In the contemporary era, diabetes has emerged as a
pervasive and critical chronic illness. The global prevalence of
diabetes is projected to increase to 643 million by 2030 and 783
million by 2045, representing a significant threat to human life
and health (1). Chronic diabetic
wounds represent a significant complication of diabetes that also
presents a substantial global public health challenge. When
patients with diabetes develop foot ulcers, they are often at risk
of subsequent osteomyelitis, amputation and even mortality
(2). The global prevalence of
diabetic foot is 6.3% and the risk of developing foot ulcers ranges
between 19–34% among patients with diabetes (3). Owing to the challenges associated
with wound healing in patients with diabetes, as well as the
elevated morbidity and mortality rates in the population, this
problem has received increasing attention in the biomedical
field.
A growing body of evidence indicates that
macrophages play a crucial role in the process of diabetic wound
healing. During normal wound healing, there is a gradual transition
in the macrophage phenotype, shifting from the initial M1 phenotype
associated with the acute response, to the later M2 phenotype that
promotes healing. However, in diabetic individuals, this phenotypic
macrophage imbalance hinders the resolution of inflammation,
resulting in persistent nonhealing of wounds (4). Accordingly, the modulation of
macrophage polarization has become a key objective in the
management of diabetic wounds.
The advent of novel high-throughput technologies has
revealed epigenetic mechanisms governing gene expression in
macrophages, which play a pivotal role in modulating their
plasticity (5). The present review
aimed to elucidate the mechanisms behind macrophage involvement in
diabetic wounds and the epigenetic factors that govern macrophage
polarization in diabetic wounds, with the aim of furthering our
understanding of the pathogenesis behind diabetic wounds and
identifying potential targets for clinical interventions.
Macrophages, a vital element of the immune system,
are instrumental in maintaining homeostasis, preserving tissue
integrity and regulating inflammatory processes (6–8). The
distinguishing features of macrophages include their functional
plasticity and diversity (9).
Macrophage polarization is characterized by distinct functional and
phenotypic characteristics in response to local microenvironmental
stimuli. Macrophages can be conventionally categorized as M1 or M2
(10–12). M1 macrophages represent a
pro-inflammatory phenotype. They are also referred to as classical
macrophages, can initiate inflammation and possess potent
antimicrobial properties. However, they also carry the potential to
induce tissue damage. By contrast, M2 macrophages, also known as
alternatively activated macrophages, exhibit an anti-inflammatory
phenotype and facilitate a state that is conducive to
anti-inflammatory responses and tissue repair (13,14).
Macrophage polarization determines the fate of organs and tissues
during inflammation or injury. In the preliminary phases of an
inflammatory response, macrophage polarization into the M1 form is
initiated through the classical pathway by lipopolysaccharide
(LPS), a key outer membrane component in gram-negative bacteria.
Moreover, cytokines such as interferon (IFN)-γ, tumor necrosis
factor (TNF) and granulocyte-macrophage colony-stimulating factor
also contribute to macrophage activation. M1 macrophages actively
secrete a variety of pro-inflammatory cytokines, including
interleukin (IL)-6, TNF, IL-1β, IFN-β, IL-12 and IL-23 and nitric
oxide. These substances work in conjunction to facilitate rapid and
efficient physiological responses to infection and tissue damage
(15,16). It is also noteworthy that M1
macrophages can regulate immune response with the help of Th1 and
Th17 cells. Through complex cell signaling and molecular
interactions, they can help to efficiently eliminate pathogens and
maintain the immune balance within the body. Prolonged polarization
of macrophages in the M1 state may lead to tissue damage (17). In the advanced stages of the
inflammatory response, M2 macrophages are activated through
alternative pathways, resulting in the production of various
anti-inflammatory cytokines. This facilitates the amelioration of
inflammatory responses and promotes tissue damage repair (18). Based on the corresponding functions
induced by various stimulants, M2 macrophages can be categorized
into four distinct subtypes: M2a, M2b, M2c and M2d. T-helper (Th)2
cells and M2 macrophages can engage in reciprocal interactions to
maintain immune homeostasis. Stimulated by the IL-4 and IL-13
secreted by Th2 cells, M2a macrophages, also known as wound-healing
macrophages, express markers such as the mannose receptor (MR or
CD206) and decoy IL-1 receptor (IL-1R). They secrete
anti-inflammatory factors including IL-10, transforming growth
factor (TGF)-β, arginase-1 (Arg-1), C-C motif chemokine ligand
(CCL)17, CCL18, CCL22 and CCL24 to facilitate the resolution of
chronic inflammation and accelerate the process of wound healing
(19–22). Specifically, M2a macrophages
secrete IL-10, which in turn can suppress the activity of Th1 cells
and other pro-inflammatory cells, indirectly maintaining the
relative advantage of Th2 cells and promoting Th2-type immune
responses, thus playing a role in wound healing and tissue
remodeling (23). M2b macrophages,
which have also been described as regulatory macrophages, are
primarily triggered by the presence of immune complexes, Toll-like
receptor (TLR) agonists, LPS, or IL-1β. They express various
surface protein markers, including CD86, IL-10R, IL-12R and TNF
superfamily member 14 (TNFSF14) (24–26).
M2b macrophages have been observed to exhibit dual regulatory
functions characterized by the production of pro-inflammatory
factors (IL-1β, IL-6 and TNF-α), as well as high expression of the
anti-inflammatory factor IL-10 and low expression of IL-12.
Consequently, they can also serve to inhibit immune-inflammatory
responses (20,23,25).
M2c cells, also known as inactivated macrophages, are primarily
stimulated by glucocorticoids, IL-10 and TGF-β. Their main surface
protein markers include CD163, CD206, TLR-1 and TLR-8. These
macrophages secrete IL-10, TGF-β, CCL13, CCL16, CCL18 and other
cytokines that contribute to immunosuppression and the phagocytosis
of apoptotic cells (27,28). Tumor-associated M2d macrophages can
be induced by IL-6 or a combination of TLR ligands and A2a
adenosine receptor agonists. Their surface markers include IL-10R
and IL-12R. M2d macrophages are capable of secreting cytokines that
facilitate angiogenesis and tumorigenesis, including IL-10, IL-12,
TNF-α, TGF-β and vascular endothelial growth factor (VEGF)
(29–32). Despite their opposing functions, M1
and M2 macrophages can transition from one phenotype to the other
in specific states to collectively maintain the dynamic equilibrium
of the immune system. Additionally, changes in extracellular matrix
(ECM) composition, signaling by other immune cells and metabolic
state changes can affect macrophage plasticity. Changes in ECM
components such as collagen and fibronectin can interact with
macrophage cell surface receptors to regulate their activation
states (33). Other immune cells,
such as B cells and dendritic cells, release signaling molecules
that can influence the polarization direction and function of
macrophages. Metabolic state changes such as hypoxia and nutrient
imbalance can prompt macrophages to adjust their metabolic
pathways, thereby affecting their phenotypes and functions
(34). These factors enable
macrophages to play different roles in complex environments, thus
assisting in both immune regulation and tissue repair (6).
The process of wound healing is a complex biological
phenomenon that occurs in four distinct, overlapping and highly
regulated stages: Hemostasis, inflammation, proliferation and
remodeling (35,36). Each requires a particular sequence,
timing and duration at an optimal intensity to achieve effective
wound healing. Disruption of any of these stages can result in
delayed wound healing. Hemostasis, the initial phase, involves
three processes: Vasoconstriction, primary hemostasis and secondary
hemostasis. Following injury, blood vessels in the vicinity quickly
constrict to minimize the bleeding caused by damage to the
microvascular system. Primary and secondary hemostasis occur via
two parallel and mechanically linked pathways (37). After a wound is sustained and blood
vessels rupture, the endo-subcutaneous thrombotic matrix is
exposed, which initiates primary hemostasis. This process includes
platelet aggregation and the formation of platelet thrombi
(38). Secondary hemostasis
entails activation of the clotting cascade, which culminates in the
conversion of soluble fibrinogen into insoluble fibrin chains that
collectively constitute the fibrin network. The platelet plug
adheres to the fibrin network, forming a clot that stops bleeding
and releases complement and growth factors, including TGF-β,
platelet-derived growth factor (PDGF), fibroblast growth factor
(FGF) and epidermal growth factor (EGF). Furthermore, it serves as
a provisional scaffold for infiltrating cells, which are vital for
the process of wound healing (39–41).
It has been identified that macrophages play a critical role in
this part of wound healing (42).
Although not directly involved in the hemostatic phase, they are
essential for coordinating various transitions that take place
among the subsequent phases of inflammation, proliferation and
remodeling.
Once hemostasis is achieved, the wound enters an
inflammatory phase characterized by persistent infiltration of
neutrophils, macrophages and lymphocytes (43). Neutrophils are of particular
importance during the initial stages of inflammation, primarily
functioning in an anti-infective capacity (44). Macrophages are critical regulatory
cells involved in inflammatory responses. Following tissue injury,
resident dermal macrophages are the earliest responders, initiating
an inflammatory response by releasing hydrogen peroxide that leads
to the sequential recruitment of neutrophils and monocytes to the
site. The recruited monocytes then further differentiate into
macrophages (45). The TLR family
plays a pivotal role in macrophage polarization, with particular
emphasis on TLR2 and TLR4. TLR2 recognizes diverse
pathogen-associated molecular patterns (PAMPs) and
damage-associated molecular patterns, whereas TLR4 predominantly
identifies LPS. Upon binding to their respective ligands, TLR2 and
4 recruit myeloid differentiation primary response 88 (MyD88),
leading to its activation, as well as the subsequent activation of
TNF receptor-associated factor 6 (TRAF6). TRAF6 activation results
in nuclear translocation of NF-κB, which initiates the
transcription of pro-inflammatory cytokines and other genes, as
well as activation of the MAPK pathway (46–48).
These pro-inflammatory cytokines and their associated transcription
factors collectively drive macrophage polarization toward the M1
phenotype, resulting in an early-stage inflammatory response
involving pathogen clearance and tissue repair. Owing to their
unique recognition mechanism, M1 macrophages can accurately
identify PAMPs on the surface of bacteria or fungi, thereby forming
phagocytic lysosomes. M1 macrophages then release potent
antibacterial mediators such as reactive oxygen species (ROS) and
active nitrogen into the surrounding environment to effectively
eliminate pathogens (49,50). The excessive and uncontrolled
release of inflammatory cytokines has a significant detrimental
effect on the ability of the body to repair damaged tissues. These
cytokines exacerbate the effects of ROS on tissues and prolong the
inflammatory response, which should have resolved itself, thus
impeding essential tissue regeneration processes (51,52).
Macrophages are also involved in the clearance of cellular debris
and apoptotic neutrophils (53).
In the later stages of the inflammatory response, TLR-induced
inflammation is gradually attenuated, allowing for continued wound
healing (48). At this stage,
macrophages typically undergo a gradual transition to the M2
phenotype and secrete substantial amounts of anti-inflammatory
cytokines, which strongly facilitate the gradual resolution of
inflammation and tissue healing.
The neuroimmune axis plays an important role in the
healing process. Complex interactions occur between
neurotransmitters released by nerve fibers and immune cells. For
example, neuropeptides such as Substance P (SP) can increase the
levels of inflammatory factors such as TGF-β, TNF-α, IL-1β, IL-2,
IL-8 and IL-6 that are released by dendritic cells, T cells,
neutrophils and macrophages (54).
SP also ensures extravasation, migration and subsequent
accumulation of white blood cells at the site of injury, thereby
creating an inflammatory microenvironment that further ensures the
proliferation and angiogenesis of endothelial cells and promotes
wound healing (55).
The transition from M1 to M2 macrophages indicates
the onset of the proliferative phase of wound healing, which
involves angiogenesis and re-epithelialization (40,41).
A prominent characteristic of this phase is the extensive
activation of endothelial cells, fibroblasts, keratinocytes and
macrophages (56). As mesenchymal
cells, fibroblasts are widely distributed throughout a range of
tissues and play a pivotal role in the promotion of granulation
tissue formation and the replacement of transient stroma during the
proliferative process (57).
Fibroblasts derived from different sources are mobilized to the
wound site, where they proliferate to bridge gaps in the wound and
facilitate the generation of new ECM (58). Initially, vascularized ECM
transforms into granulation tissue and keratinocyte-represented
epithelial cells undergo proliferation and migration toward the
granulation tissue, thereby facilitating re-epithelialization. The
anti-inflammatory and proliferative characteristics of M2
macrophages have been well documented. M2 macrophages possess the
capacity to increase levels of anti-inflammatory factors, lower
levels of pro-inflammatory ones and produce a considerable quantity
of growth factors, including PDGF, EGF, VEGF, TGF-β1 and
insulin-like growth factor 1. These growth factors facilitate cell
proliferation, the formation of granulation tissue and angiogenesis
(59–62). M2a macrophages not only effectively
inhibit the inflammatory response and reduce inflammation-induced
tissue damage but also actively promote the normal development and
functional maintenance of blood vessels, thereby providing the
necessary conditions for wound healing (63). It has also been demonstrated that
M2c macrophages can facilitate the migration of vascular
endothelial cells and enhance angiogenesis (64). Additionally, M2c macrophages
secrete matrix metalloproteinase (MMP)-9 to recruit blood vessels
and blood-derived stem cells to the injury site. This promotes
angiogenesis, phagocytosis of wound debris and the deposition of
ECM components (63,65).
The final stage of wound healing, known as
remodeling or regression, is a complex and protracted process
involving tissue restructuring and the enhancement of wound
strength. This phase involves ECM reorganization, wound contraction
and scar maturation (66).
Fibroblasts facilitate wound contraction by differentiating into
myofibroblasts, which further promote wound contraction. New
collagen scaffolds are formed during tissue repair and
reconstruction. If the original ECM is not degraded in a timely
manner, new tissue formation is impeded. MMPs target the
degradation of existing ECM components, thereby creating favorable
conditions for the integration of new collagen scaffolds and tissue
remodeling. Collagen I replaces collagen III as the primary ECM
component, leading to remodeling and increased tensile strength.
The main producers of MMPs are macrophages and the naturally
occurring tissue inhibitors of metalloproteinases are responsible
for controlling their activity (67). During tissue remodeling, the
population of macrophages decreases and the remaining ones are
involved in regulating collagen and the ECM (9,61,68,69).
In addition to MMP release, M2 macrophages also regulate collagen
turnover via the mannose receptor (70).
