
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar
|
|
70
|
Broers JLV and Ramaekers FCS: The role of
the nuclear lamina in cancer and apoptosis. Adv Exp Med Biol.
773:27–48. 2014. View Article : Google Scholar
|
|
71
|
Broers JL, Machiels BM, Kuijpers HJ,
Smedts F, van den Kieboom R, Raymond Y and Ramaekers FC: A- and
B-type lamins are differentially expressed in normal human tissues.
Histochem Cell Biol. 107:505–517. 1997. View Article : Google Scholar
|
|
72
|
Kaufmann SH, Mabry M, Jasti R and Shaper
JH: Differential expression of nuclear envelope lamins A and C in
human lung cancer cell lines. Cancer Res. 51:581–566. 1991.
|
|
73
|
Guinde J, Frankel D, Perrin S, Delecourt
V, Lévy N, Barlesi F, Astoul P, Roll P and Kaspi E: Lamins in lung
cancer: Biomarkers and Key factors for disease progression through
miR-9 regulation? Cells. 7:782018. View Article : Google Scholar
|
|
74
|
Machiels BM, Broers JL, Raymond Y, de Ley
L, Kuijpers HJ, Caberg NE and Ramaekers FC: Abnormal A-type lamin
organization in a human lung carcinoma cell line. Eur J Cell Biol.
67:328–335. 1995.
|
|
75
|
Capo-chichi CD, Cai KQ, Smedberg J,
Ganjei-Azar P, Godwin AK and Xu XX: Loss of A-type lamin expression
compromises nuclear envelope integrity in breast cancer. Chin J
Cancer. 30:415–425. 2011. View Article : Google Scholar
|
|
76
|
Harris H: Concerning the origin of
malignant tumours by Theodor Boveri. Translated and annotated by
Henry Harris. Preface. J Cell Sci. 121 (Suppl 1):S5–S6. 2008.
|
|
77
|
Holland AJ and Cleveland DW: Boveri
revisited: Chromosomal instability, aneuploidy and tumorigenesis.
Nat Rev Mol Cell Biol. 10:478–487. 2009. View Article : Google Scholar
|
|
78
|
Roschke AV, Tonon G, Gehlhaus KS, McTyre
N, Bussey KJ, Lababidi S, Scudiero DA, Weinstein JN and Kirsch IR:
Karyotypic complexity of the NCI-60 drug-screening panel. Cancer
Res. 63:8634–8647. 2003.
|
|
79
|
Thompson SL, Bakhoum SF and Compton DA:
Mechanisms of chromosomal instability. Curr Biol. 20:R285–R295.
2010. View Article : Google Scholar
|
|
80
|
Sen S: Aneuploidy and cancer. Curr Opin
Oncol. 12:82–88. 2000. View Article : Google Scholar
|
|
81
|
Ozols RF, Bookman MA, Connolly DC, Daly
MB, Godwin AK, Schilder RJ, Xu X and Hamilton TC: Focus on
epithelial ovarian cancer. Cancer Cell. 5:19–24. 2004. View Article : Google Scholar
|
|
82
|
Agarwal R and Kaye SB: Ovarian cancer:
Strategies for overcoming resistance to chemotherapy. Nat Rev
Cancer. 3:502–516. 2003. View Article : Google Scholar
|
|
83
|
Capo-Chichi CD, Cai KQ and Xu XX:
Overexpression and cytoplasmic localization of caspase-6 is
associated with lamin A degradation in set of ovarian cancers.
Biomarker Res. 6:302018. View Article : Google Scholar
|
|
84
|
Agrelo R, Setien F, Espada J, Artiga MJ,
Rodriguez M, Pérez-Rosado A, Sanchez-Aguilera A, Fraga MF, Piris MA
and Esteller M: Inactivation of the lamin A/C gene by CpG island
promoter hypermethylation in hematologic malignancies, and its
association with poor survival in nodal diffuse large B-cell
lymphoma. J Clin Oncol. 23:3940–3947. 2005. View Article : Google Scholar
|
|
85
|
Maresca G, Natoli M, Nardella M, Arisi I,
Trisciuoglio D, Desideri M, Brandi R, D'Aguanno S, Nicotra MR,
D'Onofrio M, et al: LMNA knock-down affects differentiation and
progression of human neuroblastoma cells. PLoS One. 7:e455132012.