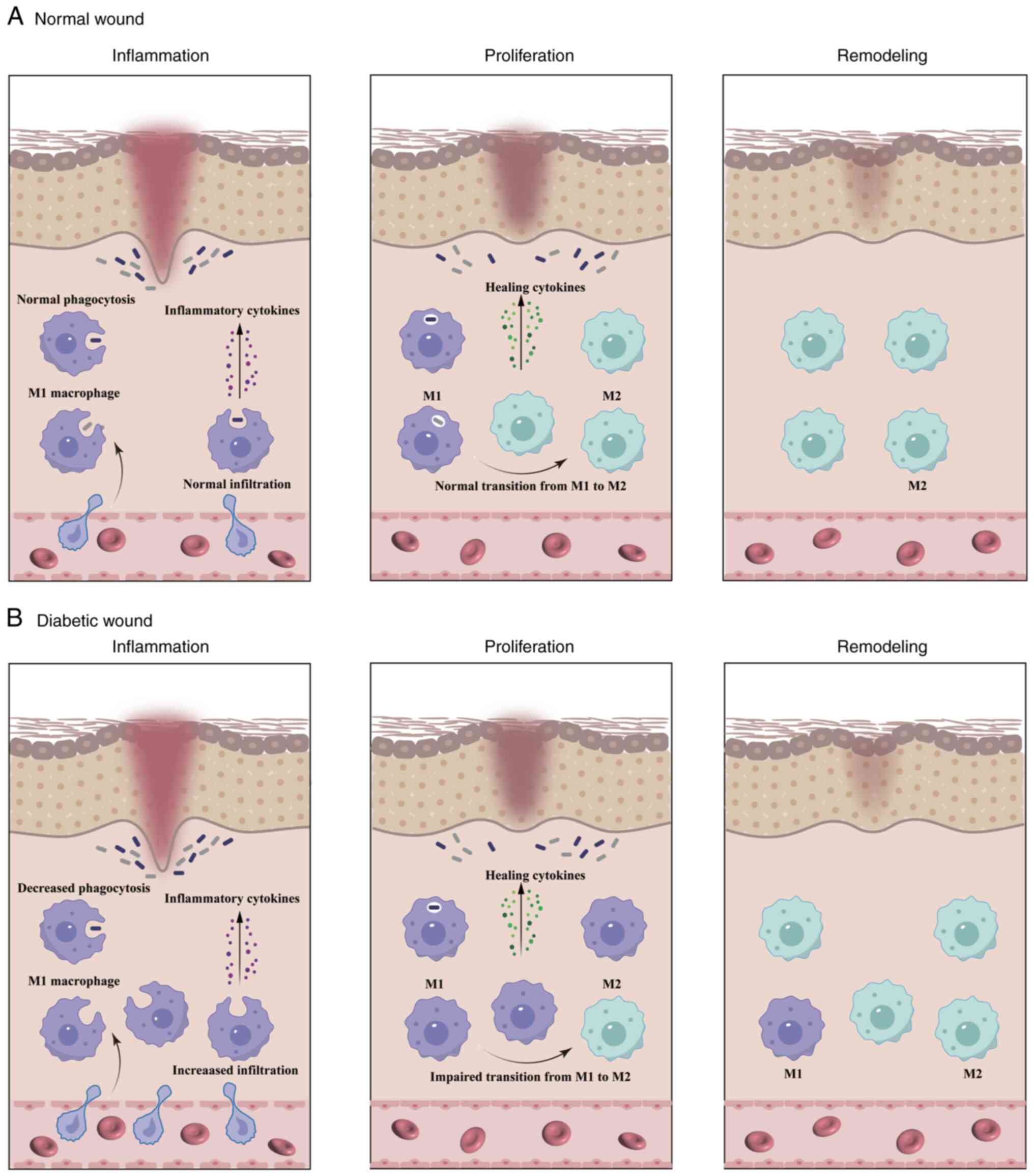
The wound-healing process in diabetes is halted at a
specific stage, typically the inflammatory phase, and fails to
advance further. Dysregulated macrophage phenotypes and functions
represent critical factors that contribute to persistent
non-healing in diabetic wounds (Fig.
1). These findings indicate that the diabetic microenvironment
stimulates an increased production of hematopoietic stem cells in
the bone marrow, which then differentiate into a greater number of
monocytes entering the peripheral blood. These monocytes then
migrate to the wound and transform into M1 macrophages, which are
characterized by IL-1β and TNF-α markers. This exacerbates the
inflammatory response at the wound site and impedes normal wound
healing (71). Barman et al
(72) corroborate this conclusion,
demonstrating a higher number of monocyte infiltrations in the
wounds of mice with type 2 diabetes mellitus (T2D) compared with
healthy mice. Elevated TLR2 and 4 levels in diabetic wounds have
been shown to result in sustained activation of pro-inflammatory
signals and the persistence of M1 macrophages, thereby impeding
progression to the next healing stage (73–75).
Macrophages in diabetic mice display a sustained increase in
M1-like macrophage markers, including nitric oxide synthase 2,
TNF-α, IL-1β and MMP9, and decreased M2-like macrophage markers
such as Arginase 1, CD206 and CD36 (76,77).
Similarly, diabetic foot ulcers in humans show increased expression
of M1-like markers such as CD68 and IL-1β and decreased expression
of M2-like markers such as CD163, CD206 and Arg-1 (76,78).
Persistent hyperglycemia and oxidative stress have been shown to
synergistically exacerbate the polarization propensity of M1
macrophages, leading to sustained secretion of potent
pro-inflammatory mediators that inflict severe damage to wounded
tissues (42,56). Therefore, the persistence of the
pro-inflammatory M1 phenotype and deficiency of M2-type macrophages
in diabetic wounds may contribute to an unbridled pro-inflammatory
microenvironment (79).
Furthermore, macrophages in diabetic wounds exhibit reduced
bactericidal and phagocytic activities, impairing their ability to
effectively eliminate dead tissue and pathogens. This results in
prolonged inflammation that further complicates the healing process
(80,81). Patients with diabetes often have
neuropathy and immune dysfunction as well, resulting in disorders
in neural-immune interactions that can impair tissue healing.
Neuropeptides such as SP and calcitonin gene-related peptide (CGRP)
interact with cytokines released by immune cells to form a
neural-immune axis. For example, the reduced release of SP by nerve
fibers in diabetic wounds may cause a decrease in the levels of
pro-inflammatory cytokines released by immune cells, affecting the
initiation of the early inflammatory response in wounds (82). Concurrently, the reduction in CGRP
levels in the diabetic state may interfere with its ability to
polarize macrophages to a pro-repair phenotype, leading to delayed
wound healing (83,84).
Initiation of the proliferative phase in diabetic
wounds presents a complex challenge and represents a significant
factor contributing to the complexity of wound repair progression.
In diabetic wounds, macrophages persist in the pro-inflammatory M1
phenotype. This promotes an inflammatory response and inhibits the
initiation of tissue proliferation, ultimately leading to impaired
wound healing (84–86). A hyperglycemic environment impedes
the transition of macrophages from the M1 to the M2 phenotype,
resulting in compromised proliferation and migration of endothelial
cells and fibroblasts. This ultimately leads to impaired
angiogenesis, collagen deposition and wound healing (87). The dysregulation of macrophage
activity in diabetic wounds results in a reduction in VEGF-A and
VEGF receptor 1 signaling, leading to impaired angiogenesis and
delayed wound healing (88,89).
Although the complex process of diabetic wound
healing is multifaceted and involves a diverse array of cellular,
molecular and physiological mechanisms related to macrophages, it
primarily correlates with the hyperactivation of M1 macrophages and
impaired transformation from M1 to M2 macrophages (4,79).
The modulation of macrophage polarization has proven to be a
promising avenue for enhancing diabetic wound healing owing to the
pivotal role of macrophages in the process (90).
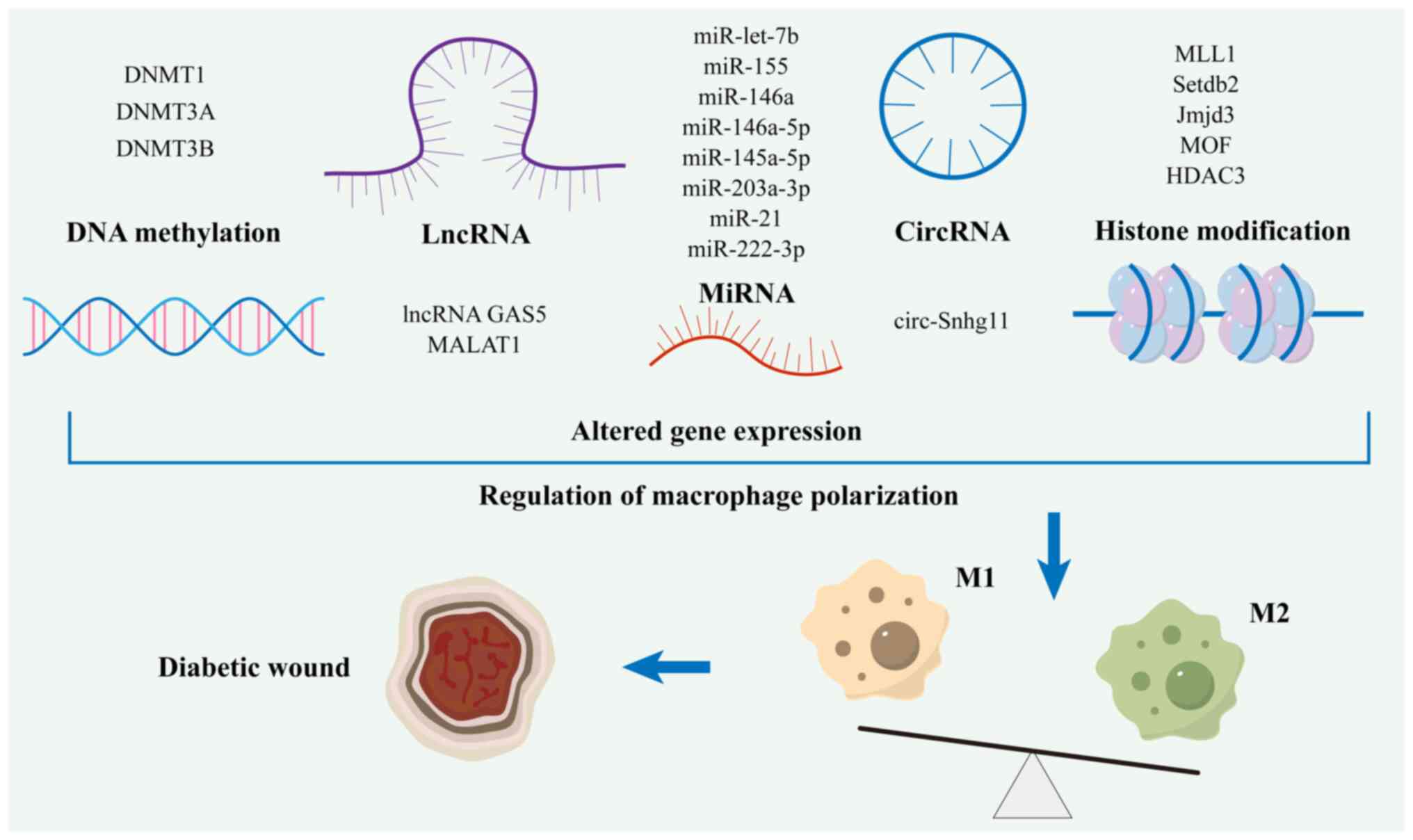
Epigenetic modifications are dynamic and inheritable
changes that occur in the genome without altering DNA sequences
(91). Investigating epigenetics
is crucial for understanding the pathogenesis of diseases and the
effect of environmental factors on gene expression. These
alterations primarily involve DNA methylation, histone
modification, chromatin remodeling and non-coding RNAs (ncRNAs)
RNAs, all of which contribute to the modulation of gene expression
(92). A growing body of research
has highlighted the pivotal role of epigenetics in controlling
macrophage phenotypes. A number of studies have demonstrated that
epigenetic modifications in macrophages are closely associated with
the pathogenesis of T2D and its associated complications, such as
diabetic wounds (Fig. 2) (93,94).
DNA methylation is a stable, widespread and abundant
form of epigenetic modification that markedly affects gene
expression (95). It plays a
pivotal role in a number of biological processes and diseases. For
example, during cell differentiation and development, DNA
methylation precisely regulates the expression of specific genes,
thereby determining cell fate and function (96). With regard to metabolic regulation,
DNA methylation can regulate the expression of genes associated
with metabolism, thereby participating in the occurrence and
development of metabolic diseases such as obesity and diabetes.
This process is primarily catalyzed by DNA methyltransferase
(DNMT), which uses S-adenosylmethionine as a methyl donor,
selectively adding methyl groups to the cytosines of two DNA
nucleotides, predominantly forming 5-methylcytosine and a minor
quantity of N6-methylpurine (97–99).
The regulation of DNA methylation is primarily governed by the DNMT
family, which comprises DNMT1, DNMT2, DNMT3A and DNMT3B (100,101). During cell division, newly
synthesized DNA strands contain hemimethylated sites. DNMT1
accurately recognizes these sites and is responsible for
maintaining methylation during DNA replication. DNMT3A and DNMT3B
are enzymes that participate in de novo DNA methylation and
are responsible for regulating the methylation patterns of the
genome (102–104). Hypermethylation of the promoter
prevents the binding of transcription factors or recruits
inhibitory complexes, resulting in the shutdown of gene expression,
also known as gene silencing. By contrast, hypomethylation promotes
gene expression (105,106).
DNA demethylation is a precisely regulated
biological process that involves the removal of methyl groups from
DNA molecules, thereby modifying their methylation status. There
are two types of DNA demethylation in mammals: Active and passive
(107). The ten-eleven
translocation protein family plays an essential role in active DNA
demethylation by catalyzing the oxidation of methylated cytosines.
This process is followed by repair via the base excision repair
pathway (108). Passive
demethylation can be described as a failure of the methylation
maintenance mechanism during semi-conservative DNA replication. As
a result, DNMT1 is unable to fully methylate the 5-C site on the
strand, leading to a reduction in genome-wide methylation over time
(109,110). DNA demethylation reverses the
methylation pattern, signifying gene activation. DNA methylation
and demethylation play equally crucial roles and the dynamic
equilibrium between them determines the ultimate epigenetic
methylation pattern in the cell (111).
A growing body of research has demonstrated that
abnormal DNA methylation patterns regulate macrophage expression
and affect diabetic wound healing (Table I). Notch1, PU.1 and Krüppel-like
factor 4 are needed to promote monocyte differentiation and
macrophage polarization. The hematopoietic stem cells of mice with
T2D exhibit NADPH oxidase 2 (NOX-2)-induced oxidative stress,
leading to increased expression of DNMT1 via the downregulation of
let-7d-3p microRNA (miRNAs/miRs), thereby promoting methylation of
the promoters of the three aforementioned genes and inhibiting
their expression. This mechanism reduces macrophage infiltration
into wounds and enhances the tendency of M1 macrophages to
polarize, thereby promoting an inflammatory response while
inhibiting wound repair (112).
The promoters of M1-specific genes (Cfb, Serping1 and Tnfsf15) in
macrophages isolated from the ischemic muscles of hyperlipidemic
mice and those with T2D exhibit significant hypomethylation,
resulting in the upregulation of M1 gene expression and promotion
of M1 macrophage polarization. By contrast, the promoters of M2
macrophage-specific genes (Nrp1, Cxcr4, Plxnd1, Arg1, Cdk18 and
Fes) were found to be markedly hypermethylated, leading to
downregulation of M2 gene expression and hindering of M2
polarization in macrophages. This demonstrates that changes to DNA
methylation can alter the polarization of macrophages in diabetic
ischemic muscles, thereby exerting a regulatory effect on gene
expression (113). Davis et
al (114) found that
cyclooxygenase-2/prostaglandin E2 (Cox-2/PGE2) was upregulated in
macrophages from both human and mouse diabetic wounds and could
regulate the downstream macrophage-mediated inflammatory response.
Their study demonstrated that TGF-β1 was able to induce miR-29b
expression in diabetic wound macrophages. Furthermore, miR-29b was
able to downregulate the expression of DNMT3b, leading to a
hypomethylated state of the Cox-2 promoter that led to increased
Cox-2/PGE2 production. This mechanism promotes the polarization of
macrophages toward the M1 phenotype, resulting in persistent and
unabated local inflammation at the wound site, thereby impeding the
tissue repair process following injury (114). Therefore, based on research into
epigenetic modifications, the expression of this gene can be
inhibited by targeting hypermethylation of the Cox-2 promoter. This
intervention has the potential to reverse the inflammatory
macrophage phenotype and contribute positively to diabetic wound
repair. These findings suggest that aberrant DNA methylation
patterns, characterized by both hypermethylation and
hypomethylation, may contribute to the increased infiltration of
pro-inflammatory macrophages into diabetic wounds, thereby
promoting inflammatory responses and impeding wound healing.
Consequently, targeting abnormal DNA methylation patterns to
reverse the macrophage phenotype holds promise as a potential
therapeutic strategy for enhancing diabetic wound recovery.
The nucleosome, composed of ~146 bp of DNA wrapped
around histone octamers (H2A, H2B, H3 and H4), serves as the basic
structural element of chromatin (115). Histone modifications such as
acetylation, methylation, phosphorylation, polymerization and
ubiquitination play crucial roles in shaping chromatin structure,
maintaining nucleosome stability and regulating gene transcription
(116). Histone modification
primarily affects arginine, lysine, serine, threonine and tyrosine
residues in the N-terminal tails of histone proteins (117). Methylation and acetylation are
the most widely studied types of histone modifications. Histone
methylation is catalyzed by histone methyltransferases. This
modification typically targets lysine and arginine residues on
histones and demethylases actively remove the resulting methylation
marks. Lysine methylation is a reliable indicator of gene
expression control. For example, lysine methylation at position 4
of H3 promotes transcriptional activation, whereas the same
modification at positions 9 and 27 tends to inhibit transcription
(118). Histone acetylation is a
dynamic modification that occurs primarily at the relatively
conserved N-terminal lysine positions of H3 and H4. This process is
mediated by the coordination between histone acetyltransferases and
histone deacetylases. Lysine acetylation often leads to
transcriptional activation and deacetylation during gene silencing
(119,120). Histone modification has emerged
as an attractive target for regulating macrophage phenotypes in a
number of diseases. For instance, SET and MYND domain-containing
protein (SMYD)3, a histone lysine methyltransferase of the SMYD
family, promotes macrophage conversion from M1 to M2 by activating
the tricarboxylic acid cycle and regulating the transcriptional
activities of metabolic enzymes such as citrate synthase, succinate
dehydrogenase complex subunit C and pyruvate carboxylase (121). Enhancer of zeste homolog 2
functions as a histone methyltransferase that induces
trimethylation of lysine 27 residue of histone H3 (H3K27)
(H3K27me3) to modulate the polarization of liver macrophages from
M2 to M1. This process contributes to the development and
progression of autoimmune hepatitis and autoimmune reactions
(122).
Histone modification has been shown to potentially
regulate macrophage phenotypes in diabetic wounds (Table II). Mixed-lineage leukemia 1
(MLL1) serves as a biomarker of inflammation and a key factor in
macrophage activation (123).
During the early inflammatory phase of normal wound healing, MLL1
is upregulated in macrophages. MLL1 is also a histone
methyltransferase specific to the lysine 4 residue of histone H3
(H3K4). MLL1 elevates H3K4me3 at the NF-κB binding site, initiating
pro-inflammatory macrophage-mediated wound inflammatory storms that
impede wound healing. The timing of MLL1 expression in prediabetic
wound macrophages corresponds to the temporal changes in
inflammation levels in prediabetic mouse models, with early
reduction and late elevation (124). Similarly, research has indicated
that MLL1 mediates the alteration of H3K4me3 on the TLR4 promoter
in macrophages within diabetic wounds, subsequently activating the
transcription of the TLR4 gene and facilitating the polarization of
M1 macrophages. This can result in the dysregulation of
inflammation and impairment of wound healing in diabetes (74). This suggests that MLL1, a potential
therapeutic target for diabetic refractory wounds, plays an
essential role in determining the wound macrophage phenotype
through histone modifications. SET domain bifurcated 2 (Setdb2)
functions as a histone methyltransferase that specifically targets
histone H3 lysine 9 (H3K9) for methylation, thereby modulating the
chromatin structure and silencing gene expression (125). Kimball et al (126) found that Setdb2 can act as a
brake on the inflammatory response in normal wounds. By contrast,
Setdb2 expression is markedly reduced in diabetic wounds, thus
allowing unrestricted expression of inflammatory genes. Following a
typical wound injury, the elevation of Setdb2 expression in
macrophages results in an augmentation of H3K9me3 levels at the
NF-κB binding site on the promoter of the gene that stimulates
inflammation. This phenomenon culminates in the suppression of gene
transcription, thereby facilitating the attenuation of wound
inflammation and the transition to the proliferative stage. The
regulation of Setdb2 expression in wound macrophages is controlled
by IFN-β, which targets the JAK/STAT1 pathway at the end of the
inflammatory phase. However, disruption of the IFN-β-Setdb2
regulatory axis within diabetic wound macrophages results in a
failure of the phenotype transition necessary for macrophage
repair, which in turn delays wound healing (126). Setdb2, with its powerful function
of modulating macrophage plasticity, may represent a promising new
target for treating refractory diabetic wounds.
H3K27me3 in the promoter region inhibits gene
transcription and effectively represses gene expression. Jumonji
domain-containing protein-3 (Jmjd3) is a histone demethylase that
is specific to H3K27. Upregulation of Jmjd3 leads to the inhibition
of histone methylation and activation of gene transcription. It has
been shown that palmitate stimulation can increase the expression
of Jmjd 3, thereby inducing macrophages to enter a pro-inflammatory
state and upregulating inflammatory cytokine levels by removing the
inhibitory H3K27me3 marker on the promoter of the NF-κB regulatory
gene. Inhibitors of Jmjd3 may suppress the expression of NF-κB
inflammatory genes via histone modification pathways and enhance
diabetic wound healing by modulating the macrophage phenotype
(127). M1 macrophages secrete
pro-inflammatory factors, such as IL-12, to initiate an
inflammatory immune response. Gallagher et al (78) discovered that a significant
increase in Jmjd3 expression on the IL-12 promoters of diabetic
wound macrophages results in the specific removal of methyl groups
from H3K27me3 by Jmjd3, effectively eliminating the inhibitory
function of H3K27me3. This leads to an increased expression of
IL-12 and continuous activation of pro-inflammatory macrophages,
which keeps wounds in the inflammatory phase and makes the
transition to the proliferative phase more difficult, resulting in
delayed wound healing. However, the H3K27 demethylase inhibitor
GSK-J4 enhances H3K27me3-mediated inhibition of the IL-12 promoter
and effectively reverses IL-12 expression in the macrophages
(78). Given these mechanisms,
targeting histone demethylases to regulate macrophage-mediated
inflammation may represent a novel approach to correcting diabetic
wound healing.
Males absent on the first (MOF) is a histone
acetyltransferase that selectively acetylates H4K16 to enhance gene
transcription (128,129). High expression of inflammatory
cytokines mediated by NF-κB in diabetic wound macrophages leads to
chronic inflammation and impedes transition to the proliferative
phase. MOF has been shown to be overexpressed in macrophages within
diabetic wounds, where it facilitates the expression of
inflammatory genes by acetylating histone H4K16 on NF-κB-mediated
inflammatory gene promoters, thereby contributing to the delayed
healing of diabetic wounds. The expression of MOF in macrophages is
regulated by TNF-α. Inhibition of TNF-α has been shown to reduce
the level of MOF in macrophages and suppress the inflammatory
response at the wound site, thereby facilitating diabetic wound
repair. Targeting MOF with TNF-α has the potential to reverse the
pro-inflammatory macrophage phenotype in diabetic wounds, thereby
representing a promising avenue for chronic diabetic wound
treatment (130). Histone
deacetylase 3 (HDAC3) has been shown to trigger the inflammatory
response and suppress the anti-inflammatory phenotype in
macrophages (131–133). HDAC3 expression is upregulated in
both human and mouse diabetic wounds, when compared with
corresponding levels in normal wounds (134–136). BG45 is a selective HDAC3
inhibitor that has been proven to effectively reverse the
macrophage phenotype, reduce the expression of pro-inflammatory
factors secreted by M1 macrophages and increase the levels of
anti-inflammatory pro-healing factors secreted by M2 macrophages.
Furthermore, BG45 treatment also decreases the number of
neutrophils and macrophages that infiltrate the diabetic wounds.
Overall, BG45 facilitates wound healing by enhancing the M2
phenotype (Arg-1, CD206 macrophages) and promoting the expression
of wound-healing markers such as CD31, VEGF and colligation-1A,
while inhibiting the expression of IL-1β. It was hypothesized that
HDAC3 delays wound healing by inhibiting the phenotypic switch from
M1 to M2, thus negatively regulating angiogenesis and increasing
the infiltration of both neutrophils and macrophages. Therefore,
the development of corresponding regulators of histone
modifications has significant potential for advancing diabetic
wound healing (136). Regulation
of the macrophage phenotype in diabetic wounds and the promotion or
delay of wound healing can be achieved through histone
modifications such as methylation, demethylation, acetylation and
deacetylation. Therefore, the development of corresponding
regulators of histone modifications holds significant potential for
advancing diabetic wound healing.
ncRNAs have emerged as potential novel biomarkers.
These molecules are transcribed from the genome, do not encode
proteins and are involved in regulating the expression levels of
certain genes that are crucial for orderly cell differentiation and
development (137,138). These RNAs have markedly enhanced
our understanding of gene expression regulatory networks and offer
novel targets and foundations for disease diagnosis, treatment and
prevention. In recent years, advancements in high-throughput
sequencing technology and bioinformatics have revealed the
functions and mechanisms of ncRNAs. Among these, the primary
components consist of long non-coding RNAs (lncRNAs), miRNAs and
circular RNAs (circRNAs). Accumulating evidence indicates that
ncRNAs play an instrumental role in influencing the phenotypes of
macrophages and offer a potential avenue for therapies targeting
diabetic wounds (Table III).
lncRNAs are an important part of gene regulatory
networks that can affect gene expression in various ways, including
chromatin remodeling, transcriptional regulation and
post-transcriptional regulation. It was previously hypothesized
that lncRNAs were simply byproducts of transcription (139). It has since become evident that
they perform various regulatory functions related to chromatin,
cytoplasmic mRNA, membraneless nucleosomes and signaling pathways
(140). These RNA molecules
regulate various physiological processes such as immunity,
inflammation, proliferation, cell differentiation and cell survival
(141). They have emerged as
critical regulators of gene expression in a number of human
diseases (142). lncRNAs
influence the inflammatory response by regulating macrophage
polarization, making them significant factors in diabetic wound
healing. Hu et al (143)
discovered a significant increase in the expression of the GAS5
LncRNA in diabetic wounds and human diabetic skin. Further
investigations revealed that the overexpression of GAS5 leads to a
considerable rise in the expression levels of mRNA markers related
to the M1 macrophage phenotype, including inducible nitric oxide
synthase (iNOS), TNFα and IL-1β. Conversely, there was no effect on
the expression levels of M2 macrophage marker RNAs such as Arg1 and
Mrc1. Finally, it was observed that GAS5 knockout boosted diabetic
wound healing. This indicates that GAS5 affects wound healing by
stimulating the activation of M1 pro-inflammatory macrophages,
potentially by inducing the expression of STAT1 (143). Metastasis-associated lung
adenocarcinoma transcript 1 (MALAT1) is a lncRNA that functions as
a transcriptional regulator of numerous genes, including those
involved in cancer metastasis, cell migration and cell cycle
regulation. MALAT1 functions as a competitive RNA by binding
diverse miRNAs to execute biological processes. For instance, it
upregulates MFGE8 expression by competitively binding to
miR-1914-3p, thereby inhibiting TGFB1 and SMAD3, promoting M2
macrophage polarization, enhancing macrophage phagocytosis and
reducing apoptosis, thus promoting diabetic wound healing (144).
Although there is limited research on the role of
lncRNAs in diabetic wounds, their ability to induce the
polarization of pro-inflammatory M1 macrophages and regulate the
polarization of pro-healing M2 macrophages suggests that targeting
lncRNAs to modulate macrophage phenotypes holds promise for
treating diabetic wounds. However, the precise mechanisms of action
of lncRNAs remain unclear, warranting further exploration through
additional studies.
The miRNA class of ncRNAs is involved in a variety
of physiological and pathological processes, including metabolism,
proliferation, apoptosis, differentiation and development. They
also act as potent gene regulators of a wide range of cellular
activities (145,146). Increasing attention is being paid
to the role of miRNAs in terms of regulating macrophage plasticity
and polarization (147). For
example, it has been shown that miR-155 expression significantly
influences the polarization of M1 macrophages (148). One group observed miR-155
overexpression in the skin of diabetic mice (149). Local inhibition of miR-155
decreases wound infiltration by T cells and macrophages, leading to
improved tissue inflammation and accelerated healing of diabetic
wounds (150). However, the
precise mediating pathways behind this phenomenon merit further
investigation. The expression of miR-146a has been found to be
reduced in the macrophages of patients with diabetes. TLR4 is a
target gene that is negatively regulated by miR-146. Peng et
al (151) discovered that
overexpressing miR-146a leads to the polarization of M2 macrophages
by suppression of the TLR4/NF-κB axis, resulting in improved wound
healing of diabetic ulcers. Overexpression of miR-145a-5p inhibits
M1 macrophage polarization while promoting M2 polarization in RAW
264.7 macrophages and M2 macrophages promote inflammatory
regression and tissue remodeling by releasing growth and
anti-inflammatory cytokines, suggesting that this may be an
essential mechanism for their therapeutic effect on diabetic wound
repair (152). Hyperglycemia
induces the expression of miRNA-21, downregulating phosphatase and
tensin homolog. This indirectly induces the production of NOX2 and
ROS, thereby promoting the polarization of M1 macrophages and the
inflammatory response. This may represent another important
mechanism for treating refractory non-healing diabetic wounds
(153).
Previous studies have indicated that negatively
charged miRNAs encounter challenges in terms of penetrating cell
membranes and are susceptible to degradation and elimination in the
wound microenvironment (154,155). Exosomes are extracellular
vesicles found in various bodily fluids that facilitate the
transfer of biomolecules and represent key players in intercellular
communication. The membrane structure of exosomes protects miRNAs
from degradation by enzymes and other chemicals, allowing for
stable loading into exosomes and their subsequent transport to
target cells for expression (156). Most exosomes carry miRNAs and
their combination presents promising prospects for translational
medicine (157–159). Previous studies have shown that
exosomes isolated from relatively lean donor adipose tissues
promote the polarization of macrophages toward the M2 phenotype
(160). Xia et al
(161) found that exosomes
extracted from the adipose tissue of a relatively lean donor
hindered the expression of the Bim protein, by activating
miR-222-3p. This activation induced macrophages to convert to the
M2 phenotype, ultimately resulting in reduced inflammation and
improved wound healing (161).
Epidermal stem cells (EpiSCs) play a critical role in skin wound
repair, where they are readily available (162,163). Previous studies have established
the potential of EpiSCs to heal wounds associated with diabetes
(164,165). JAK2 and STAT3 signaling has been
shown to be a key factor regulating the polarization of macrophages
into the M2 type. Cytokine signal transduction inhibitor 3 (SOCS3)
inhibits JAK kinase activity, thereby blocking the JAK2/STAT3
pathway and serving as a primary negative regulator of its
signaling. Enrichment of miR-203a-3p in exosomes originating from
EpiSCs SOCS3 expression in macrophages. This activation triggers
the JAK2/STAT3 signaling pathway and induces the polarization of M2
macrophages, releasing anti-inflammatory factors and promoting
collagen deposition as well as angiogenesis within granulation
tissue. As a result, wound healing in diabetes is accelerated
(166). The application of
mesenchymal stromal cells (MSCs) to skin wounds has shown promise
because of their ease of collection and low risk of immune
rejection (167–169). MSCs promote skin cell migration,
blood vessel development, re-epithelialization of wounds and
granulation tissue generation, all of which ultimately accelerate
healing (170). The biological
properties of MSCs, which facilitate the transition from the
inflammatory to proliferative phases, are crucial for treating
wounds with high levels of inflammation that hinder the healing
process. Ti et al (171)
discovered that MiR-let-7b in exosomes derived from MSCs
pre-treated with LPS encourage macrophage M2 polarization by
suppressing the TLR4/NF-κB/STAT3/AKT signaling pathway, thereby
reducing inflammation and promoting diabetic skin wound healing.
Bone MSCs (BMSCs) play a major role in tissue repair, particularly
in terms of accelerating wound healing (172,173). By suppressing the expression of
TRAF6, BMSC-derived exosomes carrying miR-146a-5p stimulate the
polarization of M2 macrophages and reduce the polarization of M1
macrophages, leading to the proliferation, migration and
angiogenesis of endothelial cells. This results in improved healing
of recalcitrant diabetic wounds (174). Overall, miRNA-carrying exosomes
have shown significant potential for the treatment of chronic
diabetic wounds.
miR-29ab1 has also been shown to play a crucial role
in diabetes-associated inflammation and the transformation of
macrophage phenotypes (175).
Compared with healthy subjects, individuals with diabetes
experience a rise in ectopic miR-29ab1, alongside an increase in M1
polarization and elevated levels of IL-1β and TNF-α in skin wounds.