View Article : Google Scholar
|
|
86
|
Venables RS, McLean S, Luny D, Moteleb E,
Morley S, Quinlan RA, Lane EB and Hutchison CJ: Expression of
individual lamins in basal cell carcinomas of the skin. Br J
Cancer. 84:512–519. 2001. View Article : Google Scholar
|
|
87
|
Foster CR, Robson JL, Simon WJ, Twigg J,
Cruikshank D, Wilson RG and Hutchison CJ: The role of Lamin A in
cytoskeleton organization in colorectal cancer cells: A proteomic
investigation. Nucleus. 2:434–243. 2011. View Article : Google Scholar
|
|
88
|
Kong L, Schäfer G, Bu H, Zhang Y, Zhang Y
and Klocker H: Lamin A/C protein is overexpressed in
tissue-invading prostate cancer and promotes prostate cancer cell
growth, migration and invasion through the PI3K/AKT/PTEN pathway.
Carcinogenesis. 33:751–759. 2012. View Article : Google Scholar
|
|
89
|
Bell ES, Shah P, Zuela-Sopilniak N, Kim D,
Varlet AA, Morival JLP, McGregor AL, Isermann P, Davidson PM,
Elacqua JJ, et al: Low lamin A levels enhance confined cell
migration and metastatic capacity in breast cancer. Oncogene.
41:4211–4230. 2022. View Article : Google Scholar
|
|
90
|
Wang Y, Jiang J, He L, Gong G and Wu X:
Effect of lamin-A expression on migration and nuclear stability of
ovarian cancer cells. Gynecol Oncol. 152:166–176. 2019. View Article : Google Scholar
|
|
91
|
Miles FL, Pruitt FL, van Golen KL and
Cooper CR: Stepping out of the flow: Capillary extravasation in
cancer metastasis. Clin Exp Metastasis. 25:305–324. 2008.
View Article : Google Scholar
|
|
92
|
Valastyan S and Weinberg RA: Tumor
metastasis: Molecular insights and evolving paradigms. Cell.
147:275–292. 2011. View Article : Google Scholar
|
|
93
|
Roncato F, Regev O, Feigelson SW, Yadav
SK, Kaczmarczyk L, Levi N, Drago-Garcia D, Ovadia S, Kizner M,
Addadi Y, et al: Reduced Lamin A/C does not facilitate cancer cell
transendothelial migration but compromises lung metastasis. Cancers
(Basel). 13:23832021. View Article : Google Scholar
|
|
94
|
Mitchell MJ, Denais C, Chan MF, Wang Z,
Lammerding J and King MR: Lamin A/C deficiency reduces circulating
tumor cell resistance to fluid shear stress. Am J Physiol Cell
Physiol. 309:C736–C746. 2015. View Article : Google Scholar
|
|
95
|
Ferrari R, Infante E and Chavrier P:
Nucleus-invadopodia duo during cancer invasion. Trends Cell Biol.
29:93–96. 2019. View Article : Google Scholar
|
|
96
|
Osmanagic-Myers S, Dechat T and Foisner R:
Lamins at the crossroads of mechanosignaling. Genes Dev.
29:225–237. 2015. View Article : Google Scholar
|
|
97
|
Kaspi E, Frankel D, Guinde J, Perrin S,
Laroumagne S, Robaglia-Schlupp A, Ostacolo K, Harhouri K,
Tazi-Mezalek R, Micallef J, et al: Low lamin A expression in lung
adenocarcinoma cells from pleural effusions is a pejorative factor
associated with high number of metastatic sites and poor
performance status. PLoS One. 12:e01831362017. View Article : Google Scholar
|
|
98
|
Willis ND, Cox TR, Rahman-Casañs SF, Smits
K, Przyborski SA, van den Brandt P, van Engeland M, Weijenberg M,
Wilson RG, de Bruïne A and Hutchison CJ: Lamin A/C is a risk
biomarker in colorectal cancer. PLoS One. 3:e29882008. View Article : Google Scholar
|
|
99
|
Setti Boubaker N, Gurtner A, Trabelsi N,
Manni I, Ayed H, Saadi A, Zaghbib S, Naimi Z, Sahraoui G, Zouari S,
et al: The diagnostic applicability of A-type Lamin in non-muscle
invasive bladder cancer. Ann Diagn Pathol. 54:1518082021.