After using a Chinese traditional medicine hydrogel containing
chitosan and puerarin to treat diabetic wounds, Zeng et al
(176) observed that inhibiting
ectopic miR-29ab1 expression leads to a reduction in IL-1β and
TNF-α levels and inhibits the polarization of M1 macrophages within
diabetic wounds. This improves the inflammatory response and
promotes healing. Overall, miRNAs participate in several diabetic
wound-healing processes by regulating M1 vs. M2 polarization,
particularly during inflammation and are emerging as promising
therapeutic targets for managing diabetes (152,153,174).
Research on miRNAs has reached a mature state, with
significant achievements having been demonstrated in the treatment
of diabetic wounds. By targeting specific genes, miRNAs can
precisely modulate macrophage polarization, offering the potential
to enhance the healing environment within diabetic wounds.
Furthermore, miRNAs possess certain advantages such as
multi-targeting and long-lasting effects that facilitate their
broad application (177,178). At the same time, the intricate
regulatory network between miRNAs and genes poses a significant
challenge to research and applications. Moreover, miRNAs can
inadvertently regulate unexpected targets, potentially causing
adverse reactions (179).
Exosomes, which serve as efficient carriers for intercellular
communication, can facilitate the targeted delivery of miRNAs to
improve the bioavailability of therapeutic molecules (180). However, there are still
challenges associated with exosomes, including their limited
capacity and the complexities of separation and purification, that
warrant further research before they can be resolved (181).
CircRNAs are a cohort of biologically active ncRNAs
that exist in a closed-loop form (182). Most circRNAs contain multiple
miRNA-binding sites and function as miRNA sponges within cells.
Some have also been identified as competitive endogenous RNAs
(ceRNAs). These ceRNAs compete for miRNA-binding sites, leading to
reduced miRNA activity and consequent effects on gene expression
and protein synthesis (183–185). Increasing evidence has linked
circRNAs to the mechanisms underlying various diseases, including
diabetic ulcers (186,187). The expression of circRNA Snhg11
decreases significantly under hyperglycemic conditions. Compared
with healthy controls, patients with diabetes have been shown to
have increased expression of miR-144-3p, which is the downstream
target of circRNA Snhg11 (188).
Exosomes derived from adipose stem cells (ADSCs) can enhance
fibroblast migration and proliferation, as well as collagen
synthesis, to accelerate skin wound healing (189). The high expression of circRNA
Snhg11 in exosomes derived from hypoxic-preconditioned ADSCs acts
as a sponge for miR-144-3p, thereby augmenting HIF-1α expression,
facilitating angiogenesis and M2 macrophage polarization,
suppressing hyperglycemia-induced endothelial cell damage and
expediting diabetic wound healing (190).
Overall, circRNAs represent potential targets for
treating diabetic foot ulcers and have a range of potential
applications. However, relatively few studies have been conducted
on these molecules to date. Further studies are therefore warranted
to explore their regulatory mechanisms.
In the diabetic environment, various epigenetic
modifications, including DNA methylation, histone modification and
regulation of ncRNAs, may interact in complex ways to modulate
macrophage polarization by influencing the expression of specific
genes, thereby either promoting or delaying wound healing. A number
of studies have shown that ncRNAs can regulate the expression and
activity of DNMT through various mechanisms (191,192). For instance, certain miRNAs can
directly target DNMT mRNA, inhibit its translation, or facilitate
its degradation to reduce its expression. This reduction may result
in the hypomethylation of specific gene promoter regions and impact
gene expression and cellular function. Davis et al (114) discovered that, in diabetic
wounds, TGF-β1 can stimulate miR-29b expression within local
macrophages to inhibit DNMT3B-mediated hypermethylation of the
Cox-2 promoter, thereby enhancing the production of Cox-2/PGE2 and
leading to a sustained pro-inflammatory phenotype and impaired
macrophage function that impacts wound healing. A wide range of
diverse inter-regulatory relationships exist among ncRNAs.
Specifically, lncRNAs and circRNAs have been the focus of extensive
research into the regulatory mechanisms that involve miRNAs.
lncRNAs can function as molecular sponges for miRNAs, inhibiting
their activity, or they can bind with miRNAs to form RNA-induced
silencing complexes that participate in gene expression regulation.
Closed-loop circRNAs can act as ceRNAs for miRNAs, thereby
preventing miRNAs from inhibiting target genes. These reciprocal
regulations play pivotal roles in biological processes and their
aberrant control is intricately linked to disease progression.
MALAT1 competitively binds to miR-1914-3p, while circRNA Snhg11
functions as a sponge for miR-144-3p (144,190). Both of these interactions have
the potential to modulate the macrophage phenotype by regulating
the expression of target genes, thereby playing a crucial role in
diabetic wound healing.
Under the influence of different microenvironments,
epigenetics can activate or suppress various signaling pathways, to
modulate the differentiation of macrophages into distinct
phenotypes. Consequently, the signaling pathway serves as a
mediator to some extent. These signaling pathways interact to form
an intricate network that collectively regulates macrophage
polarization. A thorough exploration of the signaling pathways
associated with macrophage polarization is advantageous for
enhancing our understanding of the function and biological activity
of macrophages and for identifying new targets and strategies for
disease treatment.
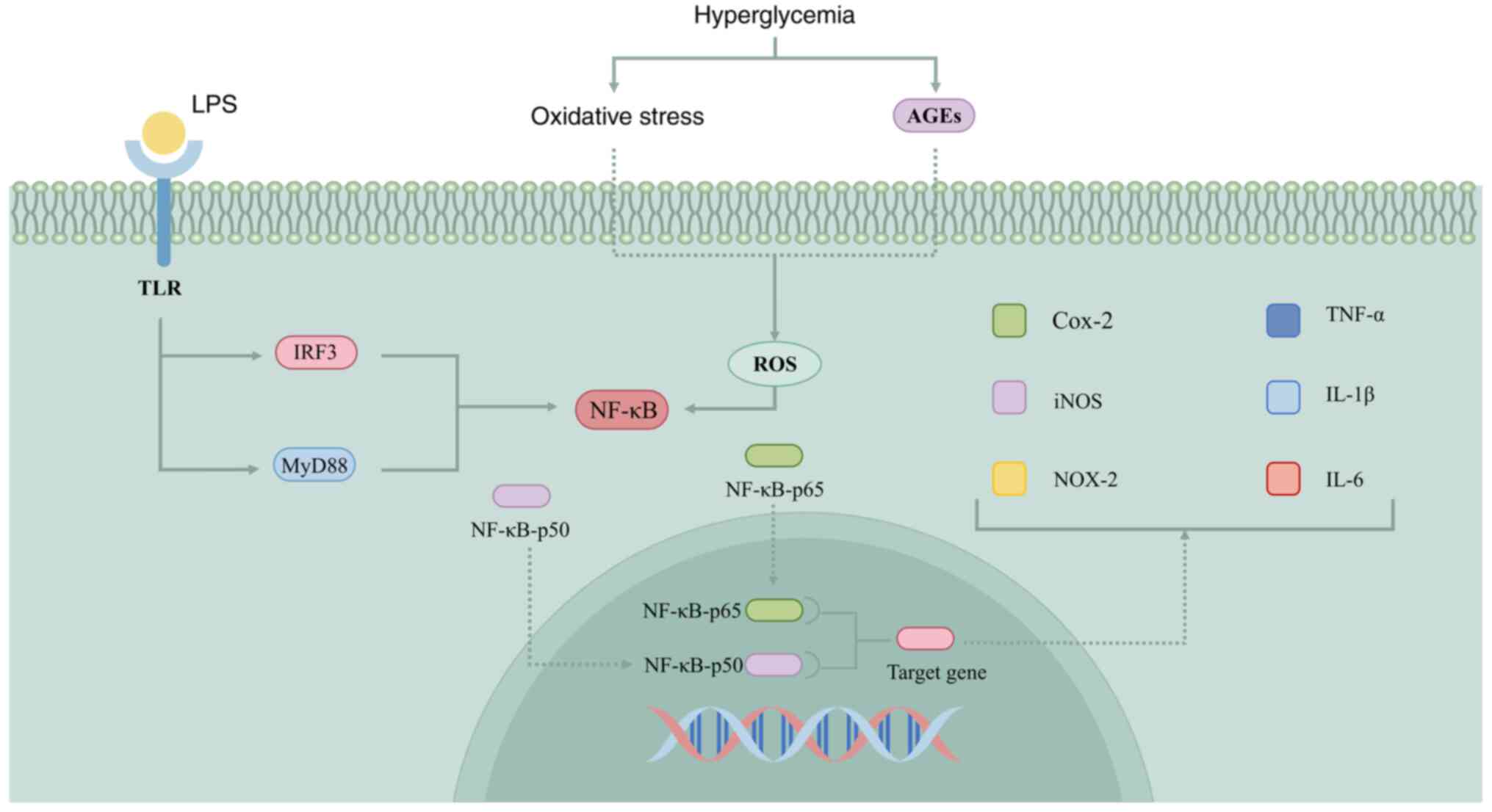
The NF-κB signaling pathway is a canonical pathway
that governs macrophage polarization (Fig. 3). NF-κB has been implicated in the
pathogenesis of numerous immune and inflammatory diseases, where it
binds to specific gene sequences to initiate gene expression
processes, thereby facilitating the production and release of a
range of pro-inflammatory mediators (193). TLR serves as a crucial sentinel
within the immune system, detecting pathogen invasion and
exhibiting expression on a variety of immune and non-immune cell
surfaces. It specifically recognizes substances released by
invading bacteria and viruses (for example, LPS, mannose, teichoic
acid and peptidoglycans) and regulates inflammation and other
innate immune responses. TLRs on macrophage surfaces bind to LPS
and activate the classical NF-κB pathway through either the
MyD88-dependent or the interferon regulatory factor 3 pathway
(194). The NF-κB-p65 and
NF-κB-p50 proteins are activated and translocated to the nucleus,
where they bind to target genes that lead to the release of
inflammatory cytokines such as TNF-α, IL-1β, IL-6, COX-2, iNOS and
NOX-2 (195). They also play a
role in regulating the polarization of macrophages toward the M1
type (196).
Excessive ROS can activate intracellular metabolic
pathways such as NF-κB. Hyperglycemic environments may induce
excessive oxidative stress in wounds, disrupting the balance
between oxidative and antioxidant activities. This can result in
the overproduction of ROS and hinder the healing process in
diabetic wounds (197,198). Furthermore, the production of
pro-oxidant advanced glycation end products (AGEs) in patients with
diabetes also stimulates the accumulation of excessive ROS, leading
to the activation of the nuclear transcription factor NF-κB
(199). Elevated AGE levels can
also induce heightened inflammation by modulating macrophage M1
polarization, impeding the transition of diabetic wounds from the
inflammatory to the proliferative stage and delaying wound healing
(200).
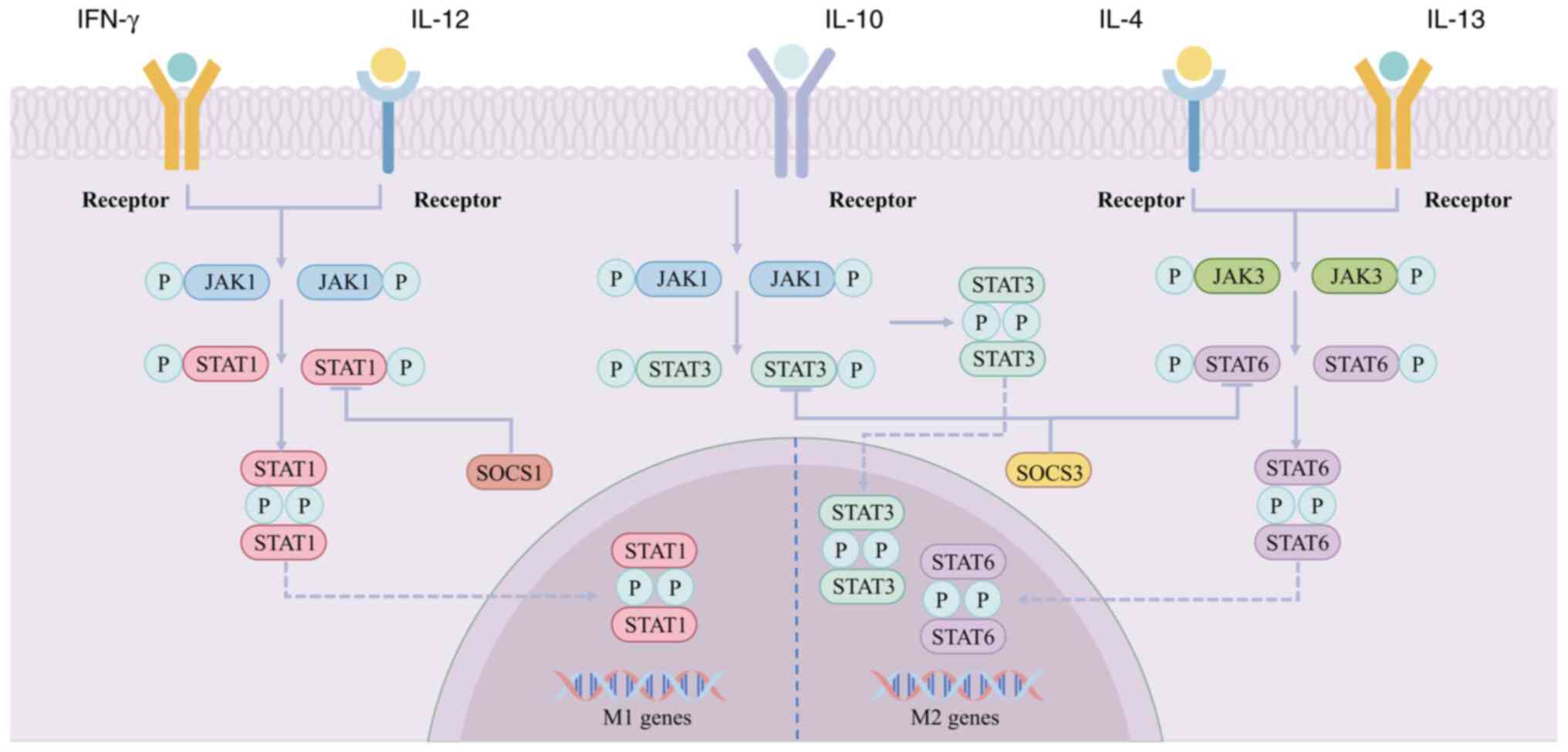
The JAK/STAT pathway is primarily responsible for
facilitating the signal transduction of cytokine receptors and
plays a vital role in a number of biological processes, including
cellular growth and proliferation, inflammatory responses, immune
regulation and the modulation of neural function (201). In addition, it serves as a
significant signaling pathway for macrophage polarization (Fig. 4). The presence of cytokines, growth
factors and chemokines in diabetic wounds can trigger activation of
the JAK-STAT pathway. JAK refers to a family of tyrosine kinases
comprising four types: JAK1, JAK2, JAK3 and TYK2. The STAT protein
family comprises seven major members: STAT1, STAT2, STAT3, STAT4,
STAT5a, STAT5b and STAT6. These proteins are activated by various
cytokines and exhibit diverse biological effects. Among these,
STAT1, STAT3 and STAT6 are the primary members of the STAT family
that regulate macrophage polarization and the inflammatory
response. Specifically, IFN-γ and IL-12 bind to their respective
receptors, leading to the activation of JAK and subsequent
phosphorylation of STAT1. This process results in the polarization
of M1-type macrophages, which in turn leads to the production of a
multitude of pro-inflammatory factors that initiate and amplify the
inflammatory response of the body against pathogen invasion
(202). IL-4 and IL-13 both
stimulate the M2 polarization of macrophages via the JAK3/STAT6
pathway, whereas IL-10 promotes the M2 polarization of macrophages
through the JAK1/STAT3 pathway (203,204).
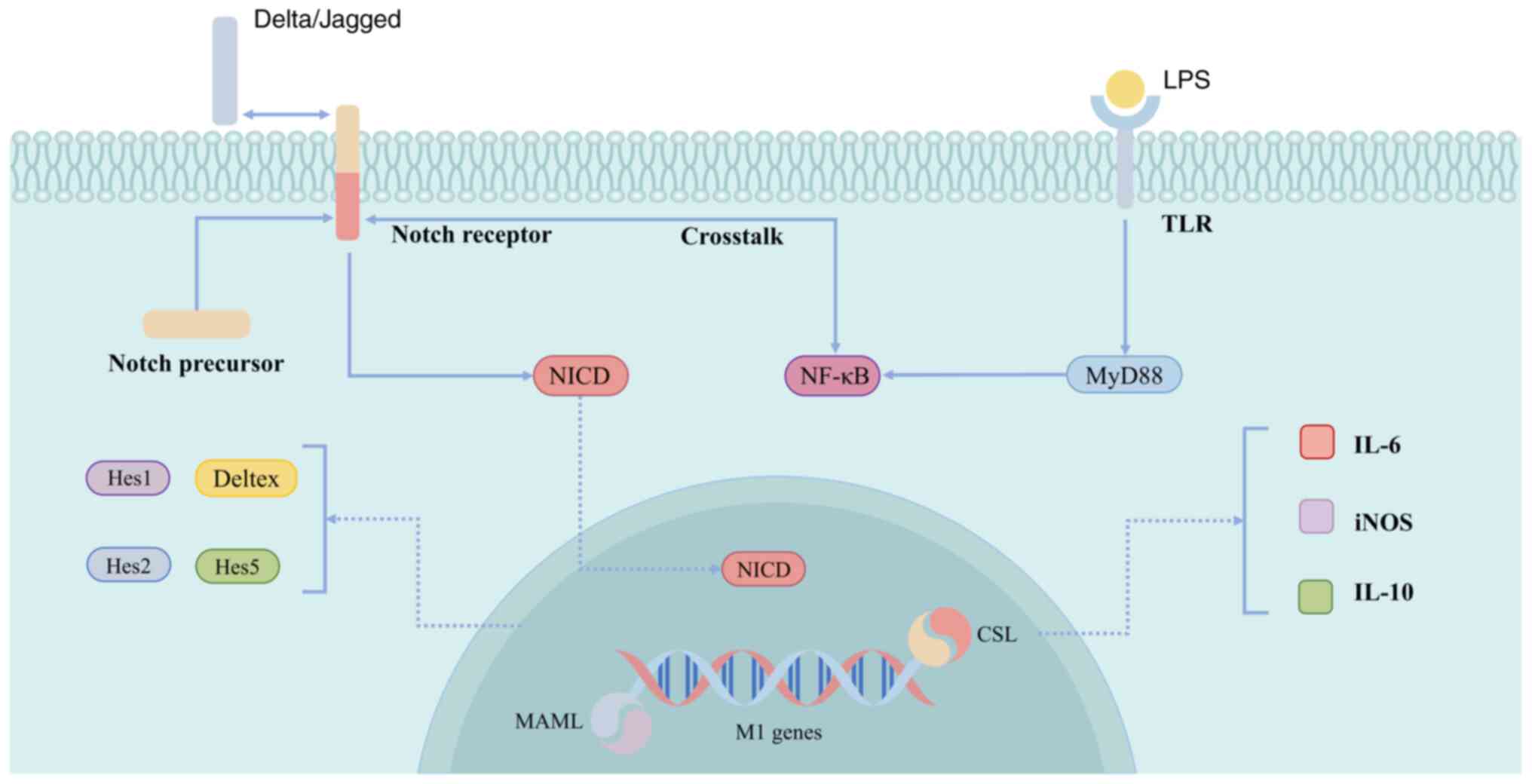
The Notch signaling pathway is a highly conserved
mechanism that exerts a significant influence on the development of
various biological organs and tissues, regulating processes such as
cell differentiation, tissue homeostasis maintenance, immune
function and neurodevelopment (209). The Notch receptor, a type I
transmembrane protein comprising Notch1, 2, 3 and 4, is located on
the plasma membrane in its inactive state. Its ligands are members
of the DSL protein family, encompassing Jagged (JAG) 1, JAG2,
delta-like (DLL)1, DLL3 and DLL4. The Notch signaling pathway is a
classical signaling pathway involved in macrophage polarization.
When JAG1 binds to a Notch receptor (1–4), it
leads to cleavage of the receptor, resulting in the translocation
of the Notch intracellular domain (NICD) to the nucleus. Once
there, the NICD forms complexes with the DNA-binding protein
Suppressor of Hairless, Lag-1 and the coactivator master-mind-like
family, leading to the transcriptional activation of downstream
target genes such as Hes1, 2 and 5. This process promotes M1
macrophage polarization and induces the expression of related
genes.
Activation of the Notch signaling pathway via the
TLR signaling cascade can also modulate the polarization of
mononuclear macrophages toward the pro-inflammatory M1 phenotype.
The expression of Notch1 is upregulated by LPS through
MyD88-dependent or-independent pathways, leading to the activation
of the downstream genes Hes1 and Deltex (124). Activation of the Notch signaling
pathway can upregulate IL-6 and iNOS secretion, downregulate IL-10
production and induce macrophage polarization toward M1.
Furthermore, the Notch signaling pathway can enhance inflammatory
responses through synergistic interactions with the NF-κB signaling
pathway. In one study, Notch1 receptor expression was found to be
significantly increased in M1 macrophages. The reversal of Notch1
receptor inhibition was shown to induce a shift from the M1
macrophage phenotype to the M2 one, thereby initiating an
anti-inflammatory response (210). Given the pivotal role of the
Notch pathway in modulating the phenotypes of wound macrophages and
the inflammatory response in vivo (Fig. 5), it is evident that targeting
Notch signaling pathway regulation represents a crucial direction
for the treatment of diabetic foot ulcers.
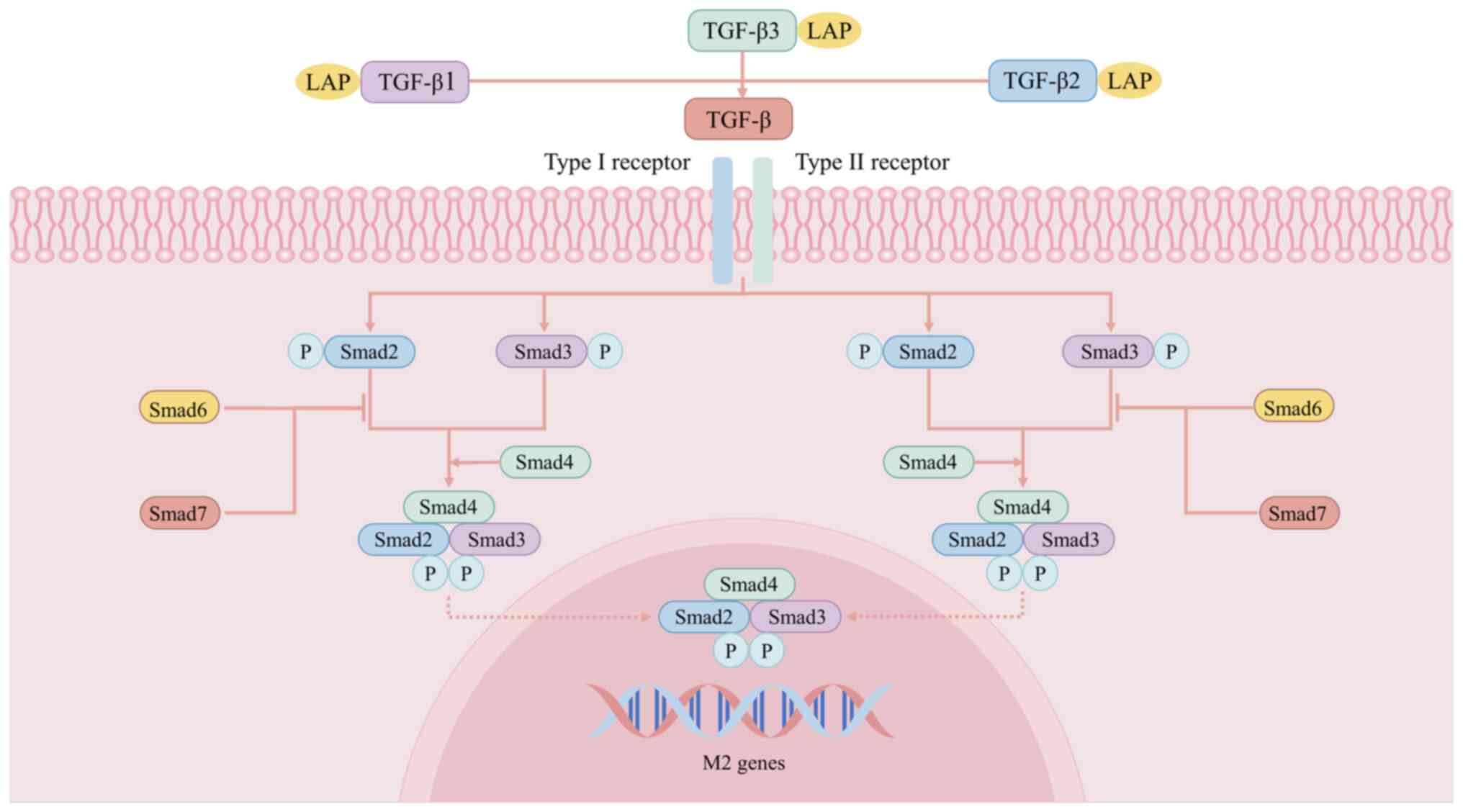
The TGF-β/Smad signaling pathway can be involved in
the regulation of cell growth and differentiation, regulation of
the ECM, immune regulation, wound healing, organ fibrosis and other
processes. The regulation of the TGF-β/Smad pathway in macrophage
polarization has received increasing attention in recent years
(Fig. 6). There are three isomers
of TGF-β: TGF-β1, TGF-β2 and TGF-β3. Smad proteins (Smad1-9) play
crucial roles as downstream signaling molecules in the TGF-β
pathway. TGF-β initially interacts with the type II receptor and
subsequently forms a receptor complex by binding to the type I
receptor, leading to phosphorylation and activation of the type I
receptor domain. Activated type I receptors initiate intracellular
signal transduction by phosphorylating the C-terminus of specific
receptor-regulated SMADs (R-SMADs) (211,212). Under the action of SMAD4, one
common Smad forms heteropolymers with two R-Smads (SMAD2 and/or
SMAD3), which then translocate to the nucleus and initiate the
transcription of target genes (213). These SMADs also induce the
transcription of inhibitory SMADs (SMAD6 and SMAD7), thereby
initiating negative feedback loops that suppress signaling.
Research has consistently demonstrated a correlation between M2
macrophage polarization and increased TGF-β pathway activation. For
instance, growth differentiation factor 3 from the TGF-β
superfamily has been shown to inhibit M1 polarization and
facilitate M2 polarization through the promotion of Smad2 and Smad3
phosphorylation (214). The
TGF-β/Smad signaling pathway can also facilitate the activation of
the PI3K pathway to induce the polarization of M2 macrophages.
Therefore, promoting diabetic wound healing by modulating the
TGF-β/Smad signaling pathway to activate M2 polarization in
macrophages may also represent a promising therapeutic
direction.
The present review provides an overview of the
current epigenetic regulation of macrophage polarization in
diabetic wounds. From the perspective of macrophages, the challenge
of diabetic wound healing primarily stems from the excessive
activation of M1 macrophages and hindered transition from the M1
state to the M2 one (76,81,215). Various epigenetic modifications,
including DNA methylation, histone modification and ncRNA
modification, can modulate the functional behavior of macrophages
within the diabetic wound microenvironment by regulating gene
expression, thereby affecting wound healing. Targeting epigenetic
modifications to modulate macrophage phenotype and function has
recently emerged as a promising therapeutic strategy for diabetic
wound healing.
However, current research on the matter would
benefit greatly from further exploration and refinement. Despite
the powerful regulatory ability of epigenetics on gene expression,
this field is still in its early stages, particularly with regard
to lncRNAs and circRNAs, which have received comparatively little
attention. The precise mechanisms behind this form of regulation
warrant further investigation. While existing studies have
primarily focused on individual epigenetic modifications or
regulatory pathways, these modifications actually form a complex
network of interactions in vivo that complicate the
regulatory process and present challenges in terms of clinical
applications. Furthermore, considering that current research has
been limited to animal and cell models, it is imperative to conduct
additional clinical studies to facilitate the translation of basic
research into clinical applications (113,149).
Currently, research on macrophages in diabetic
wounds primarily focuses on the inflammatory response. Notably,
epigenetic factors may also influence additional mechanisms of
diabetic wound healing through macrophage polarization, including
angiogenesis, re-epithelialization and ECM remodeling. However,
there is still a scarcity of studies addressing these dimensions to
provide a comprehensive overview; therefore, future investigations
are expected to explore this process in greater depth. The healing
of diabetic wounds represents a complex process involving multiple
cell types, cytokines and signaling pathways. The epigenetic
regulation of macrophage polarization represents just one aspect of
the process. Its synergistic relationships with other relevant
factors such as glycemic control, neurovascular function and
overall immune system regulation remain unclear. Future research
should emphasize the comprehensive analysis of epigenetic
regulatory networks, foster interdisciplinary collaboration,
develop more precise and effective treatment methods and actively
promote clinical applications to provide practical treatment
options for patients with diabetic wounds, while improving their
quality of life and prognoses.
Not applicable.
The present study was funded by grants from the Construction
Project for the Tianjin Health Commission Institute of Integrated
Traditional Chinese and Western Medicine (grant no. 202455) and the
Tianjin Municipal Health Commission project (grant no.
2023135).
Not applicable.
The manuscript was written by JS. Images were
obtained by YW and YC. The manuscript was edited and revised by XS
and ZZ, respectively. All authors read and approved the final
manuscript. Data authentication is not applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Reed J, Bain S and Kanamarlapudi V: A
review of current trends with type 2 diabetes epidemiology,
aetiology, pathogenesis, treatments and future perspectives.
Diabetes Metab Syndr Obes Targets Ther. 14:3567–3602. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ramanujam CL and Zgonis T: Salvage of
Charcot foot neuropathy superimposed with osteomyelitis: A case
report. J Wound Care. 19:pp. 485–487. 2010, View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Armstrong DG, Boulton AJM and Bus SA:
Diabetic Foot Ulcers and Their Recurrence. N Engl J Med.
376:2367–2375. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Louiselle AE, Niemiec SM, Zgheib C and
Liechty KW: Macrophage polarization and diabetic wound healing.
Transl Res J Lab Clin Med. 236:109–116. 2021.
|
|
5
|
den Dekker A, Davis FM, Kunkel SL and
Gallagher KA: Targeting epigenetic mechanisms in diabetic wound
healing. Transl Res J Lab Clin Med. 204:39–50. 2019.PubMed/NCBI
|
|
6
|
Wynn TA, Chawla A and Pollard JW:
Macrophage biology in development, homeostasis and disease. Nature.
496:445–455. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gordon S, Plüddemann A and Martinez
Estrada F: Macrophage heterogeneity in tissues: Phenotypic
diversity and functions. Immunol Rev. 262:36–55. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mantovani A, Biswas SK, Galdiero MR, Sica
A and Locati M: Macrophage plasticity and polarization in tissue
repair and remodelling. J Pathol. 229:176–185. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mahdavian Delavary B, van der Veer WM, van
Egmond M, Niessen FB and Beelen RHJ: Macrophages in skin injury and
repair. Immunobiology. 216:753–762. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wen JH, Li DY, Liang S, Yang C, Tang JX
and Liu HF: Macrophage autophagy in macrophage polarization,
chronic inflammation and organ fibrosis. Front Immunol.
13:9468322022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xie D and Ouyang S: The role and
mechanisms of macrophage polarization and hepatocyte pyroptosis in
acute liver failure. Front Immunol. 14:12792642023. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shapouri-Moghaddam A, Mohammadian S,
Vazini H, Taghadosi M, Esmaeili SA, Mardani F, Seifi B, Mohammadi
A, Afshari JT and Sahebkar A: Macrophage plasticity, polarization,
and function in health and disease. J Cell Physiol. 233:6425–6440.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kelly B and O'Neill LA: Metabolic
reprogramming in macrophages and dendritic cells in innate
immunity. Cell Res. 25:771–784. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Martin KE and García AJ: Macrophage
phenotypes in tissue repair and the foreign body response:
Implications for biomaterial-based regenerative medicine
strategies. Acta Biomater. 133:4–16. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kolliniati O, Ieronymaki E, Vergadi E and
Tsatsanis C: Metabolic regulation of macrophage activation. J
Innate Immun. 14:51–68. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gordon S and Martinez FO: Alternative
activation of macrophages: Mechanism and functions. Immunity.