View Article : Google Scholar
|
|
100
|
Wazir U, Ahmed MH, Bridger JM, Harvey A,
Jiang WG, Sharma AK and Mokbel K: The clinicopathological
significance of lamin A/C, lamin B1 and lamin B receptor mRNA
expression in human breast cancer. Cell Mol Biol Lett. 18:595–611.
2013. View Article : Google Scholar
|
|
101
|
Alhudiri IM, Nolan CC, Ellis IO, Elzagheid
A, Rakha EA, Green AR and Chapman CJ: Expression of Lamin A/C in
early-stage breast cancer and its prognostic value. Breast Cancer
Res Treat. 174:661–668. 2019. View Article : Google Scholar
|
|
102
|
Smith ER, George SH, Kobetz E and Xu XX:
New biological research and understanding of Papanicolaou's test.
Diagn Cytopathol. 46:507–515. 2018. View Article : Google Scholar
|
|
103
|
Dekker E, Tanis PJ, Vleugels JLA, Kasi PM
and Wallace MB: Colorectal cancer. Lancet. 394:1467–1480. 2019.
View Article : Google Scholar
|
|
104
|
Thompson SL and Compton DA: Proliferation
of aneuploid human cells is limited by a p53-dependent mechanism. J
Cell Biol. 188:369–381. 2010. View Article : Google Scholar
|
|
105
|
Saarinen I, Mirtti T, Seikkula H, Boström
PJ and Taimen P: Differential predictive roles of A- and B-type
nuclear lamins in prostate cancer progression. PLoS One.
10:e01406712015. View Article : Google Scholar
|
|
106
|
Meaburn KJ and Misteli T: Assessment of
the utility of gene positioning biomarkers in the stratification of
prostate cancers. Front Genet. 10:10292019. View Article : Google Scholar
|
|
107
|
Grozescu T and Popa F: Prostate cancer
between prognosis and adequate/proper therapy. J Med Life. 10:5–12.
2017.
|
|
108
|
Setijono SR, Park M, Kim G, Kim Y, Cho KW
and Song SJ: miR-218 and miR-129 regulate breast cancer progression
by targeting Lamins. Biochem Biophys Res Commun. 496:826–833. 2018.
View Article : Google Scholar
|
|
109
|
Chiarini F, Paganelli F, Balestra T,
Capanni C, Fazio A, Manara MC, Landuzzi L, Petrini S, Evangelisti
C, Lollini PL, et al: Lamin A and the LINC complex act as potential
tumor suppressors in Ewing Sarcoma. Cell Death Dis. 13:3462022.
View Article : Google Scholar
|
|
110
|
Snigireva AV, Vrublevskaya VV, Skarga YY,
Evdokimovskaya YV and Morenkov OS: Effect of heat shock protein 90
(Hsp90) on migration and invasion of human cancer cells in vitro.
Bull Exp Biol Med. 157:476–478. 2014. View Article : Google Scholar
|
|
111
|
Meng X, Cao J, Zheng H, Ma X, Wang Y, Tong
Y, Xie S, Lu R and Guo L: TPX2 promotes ovarian tumorigenesis by
interacting with Lamin A/C and affecting its stability. Cancer Med.
12:9738–9748. 2023. View Article : Google Scholar
|
|
112
|
Sidera K and Patsavoudi E: HSP90
inhibitors: Current development and potential in cancer therapy.