32:593–604. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mills CD and Ley K: M1 and M2 Macrophages:
The chicken and the egg of immunity. J Innate Immun. 6:716–726.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Orecchioni M, Ghosheh Y, Pramod AB and Ley
K: Macrophage Polarization: Different Gene Signatures in M1(LPS+)
vs. Classically and M2(LPS-) vs. Alternatively Activated
Macrophages. Front Immunol. 10:10842019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wynn TA and Vannella KM: Macrophages in
tissue repair, regeneration, and fibrosis. Immunity. 44:450–462.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mantovani A, Sica A, Sozzani S, Allavena
P, Vecchi A and Locati M: The chemokine system in diverse forms of
macrophage activation and polarization. Trends Immunol. 25:677–686.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
De Paoli F, Staels B and Chinetti-Gbaguidi
G: Macrophage phenotypes and their modulation in atherosclerosis.
Circ J. 78:1775–1781. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang L, Zhang S, Wu H, Rong X and Guo J:
M2b macrophage polarization and its roles in diseases. J Leukoc
Biol. 106:345–358. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mosser DM and Edwards JP: Exploring the
full spectrum of macrophage activation. Nat Rev Immunol. 8:958–969.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Matsui H, Sopko NA, Hannan JL, Reinhardt
AA, Kates M, Yoshida T, Liu X, Castiglione F, Hedlund P, Weyne E,
et al: M1 macrophages are predominantly recruited to the major
pelvic ganglion of the rat following cavernous nerve injury. J Sex
Med. 14:187–195. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yue Y, Yang X, Feng K, Wang L, Hou J, Mei
B, Qin H, Liang M, Chen G and Wu Z: M2b macrophages reduce early
reperfusion injury after myocardial ischemia in mice: A predominant
role of inhibiting apoptosis via A20. Int J Cardiol. 245:228–235.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lefèvre L, Lugo-Villarino G, Meunier E,
Valentin A, Olagnier D, Authier H, Duval C, Dardenne C, Bernad J,
Lemesre JL, et al: The C-type lectin receptors dectin-1, MR, and
SIGNR3 contribute both positively and negatively to the macrophage
response to Leishmania infantum. Immunity. 38:1038–1049. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zizzo G, Hilliard BA, Monestier M and
Cohen PL: Efficient clearance of early apoptotic cells by human
macrophages requires ‘M2c’ polarization and MerTK induction. J
Immunol. 189:3508–3520. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Martinez FO, Sica A, Mantovani A and
Locati M: Macrophage activation and polarization. Front Biosci J
Virtual Libr. 13:453–461. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang Z, Lin S, Feng W, Liu Y, Song Z, Pan
G, Zhang Y, Dai X, Ding X, Chen L and Wang Y: A potential
therapeutic target in traditional Chinese medicine for ulcerative
colitis: Macrophage polarization. Front Pharmacol. 13:9991792022.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nikovics K, Morin H, Riccobono D,
Bendahmane A and Favier AL: Hybridization-chain-reaction is a
relevant method for in situ detection of M2d-like macrophages in a
mini-pig model. FASEB J. 34:15675–15686. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Atri C, Guerfali FZ and Laouini D: Role of
human macrophage polarization in inflammation during infectious
diseases. Int J Mol Sci. 19:18012018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang Q, Ni H, Lan L, Wei X, Xiang R and
Wang Y: Fra-1 protooncogene regulates IL-6 expression in
macrophages and promotes the generation of M2d macrophages. Cell
Res. 20:701–712. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Frantz C, Stewart KM and Weaver VM: The
extracellular matrix at a glance. J Cell Sci. 123((Pt 24)):
4195–4200. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Semenza GL: Hypoxia-inducible factors in
physiology and medicine. Cell. 148:399–408. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wallace HA, Basehore BM and Zito PM: Wound
Healing Phases. StatPearls. StatPearls Publishing; Treasure Island
(FL): 2023
|
|
36
|
Huang CJ, Pu CM, Su SY, Lo SL, Lee CH and
Yen YH: Improvement of wound healing by capsaicin through
suppression of the inflammatory response and amelioration of the
repair process. Mol Med Rep. 28:1552023. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Furie B and Furie BC: Mechanisms of
Thrombus Formation. N Engl J Med. 359:938–949. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Goto S: Blood constitution: platelet
aggregation, bleeding, and involvement of leukocytes. Rev Neurol
Dis. 5 (Suppl 1):S22–S27. 2008.PubMed/NCBI
|
|
39
|
Guo S and Dipietro LA: Factors affecting
wound healing. J Dent Res. 89:219–229. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Rodrigues M, Kosaric N, Bonham CA and
Gurtner GC: Wound Healing: A cellular perspective. Physiol Rev.
99:665–706. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ding JY, Chen MJ, Wu LF, Shu GF, Fang SJ,
Li ZY, Chu XR, Li XK, Wang ZG and Ji JS: Mesenchymal stem
cell-derived extracellular vesicles in skin wound healing: Roles,
opportunities and challenges. Mil Med Res. 10:362023.PubMed/NCBI
|
|
42
|
Wu X, He W, Mu X, Liu Y, Deng J, Liu Y and
Nie X: Macrophage polarization in diabetic wound healing. Burns
Trauma. 10:tkac0512022. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Pastar I, Marjanovic J, Stone RC, Chen V,
Burgess JL, Mervis JS and Tomic-Canic M: Epigenetic regulation of
cellular functions in wound healing. Exp Dermatol. 30:1073–1089.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Rawat K and Shrivastava A: Neutrophils as
emerging protagonists and targets in chronic inflammatory diseases.
Inflamm Res. 71:1477–1488. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li M, Hou Q, Zhong L, Zhao Y and Fu X:
Macrophage Related Chronic Inflammation in Non-Healing Wounds.
Front Immunol. 12:6817102021. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Fortingo N, Melnyk S, Sutton SH, Watsky MA
and Bollag WB: Innate immune system activation, inflammation and
corneal wound healing. Int J Mol Sci. 23:149332022. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Portou MJ, Baker D, Abraham D and Tsui J:
The innate immune system, toll-like receptors and dermal wound
healing: A review. Vascul Pharmacol. 71:31–36. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Landén NX, Li D and Ståhle M: Transition
from inflammation to proliferation: A critical step during wound
healing. Cell Mol Life Sci. 73:3861–3885. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kotwal GJ and Chien S: Macrophage
differentiation in normal and accelerated wound healing. Results
Probl Cell Differ. 62:353–364. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yu H, Xiong J, Qiu J, He X, Sheng H, Dai
Q, Li D, Xin R, Jiang L, Li Q, et al: Type III Secretion Protein,
PcrV, Impairs Pseudomonas aeruginosa Biofilm Formation by
Increasing M1 Macrophage-Mediated Anti-bacterial Activities. Front
Microbiol. 11:19712020. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Won YW, Patel AN and Bull DA: Cell surface
engineering to enhance mesenchymal stem cell migration toward an
SDF-1 gradient. Biomaterials. 35:5627–5635. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yao Y, Xu XH and Jin L: Macrophage
polarization in physiological and pathological pregnancy. Front
Immunol. 10:7922019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Keewan E and Naser SA: The role of notch
signaling in macrophages during inflammation and infection:
Implication in rheumatoid arthritis? Cells. 9:1112020. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Delgado AV, McManus AT and Chambers JP:
Production of tumor necrosis factor-alpha, interleukin 1-beta,
interleukin 2, and interleukin 6 by rat leukocyte subpopulations
after exposure to substance P. Neuropeptides. 37:355–361. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Andreou I, Sun X, Stone PH, Edelman ER and
Feinberg MW: miRNAs in atherosclerotic plaque initiation,
progression, and rupture. Trends Mol Med. 21:307–318. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Deng JY, Wu XQ, He WJ, Liao X, Tang M and
Nie XQ: Targeting DNA methylation and demethylation in diabetic
foot ulcers. J Adv Res. 54:119–131. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Abbasi S, Sinha S, Labit E, Rosin NL, Yoon
G, Rahmani W, Jaffer A, Sharma N, Hagner A, Shah P, et al: Distinct
regulatory programs control the latent regenerative potential of
dermal fibroblasts during wound healing. Cell Stem Cell.
28:581–583. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Foster DS, Januszyk M, Yost KE, Chinta MS,
Gulati GS, Nguyen AT, Burcham AR, Salhotra A, Ransom RC, Henn D, et
al: Integrated spatial multiomics reveals fibroblast fate during
tissue repair. Proc Natl Acad Sci USA. 118:e21100251182021.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Cai F, Wang P, Chen W, Zhao R and Liu Y:
The physiological phenomenon and regulation of macrophage
polarization in diabetic wound. Mol Biol Rep. 50:9469–9477. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Krzyszczyk P, Schloss R, Palmer A and
Berthiaume F: The role of macrophages in acute and chronic wound
healing and interventions to promote pro-wound healing phenotypes.
Front Physiol. 9:4192018. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Vannella KM and Wynn TA: Mechanisms of
organ injury and repair by macrophages. Annu Rev Physiol.
79:593–617. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Boniakowski AE, Kimball AS, Jacobs BN,
Kunkel SL and Gallagher KA: Macrophage-Mediated Inflammation in
normal and diabetic wound healing. J Immunol. 199:17–24. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Lurier EB, Dalton D, Dampier W, Raman P,
Nassiri S, Ferraro NM, Rajagopalan R, Sarmady M and Spiller KL:
Transcriptome analysis of IL-10-stimulated (M2c) macrophages by
next-generation sequencing. Immunobiology. 222:847–856. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Jetten N, Verbruggen S, Gijbels MJ, Post
MJ, De Winther MPJ and Donners MM: Anti-inflammatory M2, but not
pro-inflammatory M1 macrophages promote angiogenesis in vivo.
Angiogenesis. 17:109–118. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Lolmede K, Campana L, Vezzoli M, Bosurgi
L, Tonlorenzi R, Clementi E, Bianchi ME, Cossu G, Manfredi AA,
Brunelli S and Rovere-Querini P: Inflammatory and alternatively
activated human macrophages attract vessel-associated stem cells,
relying on separate HMGB1- and MMP-9-dependent pathways. J Leukoc
Biol. 85:779–787. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
El Ayadi A, Jay JW and Prasai A: Current
approaches targeting the wound healing phases to attenuate fibrosis
and scarring. Int J Mol Sci. 21:11052020. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Gosain A and DiPietro LA: Aging and wound
healing. World J Surg. 28:321–326. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Cui N, Hu M and Khalil RA: Biochemical and
biological attributes of matrix metalloproteinases. Prog Mol Biol
Transl Sci. 147:1–73. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Ogle ME, Segar CE, Sridhar S and Botchwey
EA: Monocytes and macrophages in tissue repair: Implications for
immunoregenerative biomaterial design. Exp Biol Med (Maywood).
241:1084–1097. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Madsen DH, Leonard D, Masedunskas A, Moyer
A, Jürgensen HJ, Peters DE, Amornphimoltham P, Selvaraj A, Yamada
SS, Brenner DA, et al: M2-like macrophages are responsible for
collagen degradation through a mannose receptor-mediated pathway. J
Cell Biol. 202:951–966. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Kimball A, Schaller M, Joshi A, Davis FM,
denDekker A, Boniakowski A, Bermick J, Obi A, Moore B, Henke PK, et
al: Ly6CHi blood monocyte/macrophage drive chronic inflammation and
impair wound healing in diabetes mellitus. Arterioscler Thromb Vasc
Biol. 38:1102–1114. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Barman PK, Urao N and Koh TJ: Diabetes
induces myeloid bias in bone marrow progenitors associated with
enhanced wound macrophage accumulation and impaired healing. J
Pathol. 249:435–446. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Dasu MR, Thangappan RK, Bourgette A,
DiPietro LA, Isseroff R and Jialal I: TLR2 expression and
signaling-dependent inflammation impair wound healing in diabetic
mice. Lab Investig J Tech Methods Pathol. 90:1628–1636. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Davis FM, denDekker A, Kimball A, Joshi
AD, El Azzouny M, Wolf SJ, Obi AT, Lipinski J, Gudjonsson JE, Xing
X, et al: Epigenetic Regulation of TLR4 in Diabetic Macrophages
Modulates Immunometabolism and Wound Repair. J Immunol.
204:2503–2513. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Dasu MR and Martin SJ: Toll-like receptor
expression and signaling in human diabetic wounds. World J
Diabetes. 5:219–223. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Bannon P, Wood S, Restivo T, Campbell L,
Hardman MJ and Mace KA: Diabetes induces stable intrinsic changes
to myeloid cells that contribute to chronic inflammation during
wound healing in mice. Dis Model Mech. 6:1434–1447. 2013.PubMed/NCBI
|
|
77
|
Mirza RE, Fang MM, Ennis WJ and Koh TJ:
Blocking interleukin-1β induces a healing-associated wound
macrophage phenotype and improves healing in type 2 diabetes.
Diabetes. 62:2579–2587. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Gallagher KA, Joshi A, Carson WF, Schaller
M, Allen R, Mukerjee S, Kittan N, Feldman EL, Henke PK, Hogaboam C,
et al: Epigenetic changes in bone marrow progenitor cells influence
the inflammatory phenotype and alter wound healing in type 2
diabetes. Diabetes. 64:1420–1430. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Lin CW, Hung CM, Chen WJ, Chen JC, Huang
WY, Lu CS, Kuo ML and Chen SG: New horizons of macrophage
immunomodulation in the healing of diabetic foot ulcers.
Pharmaceutics. 14:20652022. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Gu XY, Shen SE, Huang CF, Liu YN, Chen YC,
Luo L, Zeng Y and Wang AP: Effect of activated autologous
monocytes/macrophages on wound healing in a rodent model of
experimental diabetes. Diabetes Res Clin Pract. 102:53–59. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Chen C, Liu T, Tang Y, Luo G, Liang G and
He W: Epigenetic regulation of macrophage polarization in wound
healing. Burns Trauma. 11:tkac0572023. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Pradhan L, Cai X, Wu S, Andersen ND,
Martin M, Malek J, Guthrie P, Veves A and Logerfo FW: Gene
expression of pro-inflammatory cytokines and neuropeptides in
diabetic wound healing. J Surg Res. 167:336–342. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Theocharidis G and Veves A: Autonomic
nerve dysfunction and impaired diabetic wound healing; the role of
neuropeptides. Auton Neurosci Basic Clin. 223:1026102020.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Lu YZ, Nayer B, Singh SK, Alshoubaki YK,
Yuan E, Park AJ, Maruyama K, Akira S and Martino MM: CGRP sensory
neurons promote tissue healing via neutrophils and macrophages.
Nature. 628:604–611. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Kim H, Wang SY, Kwak G, Yang Y, Kwon IC
and Kim SH: Exosome-Guided Phenotypic Switch of M1 to M2
Macrophages for Cutaneous Wound Healing. Adv Sci (Weinh).
6:19005132019. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Spampinato SF, Caruso GI, De Pasquale R,
Sortino MA and Merlo S: The treatment of impaired wound healing in
diabetes: looking among old drugs. Pharm (Basel). 13:602020.
|
|
87
|
Mirza R and Koh TJ: Dysregulation of
monocyte/macrophage phenotype in wounds of diabetic mice. Cytokine.
56:256–264. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Gurevich DB, Severn CE, Twomey C,
Greenhough A, Cash J, Toye AM, Mellor H and Martin P: Live imaging
of wound angiogenesis reveals macrophage orchestrated vessel
sprouting and regression. EMBO J. 37:e977862018. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Okonkwo UA, Chen L, Ma D, Haywood VA,
Barakat M, Urao N and DiPietro LA: Compromised angiogenesis and
vascular Integrity in impaired diabetic wound healing. PLoS One.
15:e02319622020. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Al Sadoun H: Macrophage phenotypes in
normal and diabetic wound healing and therapeutic interventions.
Cells. 11:24302022. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Nacev BA, Jones KB, Intlekofer AM, Yu JSE,
Allis CD, Tap WD, Ladanyi M and Nielsen TO: The epigenomics of
sarcoma. Nat Rev Cancer. 20:608–623. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Ma X, Mei S, Wuyun Q, Zhou L, Sun D and
Yan J: Epigenetics in diabetic cardiomyopathy. Clin Epigenetics.
16:522024. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Ahmed M, de Winther MPJ and Van den
Bossche J: Epigenetic mechanisms of macrophage activation in type 2
diabetes. Immunobiology. 222:937–943. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Davis FM and Gallagher KA: Epigenetic
mechanisms in monocytes/macrophages regulate inflammation in
cardiometabolic and vascular disease. Arterioscler Thromb Vasc
Biol. 39:623–634. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Yassi M, Chatterjee A and Parry M:
Application of deep learning in cancer epigenetics through DNA
methylation analysis. Brief Bioinform. 24:bbad4112023. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Wong WK, Yin B, Lam CYK, Huang Y, Yan J,
Tan Z and Wong SHD: The Interplay Between Epigenetic Regulation and
CD8+ T Cell Differentiation/Exhaustion for T Cell Immunotherapy.
Front Cell Dev Biol. 9:7832272022. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Xu F, Mao C, Ding Y, Rui C, Wu L, Shi A,
Zhang H, Zhang L and Xu Z: Molecular and enzymatic profiles of
mammalian DNA methyltransferases: structures and targets for drugs.
Curr Med Chem. 17:4052–4071. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Rabkin SW and Wong CN: Epigenetics in
Heart Failure: Role of DNA methylation in potential pathways
leading to heart failure with preserved ejection fraction.
Biomedicines. 11:28152023. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Moore LD, Le T and Fan G: DNA methylation
and its basic function. Neuropsychopharmacology. 38:23–38. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Hamidi T, Singh AK and Chen T: Genetic
alterations of DNA methylation machinery in human diseases.
Epigenomics. 7:247–265. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Vatanmakanian M, Steffan JJ, Koul S, Ochoa
AC, Chaturvedi LS and Koul HK: Regulation of SPDEF expression by
DNA methylation in advanced prostate cancer. Front Endocrinol
(Lausanne). 14:11561202023. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
De Marco K, Sanese P, Simone C and Grossi
V: Histone and DNA methylation as epigenetic regulators of DNA
damage repair in gastric cancer and emerging therapeutic
opportunities. Cancers (Basel). 15:49762023. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Zeng Y, Rong H, Xu J, Cao R, Li S, Gao Y,
Cheng B and Zhou T: DNA Methylation: An Important Biomarker and
Therapeutic Target for Gastric Cancer. Front Genet. 13:8239052022.
View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Adam S, Klingel V, Radde NE, Bashtrykov P
and Jeltsch A: On the accuracy of the epigenetic copy machine:
Comprehensive specificity analysis of the DNMT1 DNA
methyltransferase. Nucleic Acids Res. 51:6622–6633. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Smith ZD and Meissner A: DNA methylation:
Roles in mammalian development. Nat Rev Genet. 14:204–220. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Topriceanu CC, Dev E, Ahmad M, Hughes R,
Shiwani H, Webber M, Direk K, Wong A, Ugander M, Moon JC, et al:
Accelerated DNA methylation age plays a role in the impact of
cardiovascular risk factors on the human heart. Clin Epigenetics.
15:1642023. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Chen ZX and Riggs AD: DNA methylation and
demethylation in mammals. J Biol Chem. 286:18347–18353. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Ren L, Chang YF, Jiang SH, Li XH and Cheng
HP: DNA methylation modification in Idiopathic pulmonary fibrosis.
Front Cell Dev Biol. 12:14163252024. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Yano N and Fedulov AV: Targeted DNA
Demethylation: Vectors, Effectors and Perspectives. Biomedicines.
11:13342023. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Guo F, Li X, Liang D, Li T, Zhu P, Guo H,
Wu X, Wen L, Gu TP, Hu B, et al: Active and passive demethylation
of male and female pronuclear DNA in the mammalian zygote. Cell
Stem Cell. 15:447–459. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Ravichandran M, Jurkowska RZ and Jurkowski
TP: Target specificity of mammalian DNA methylation and
demethylation machinery. Org Biomol Chem. 16:1419–1435. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Yan J, Tie G, Wang S, Tutto A, DeMarco N,
Khair L, Fazzio TG and Messina LM: Diabetes impairs wound healing
by Dnmt1-dependent dysregulation of hematopoietic stem cells
differentiation towards macrophages. Nat Commun. 9:332018.
View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Babu M, Durga Devi T, Mäkinen P, Kaikkonen
M, Lesch HP, Junttila S, Laiho A, Ghimire B, Gyenesei A and
Ylä-Herttuala S: Differential promoter methylation of macrophage
genes is associated with impaired vascular growth in ischemic
muscles of hyperlipidemic and type 2 diabetic Mice: Genome-Wide
Promoter Methylation Study. Circ Res. 117:289–299. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Davis FM, Tsoi LC, Wasikowski R, denDekker
A, Joshi A, Wilke C, Deng H, Wolf S, Obi A, Huang S, et al:
Epigenetic regulation of the PGE2 pathway modulates macrophage
phenotype in normal and pathologic wound repair. JCI Insight.
5:e1384432020. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Park J, Lee K, Kim K and Yi SJ: The role
of histone modifications: From neurodevelopment to neurodiseases.
Signal Transduct Target Ther. 7:2172022. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Li Y, Hu M, Xie J, Li S and Dai L:
Dysregulation of histone modifications in bone marrow mesenchymal
stem cells during skeletal ageing: Roles and therapeutic prospects.
Stem Cell Res Ther. 14:1662023. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Fulton MD, Zhang J, He M, Ho MC and Zheng
YG: Intricate Effects of α-Amino and Lysine Modifications on
Arginine Methylation of the N-Terminal Tail of Histone H4.
Biochemistry. 56:3539–3548. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Jambhekar A, Dhall A and Shi Y: Roles and
regulation of histone methylation in animal development. Nat Rev
Mol Cell Biol. 20:625–641. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Shvedunova M and Akhtar A: Modulation of
cellular processes by histone and non-histone protein acetylation.
Nat Rev Mol Cell Biol. 23:329–349. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Allis CD and Jenuwein T: The molecular
hallmarks of epigenetic control. Nat Rev Genet. 17:487–500. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Zhu W, Liu L, Wu J, Gao R, Fu L, Yang X,
Zou Y, Zhang S and Luo D: SMYD3 activates the TCA cycle to promote
M1-M2 conversion in macrophages. Int Immunopharmacol.
127:1113292024. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Chi G, Pei JH and Li XQ: EZH2-mediated
H3K27me3 promotes autoimmune hepatitis progression by regulating
macrophage polarization. Int Immunopharmacol. 106:1086122022.
View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Schaller MA: MLL1 is central to
macrophage-mediated inflammation. Blood. 141:687–689. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Kimball AS, Joshi A, Carson WF IV,
Boniakowski AE, Schaller M, Allen R, Bermick J, Davis FM, Henke PK,
Burant CF, et al: The Histone Methyltransferase MLL1 directs
macrophage-mediated inflammation in wound healing and is altered in
a murine model of obesity and type 2 diabetes. Diabetes.
66:2459–2471. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Schliehe C, Flynn EK, Vilagos B, et al:
The methyltransferase Setdb2 mediates virus-induced susceptibility
to bacterial superinfection. Nat Immunol. 16:67–74. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Kimball AS, Davis FM, denDekker A, Joshi
AD, Schaller MA, Bermick J, Xing X, Burant CF, Obi AT, Nysz D, et
al: The Histone Methyltransferase Setdb2 Modulates Macrophage
Phenotype and Uric Acid Production in Diabetic Wound Repair.
Immunity. 51:258–271.e5. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Davis FM, denDekker A, Joshi AD, Wolf SJ,
Audu C, Melvin WJ, Mangum K, Riordan MO, Kunkel SL and Gallagher
KA: Palmitate-TLR4 signaling regulates the histone demethylase,
JMJD3, in macrophages and impairs diabetic wound healing. Eur J
Immunol. 50:1929–1940. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Taipale M, Rea S, Richter K, Vilar A,
Lichter P, Imhof A and Akhtar A: hMOF histone acetyltransferase is
required for histone H4 lysine 16 acetylation in mammalian cells.
Mol Cell Biol. 25:6798–6810. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Rea S, Xouri G and Akhtar A: Males absent
on the first (MOF): From flies to humans. Oncogene. 26:5385–5394.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
denDekker AD, Davis FM, Joshi AD, Wolf SJ,
Allen R, Lipinski J, Nguyen B, Kirma J, Nycz D, Bermick J, et al:
TNF-α regulates diabetic macrophage function through the histone
acetyltransferase MOF. JCI Insight. 5:e1323062020. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Mullican SE, Gaddis CA, Alenghat T, Nair
MG, Giacomin PR, Everett LJ, Feng D, Steger DJ, Schug J, Artis D
and Lazar MA: Histone deacetylase 3 is an epigenomic brake in
macrophage alternative activation. Genes Dev. 25:2480–2488. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Shakespear MR, Halili MA, Irvine KM,
Fairlie DP and Sweet MJ: Histone deacetylases as regulators of
inflammation and immunity. Trends Immunol. 32:335–343. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Chen X, Barozzi I, Termanini A, Prosperini
E, Recchiuti A, Dalli J, Mietton F, Matteoli G, Hiebert S and
Natoli G: Requirement for the histone deacetylase Hdac3 for the
inflammatory gene expression program in macrophages. Proc Natl Acad
Sci USA. 109:E2865–E2874. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Teena R, Dhamodharan U, Ali D, Rajesh K
and Ramkumar KM: Gene expression profiling of multiple histone
deacetylases (HDAC) and Its Correlation with NRF2-Mediated redox
regulation in the pathogenesis of diabetic foot ulcers.
Biomolecules. 10:14662020. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Cabanel M, da Costa TP, El-Cheikh MC and
Carneiro K: The epigenome as a putative target for skin repair: the
HDAC inhibitor Trichostatin A modulates myeloid progenitor
plasticity and behavior and improves wound healing. J Transl Med.
17:2472019. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Karnam K, Sedmaki K, Sharma P, Mahale A,
Ghosh B and Kulkarni OP: Pharmacological blockade of HDAC3
accelerates diabetic wound healing by regulating macrophage
activation. Life Sci. 321:1215742023. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Djebali S, Davis CA, Merkel A, Dobin A,
Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F,
et al: Landscape of transcription in human cells. Nature.
489:101–108. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Tang YB, Uwimana MMP, Zhu SQ, Zhang LX, Wu
Q and Liang ZX: Non-coding RNAs: Role in diabetic foot and wound
healing. World J Diabetes. 13:1001–1013. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Maciak P, Suder A, Wadas J, Aronimo F,
Maiuri P, Bochenek M, Pyrc K, Kula-Pacurar A and Pabis M: Dynamic
changes in LINC00458/HBL1 lncRNA expression during hiPSC
differentiation to cardiomyocytes. Sci Rep. 14:1092024. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Zhang P, Wu S, He Y, Li X, Zhu Y, Lin X,
Chen L, Zhao Y, Niu L, Zhang S, et al: LncRNA-Mediated adipogenesis
in different adipocytes. Int J Mol Sci. 23:74882022. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Zaki A, Ali MS, Hadda V, Ali SM, Chopra A
and Fatma T: Long non-coding RNA (lncRNA): A potential therapeutic
target in acute lung injury. Genes Dis. 9:1258–1268. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Hussein RM: Long non-coding RNAs: The
hidden players in diabetes mellitus-related complications. Diabetes
Metab Syndr. 17:1028722023. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Hu J, Zhang L, Liechty C, Zgheib C, Hodges
MM, Liechty KW and Xu J: Long Noncoding RNA GAS5 regulates
macrophage polarization and diabetic wound healing. J Invest
Dermatol. 140:1629–1638. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Kuang L, Zhang C, Li B, Deng H, Chen R and
Li G: Human Keratinocyte-Derived Exosomal MALAT1 promotes diabetic
wound healing by upregulating MFGE8 via microRNA-1914-3p. Int J
Nanomedicine. 18:949–970. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Saliminejad K, Khorram Khorshid HR,
Soleymani Fard S and Ghaffari SH: An overview of microRNAs:
Biology, functions, therapeutics, and analysis methods. J Cell
Physiol. 234:5451–5465. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Kmiołek T, Rzeszotarska E, Wajda A,
Walczuk E, Kuca-Warnawin E, Romanowska-Próchnicka K, Stypinska B,
Majewski D, Jagodzinski PP, Pawlik A and Paradowska-Gorycka A: The
Interplay between Transcriptional Factors and MicroRNAs as an
Important Factor for Th17/Treg Balance in RA Patients. Int J Mol
Sci. 21:71692020. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Self-Fordham JB, Naqvi AR, Uttamani JR,
Kulkarni V and Nares S: MicroRNA: Dynamic regulators of macrophage
polarization and plasticity. Front Immunol. 8:10622017. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Pasca S, Jurj A, Petrushev B, Tomuleasa C
and Matei D: MicroRNA-155 Implication in M1 polarization and the
impact in inflammatory diseases. Front Immunol. 11:6252020.
View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Moura J, Sørensen A, Leal EC, Svendsen R,
Carvalho L, Willemoes RJ, Jørgensen PT, Jenssen H, Wengel J,
Dalgaard LT and Carvalho E: microRNA-155 inhibition restores
Fibroblast Growth Factor 7 expression in diabetic skin and
decreases wound inflammation. Sci Rep. 9:58362019. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Ye J, Kang Y, Sun X, Ni P, Wu M and Lu S:
MicroRNA-155 inhibition promoted wound healing in diabetic rats.
Int J Low Extrem Wounds. 16:74–84. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Peng X, He F, Mao Y, Lin Y, Fang J, Chen
Y, Sun Z, Zhuo Y and Jiang J: miR-146a promotes M2 macrophage
polarization and accelerates diabetic wound healing by inhibiting
the TLR4/NF-κB axis. J Mol Endocrinol. 69:315–327. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Hao Y, Yang L, Liu Y, Ye Y, Wang J, Yu C,
Yan H, Xing Y, Jia Z, Hu C, et al: mmu-miR-145a-5p accelerates
diabetic wound healing by promoting macrophage polarization toward
the M2 Phenotype. Front Med. 8:7755232021. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Liechty C, Hu J, Zhang L, Liechty KW and
Xu J: Role of microRNA-21 and its underlying mechanisms in
inflammatory responses in diabetic wounds. Int J Mol Sci.
21:33282020. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Meng Z, Zhou D, Gao Y, Zeng M and Wang W:
miRNA delivery for skin wound healing. Adv Drug Deliv Rev.
129:308–318. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Boca S, Gulei D, Zimta AA, Onaciu A, Magdo
L, Tigu AB, Ionescu C, Irimie A, Buiga R and Berindan-Neagoe I:
Nanoscale delivery systems for microRNAs in cancer therapy. Cell
Mol Life Sci. 77:1059–1086. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Tang XH, Guo T, Gao XY, Wu XL, Xing XF, Ji
JF and Li ZY: Exosome-derived noncoding RNAs in gastric cancer:
functions and clinical applications. Mol Cancer. 20:992021.
View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Yang Q, Luo Y, Ge P, Lan B, Liu J, Wen H,
Cao Y, Sun Z, Zhang G, Yuan H, et al: Emodin ameliorates severe
acute pancreatitis-associated acute lung injury in rats by
modulating exosome-specific miRNA Expression Profiles. Int J
Nanomedicine. 18:6743–6761. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Ye H, Wang F, Xu G, Shu F, Fan K and Wang
D: Advancements in engineered exosomes for wound repair: Current
research and future perspectives. Front Bioeng Biotechnol.