Recent Pat Anticancer Drug Discov. 9:1–20. 2014. View Article : Google Scholar
|
|
113
|
Wang Y, Chen Q, Wu D, Chen Q, Gong G, He L
and Wu X: Lamin-A interacting protein Hsp90 is required for DNA
damage repair and chemoresistance of ovarian cancer cells. Cell
Death Dis. 12:7862021. View Article : Google Scholar
|
|
114
|
Shao X, Tarnasky HA, Lee JP, Oko R and van
der Hoorn FA: Spag4, a novel sperm protein, binds outer dense-fiber
protein Odf1 and localizes to microtubules of manchette and
axoneme. Dev Biol. 211:109–123. 1999. View Article : Google Scholar
|
|
115
|
Shoji K, Murayama T, Mimura I, Wada T,
Kume H, Goto A, Ohse T, Tanaka T, Inagi R, van der Hoorn FA, et al:
Sperm-associated antigen 4, a novel hypoxia-inducible factor 1
target, regulates cytokinesis, and its expression correlates with
the prognosis of renal cell carcinoma. Am J Pathol. 182:2191–2203.
2013. View Article : Google Scholar
|
|
116
|
Elhanati S, Kanfi Y, Varvak A, Roichman A,
Carmel-Gross I, Barth S, Gibor G and Cohen HY: Multiple regulatory
layers of SREBP1/2 by SIRT6. Cell Rep. 4:905–912. 2013. View Article : Google Scholar
|
|
117
|
Ghosh S, Liu B, Wang Y, Hao Q and Zhou Z:
Lamin A is an endogenous SIRT6 activator and promotes
SIRT6-mediated DNA repair. Cell Rep. 13:1396–1406. 2015. View Article : Google Scholar
|
|
118
|
Li H, Ge C, Zhao F, Yan M, Hu C, Jia D,
Tian H, Zhu M, Chen T, Jiang G, et al: Hypoxia-inducible factor 1
alpha-activated angiopoietin-like protein 4 contributes to tumor
metastasis via vascular cell adhesion molecule-1/integrin β1
signaling in human hepatocellular carcinoma. Hepatology.
54:910–919. 2011. View Article : Google Scholar
|
|
119
|
Zhao J, Liu B, Yang JA, Tang D, Wang X and
Chen Q: Human sperm-associated antigen 4 as a potential biomarker
of glioblastoma progression and prognosis. Neuroreport. 30:446–451.
2019. View Article : Google Scholar
|
|
120
|
Liu T, Yu J, Ge C, Zhao F, Chen J, Miao C,
Jin W, Zhou Q, Geng Q, Lin H, et al: Sperm associated antigen 4
promotes SREBP1-mediated de novo lipogenesis via interaction with
lamin A/C and contributes to tumor progression in hepatocellular
carcinoma. Cancer Lett. 536:2156422022. View Article : Google Scholar
|
|
121
|
Eferl R and Wagner EF: AP-1: A
double-edged sword in tumorigenesis. Nat Rev Cancer. 3:859–868.
2003. View Article : Google Scholar
|
|
122
|
Shaulian E and Karin M: AP-1 as a
regulator of cell life and death. Nat Cell Biol. 4:E131–E136. 2002.
View Article : Google Scholar
|
|
123
|
Ding Y, Hao K, Li Z, Ma R, Zhou Y, Zhou Z,
Wei M, Liao Y, Dai Y, Yang Y, et al: c-Fos separation from Lamin
A/C by GDF15 promotes colon cancer invasion and metastasis in
inflammatory microenvironment. J Cell Physiol. 235:4407–4421. 2020.
View Article : Google Scholar
|
|
124
|
Sharma P and Kuehn MR: SENP1-modulated
sumoylation regulates retinoblastoma protein (RB) and Lamin A/C
interaction and stabilization. Oncogene. 35:6429–6438. 2016.
View Article : Google Scholar
|
|
125
|
Elenbaas JS, Bragazzi Cunha J,
Azuero-Dajud R, Nelson B, Oral EA, Williams JA, Stewart CL and
Omary MB: Lamin A/C maintains exocrine pancreas homeostasis by
regulating stability of RB and activity of E2F. Gastroenterology.
154:1625–1629.e8. 2018. View Article : Google Scholar
|