11:13013622023. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Khalaj K, Figueira RL, Antounians L,
Lauriti G and Zani A: Systematic review of extracellular
vesicle-based treatments for lung injury: are EVs a potential
therapy for COVID-19? J Extracell Vesicles. 9:17953652020.
View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Zhao H, Shang Q, Pan Z, Bai Y, Li Z, Zhang
H, Zhang Q, Guo C, Zhang L and Wang Q: Exosomes from
adipose-derived stem cells attenuate adipose inflammation and
obesity through polarizing M2 macrophages and beiging in white
adipose tissue. Diabetes. 67:235–247. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Xia W, Liu Y, Jiang X, Li M, Zheng S,
Zhang Z, Huang X, Luo S, Khoong Y, Hou M and Zan T: Lean adipose
tissue macrophage derived exosome confers immunoregulation to
improve wound healing in diabetes. J Nanobiotechnology. 21:1282023.
View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Blanpain C and Fuchs E: Epidermal stem
cells of the skin. Annu Rev Cell Dev Biol. 22:339–373. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Ma T, Zhao Y, Shen G, Chai B, Wang W, Li
X, Zhang Z and Meng Q: Novel bilayer cell patch combining epidermal
stem cells and angiogenic adipose stem cells for diabetic wound
healing. J Control Release. 359:315–325. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Yang RH, Qi SH, Shu B, Ruan SB, Lin ZP,
Lin Y, Shen R, Zhang FG, Chen XD and Xie JL: Epidermal stem cells
(ESCs) accelerate diabetic wound healing via the Notch signalling
pathway. Biosci Rep. 36:e003642016. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Xu H, Yang H, Wang Z, Tang Q, Cao X, Chen
C, Dong Y, Xu Z, Lv D, Rong Y, et al: Epidermal stem cell derived
exosomes alleviate excessive autophagy induced endothelial cell
apoptosis by delivering miR200b-3p to Diabetic Wounds. J Invest
Dermatol. 144:1134–1147.e2. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Yang H, Xu H, Wang Z, Li X, Wang P, Cao X,
Xu Z, Lv D, Rong Y, Chen M, et al: Analysis of
miR-203a-3p/SOCS3-mediated induction of M2 macrophage polarization
to promote diabetic wound healing based on epidermal stem
cell-derived exosomes. Diabetes Res Clin Pract. 197:1105732023.
View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Hassan WU, Greiser U and Wang W: Role of
adipose-derived stem cells in wound healing. Wound Repair Regen.
22:313–325. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Markov A, Thangavelu L, Aravindhan S,
Zekiy AO, Jarahian M, Chartrand MS, Pathak Y, Marofi F, Shamlou S
and Hassanzadeh A: Mesenchymal stem/stromal cells as a valuable
source for the treatment of immune-mediated disorders. Stem Cell
Res Ther. 12:1922021. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Zou JP, Huang S, Peng Y, Liu HW, Cheng B,
Fu XB and Xiang XF: Mesenchymal stem cells/multipotent mesenchymal
stromal cells (MSCs): Potential role in healing cutaneous chronic
wounds. Int J Low Extrem Wounds. 11:244–253. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Mahjoor M, Fakouri A, Farokhi S, Nazari H,
Afkhami H and Heidari F: Regenerative potential of mesenchymal
stromal cells in wound healing: Unveiling the influence of normoxic
and hypoxic environments. Front Cell Dev Biol. 11:12458722023.
View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Ti D, Hao H, Tong C, Liu J, Dong L, Zheng
J, Zhao Y, Liu H, Fu X and Han W: LPS-preconditioned mesenchymal
stromal cells modify macrophage polarization for resolution of
chronic inflammation via exosome-shuttled let-7b. J Transl Med.
13:3082015. View Article : Google Scholar : PubMed/NCBI
|
|
172
|
Shi Y, Wang S, Zhang W, Zhu Y, Fan Z,
Huang Y, Li F and Yang R: Bone marrow mesenchymal stem cells
facilitate diabetic wound healing through the restoration of
epidermal cell autophagy via the HIF-1α/TGF-β1/SMAD pathway. Stem
Cell Res Ther. 13:3142022. View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Li L, Chen X, Wang WE and Zeng C: How to
improve the survival of transplanted mesenchymal stem cell in
ischemic heart? Stem Cells Int. 2016:96827572016. View Article : Google Scholar : PubMed/NCBI
|
|
174
|
Zhou X, Ye C, Jiang L, Zhu X, Zhou F, Xia
M and Chen Y: The bone mesenchymal stem cell-derived exosomal
miR-146a-5p promotes diabetic wound healing in mice via macrophage
M1/M2 polarization. Mol Cell Endocrinol. 579:1120892024. View Article : Google Scholar : PubMed/NCBI
|
|
175
|
Sun Y, Zhou Y, Shi Y, Zhang Y, Liu K,
Liang R, Sun P, Chang X, Tang W, Zhang Y, et al: Expression of
miRNA-29 in Pancreatic β Cells Promotes Inflammation and Diabetes
via TRAF3. Cell Rep. 34:1085762021. View Article : Google Scholar : PubMed/NCBI
|
|
176
|
Zeng X, Chen B, Wang L, Sun Y, Jin Z, Liu
X, Ouyang L and Liao Y: Chitosan@Puerarin hydrogel for accelerated
wound healing in diabetic subjects by miR-29ab1 mediated
inflammatory axis suppression. Bioact Mater. 19:653–665.
2022.PubMed/NCBI
|
|
177
|
Reda El Sayed S, Cristante J, Guyon L,
Denis J, Chabre O and Cherradi N: MicroRNA therapeutics in cancer:
Current advances and challenges. Cancers (Basel). 13:26802021.
View Article : Google Scholar : PubMed/NCBI
|
|
178
|
Bhatnagar D, Ladhe S and Kumar D:
Discerning the Prospects of miRNAs as a Multi-Target Therapeutic
and Diagnostic for Alzheimer's Disease. Mol Neurobiol.
60:5954–5974. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
179
|
Huang S, Zhou Y, Zhang Y, Liu N, Liu J,
Liu L and Fan C: Advances in MicroRNA therapy for heart failure:
Clinical trials, preclinical studies, and controversies. Cardiovasc
Drugs Ther. Jul 28–2023.(Epub ahead of print). View Article : Google Scholar
|
|
180
|
Amina SJ, Azam T, Dagher F and Guo B: A
review on the use of extracellular vesicles for the delivery of
drugs and biological therapeutics. Expert Opin Drug Deliv.
21:45–70. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
181
|
Yin W, Ma H, Qu Y, Wang S, Zhao R, Yang Y
and Guo ZN: Targeted exosome-based nanoplatform for new-generation
therapeutic strategies. Biomed Mater. 19:2024. View Article : Google Scholar
|
|
182
|
Li X, Yang L and Chen LL: The biogenesis,
functions, and challenges of circular RNAs. Mol Cell. 71:428–442.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
183
|
Zhang W, He Y and Zhang Y: CircRNA in
ocular neovascular diseases: Fundamental mechanism and clinical
potential. Pharmacol Res. 197:1069462023. View Article : Google Scholar : PubMed/NCBI
|
|
184
|
Moallemi Rad L, Sadoughi MM, Nicknam A,
Colagar AH, Hussen BM, Taheri M and Ghafouri-Fard S: The impact of
non-coding RNAs in the pathobiology of eye disorders. Int J Biol
Macromol. 239:1242452023. View Article : Google Scholar : PubMed/NCBI
|
|
185
|
Titze-de-Almeida SS and Titze-de-Almeida
R: Progress in circRNA-Targeted Therapy in Experimental Parkinson's
Disease. Pharmaceutics. 15:20352023. View Article : Google Scholar : PubMed/NCBI
|
|
186
|
Liu J, Duan P, Xu C, Xu D, Liu Y and Jiang
J: CircRNA circ-ITCH improves renal inflammation and fibrosis in
streptozotocin-induced diabetic mice by regulating the
miR-33a-5p/SIRT6 axis. Inflamm Res. 70:835–846. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
187
|
Shang B, Xu T, Hu N, Mao Y and Du X:
Circ-Klhl8 overexpression increased the therapeutic effect of EPCs
in diabetic wound healing via the miR-212-3p/SIRT5 axis. J Diabetes
Complications. 35:1080202021. View Article : Google Scholar : PubMed/NCBI
|
|
188
|
Lareyre F, Clément M, Moratal C, Loyer X,
Jean-Baptiste E, Hassen-Khodja R, Chinetti G, Mallat Z and Raffort
J: Differential micro-RNA expression in diabetic patients with
abdominal aortic aneurysm. Biochimie. 162:1–7. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
189
|
Hu L, Wang J, Zhou X, Xiong Z, Zhao J, Yu
R, Huang F, Zhang H and Chen L: Exosomes derived from human adipose
mensenchymal stem cells accelerates cutaneous wound healing via
optimizing the characteristics of fibroblasts. Sci Rep.
6:329932016. View Article : Google Scholar : PubMed/NCBI
|
|
190
|
Shi R, Jin Y, Zhao S, Yuan H, Shi J and
Zhao H: Hypoxic ADSC-derived exosomes enhance wound healing in
diabetic mice via delivery of circ-Snhg11 and induction of M2-like
macrophage polarization. Biomed Pharmacother. 153:1134632022.
View Article : Google Scholar : PubMed/NCBI
|
|
191
|
Li Y, Luo Q, Li Z, Wang Y, Zhu C, Li T and
Li X: Long Non-coding RNA IRAIN Inhibits VEGFA Expression via
Enhancing Its DNA methylation leading to tumor suppression in renal
carcinoma. Front Oncol. 10:10822020. View Article : Google Scholar : PubMed/NCBI
|
|
192
|
Wen J, Wu Y and Luo Q: DNA
methyltransferases-associated long non-coding RNA PRKCQ-AS1
regulate DNA methylation in myelodysplastic syndrome. Int J Lab
Hematol. Apr 28–2024.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
193
|
Sun SC: The non-canonical NF-κB pathway in
immunity and inflammation. Nat Rev Immunol. 17:545–558. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
194
|
Xia T, Fu S, Yang R, Yang K, Lei W, Yang
Y, Zhang Q, Zhao Y, Yu J, Yu L and Zhang T: Advances in the study
of macrophage polarization in inflammatory immune skin diseases. J
Inflamm (Lond). 20:332023. View Article : Google Scholar : PubMed/NCBI
|
|
195
|
Zhang Q, Lenardo MJ and Baltimore D: 30
Years of NF-κB: A blossoming of relevance to human pathobiology.
Cell. 168:37–57. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
196
|
Gao S, Mao F, Zhang B, Zhang L, Zhang X,
Wang M, Yan Y, Yang T, Zhang J, Zhu W, et al: Mouse bone
marrow-derived mesenchymal stem cells induce macrophage M2
polarization through the nuclear factor-κB and signal transducer
and activator of transcription 3 pathways. Exp Biol Med (Maywood).
239:366–375. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
197
|
Deng L, Du C, Song P, Chen T, Rui S,
Armstrong DG and Deng W: The role of oxidative stress and
antioxidants in diabetic wound healing. Oxid Med Cell Longev.
2021:88527592021. View Article : Google Scholar : PubMed/NCBI
|
|
198
|
Dunnill C, Patton T, Brennan J, Barrett J,
Dryden M, Cooke J, Leaper D and Georgopoulos NT: Reactive oxygen
species (ROS) and wound healing: the functional role of ROS and
emerging ROS-modulating technologies for augmentation of the
healing process. Int Wound J. 14:89–96. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
199
|
Wan L, Bai X, Zhou Q, Chen C, Wang H, Liu
T, Xue J, Wei C and Xie L: The advanced glycation end-products
(AGEs)/ROS/NLRP3 inflammasome axis contributes to delayed diabetic
corneal wound healing and nerve regeneration. Int J Biol Sci.
18:809–825. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
200
|
Kang HJ, Kumar S, D'Elia A, Dash B, Nanda
V, Hsia HC, Yarmush ML and Berthiaume F: Self-assembled
elastin-like polypeptide fusion protein coacervates as competitive
inhibitors of advanced glycation end-products enhance diabetic
wound healing. J Control Release. 333:176–187. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
201
|
O'Shea JJ, Schwartz DM, Villarino AV,
Gadina M, McInnes IB and Laurence A: The JAK-STAT pathway: impact
on human disease and therapeutic intervention. Annu Rev Med.
66:311–328. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
202
|
Liu Y, Liu Z, Tang H, Shen Y, Gong Z, Xie
N, Zhang X, Wang W, Kong W, Zhou Y and Fu Y: The N6-methyladenosine
(m6A)-forming enzyme METTL3 facilitates M1 macrophage polarization
through the methylation of STAT1 mRNA. Am J Physiol Cell Physiol.
317:C762–C775. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
203
|
Zhang MZ, Wang X, Wang Y, Niu A, Wang S,
Zou C and Harris RC: IL-4/IL-13-mediated polarization of renal
macrophages/dendritic cells to an M2a phenotype is essential for
recovery from acute kidney injury. Kidney Int. 91:375–386. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
204
|
Degboé Y, Rauwel B, Baron M, Boyer JF,
Ruyssen-Witrand A, Constantin A and Davignon JL: Polarization of
Rheumatoid Macrophages by TNF Targeting Through an IL-10/STAT3
Mechanism. Front Immunol. 10:32019. View Article : Google Scholar : PubMed/NCBI
|
|
205
|
Liu Y, Stewart KN, Bishop E, Marek CJ,
Kluth DC, Rees AJ and Wilson HM: Unique expression of suppressor of
cytokine signaling 3 is essential for classical macrophage
activation in rodents in vitro and in vivo. J Immunol.
180:6270–6278. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
206
|
Tsogbadrakh B, Ryu H, Ju KD, Lee J, Yun S,
Yu KS, Kim HJ, Ahn C and Oh KH: AICAR, an AMPK activator, protects
against cisplatin-induced acute kidney injury through the
JAK/STAT/SOCS pathway. Biochem Biophys Res Commun. 509:680–686.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
207
|
Feng L, Sun Y, Song P, Xu L, Wu X, Wu X,
Shen Y, Sun Y, Kong L, Wu X and Xu Q: Seselin ameliorates
inflammation via targeting Jak2 to suppress the proinflammatory
phenotype of macrophages. Br J Pharmacol. 176:317–333. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
208
|
Suzuki K, Meguro K, Nakagomi D and
Nakajima H: Roles of alternatively activated M2 macrophages in
allergic contact dermatitis. Allergol Int. 66:392–397. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
209
|
Bai JW, Wei M, Li JW and Zhang GJ: Notch
signaling pathway and endocrine resistance in breast cancer. Front
Pharmacol. 11:9242020. View Article : Google Scholar : PubMed/NCBI
|
|
210
|
Singla RD, Wang J and Singla DK:
Regulation of Notch 1 signaling in THP-1 cells enhances M2
macrophage differentiation. Am J Physiol Heart Circ Physiol.
307:H1634–H1642. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
211
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
212
|
Chen G, Deng C and Li YP: TGF-β and BMP
signaling in osteoblast differentiation and bone formation. Int J
Biol Sci. 8:272–288. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
213
|
Perez LG, Kempski J, McGee HM, Pelzcar P,
Agalioti T, Giannou A, Konczalla L, Brockmann L, Wahib R, Xu H, et
al: TGF-β signaling in Th17 cells promotes IL-22 production and
colitis-associated colon cancer. Nat Commun. 11:26082020.
View Article : Google Scholar : PubMed/NCBI
|
|
214
|
Wang L, Li Y, Wang X, Wang P, Essandoh K,
Cui S, Huang W, Mu X, Liu Z, Wang Y, et al: GDF3 protects mice
against sepsis-induced cardiac dysfunction and mortality by
suppression of macrophage pro-inflammatory phenotype. Cells.
9:1202020. View Article : Google Scholar : PubMed/NCBI
|
|
215
|
Louiselle AE, Niemiec SM, Zgheib C and
Liechty KW: Macrophage polarization and diabetic wound healing.
Transl Res. 236:109–116. 2021. View Article : Google Scholar : PubMed/NCBI
|