Introduction
Colorectal cancer (CRC) is one of the top three
malignant tumors worldwide. There are numerous treatment options
for early stage CRC, such as endoscopic submucosal dissection and
radical surgery; however, the prognosis of patients with advanced
CRC remains poor. Investigating more effective treatment strategies
is important for the clinical treatment of patients with advanced
CRC (1).
Exosomes are between 30 and 200 nm in diameter, with
a double-membrane disc structure. They contain various bioactive
substances, including nucleic acids and proteins (2,3).
Exosomes can be identified by observing their morphology using
transmission electron microscopy (TEM), detecting the particle size
using nanoparticle tracking analysis (NTA) and assessing the
expression levels of exosome biomarkers, including CD81, tumor
susceptibility gene 101 protein (TSG101) and calnexin (4). Tumor exosomes abundantly surround
primary tumors such as gastric cancers, exchanging and transferring
information between cells and modulating cancer progression (5–7).
Decreased expression levels of E-cadherin and increased expression
levels of vimentin are common characteristics of
epithelial-mesenchymal transition (EMT). EMT is the first step in
cancer cell metastasis, which also relies on exosomes (8,9).
Exosomes can reach distal organs and provide a microenvironment
suitable for tumor growth, enabling tumor cells to colonize the
distal organs (10).
Sphingosine kinase 1 (SphK1) is an intracellular
signaling enzyme, which regulates cancer cell migration, growth,
apoptosis, angiogenesis and other biological tumor behaviors
(11). Studies have demonstrated
that SphK1 regulates the focal adhesion kinase (FAK)/AKT/MMP axis
to promote EMT and metastasis in CRC cells (12,13).
Given its pivotal role in regulating the malignant biological
behavior of cancer cells, SphK1 is an important potential drug
target for the treatment of cancer. Recently, exosomes derived from
metastatic CRC cells have been shown to activate hepatic stellate
cells and facilitate the liver metastasis of CRC cells (14). Upregulation of α-smooth muscle
actin (α-SMA) is a key marker of hepatic stellate cell activation.
Cytokines secreted by activated hepatic stellate cells, including
TNF-α and TGF-β, can help the colonization and proliferation of
cancer cells in the liver (15–17). However, to the best of our
knowledge, it remains unknown whether exosomal SphK1 derived from
CRC cells can promote hepatic stellate cell activation.
The present study aimed to investigate whether
exosomal SphK1 from CRC cells can enhance the migration of cancer
cells and activate hepatic stellate cells to increase the viability
of CRC cells by regulating the AKT pathway. The present study
provided an experimental basis for the future clinical application
of exosomes targeting the reduction of SphK1.
Materials and methods
Blood specimens and ELISA
Ethics approval for the present study was obtained
from the Institutional Ethics Committee of Binzhou Medical
University (approval no. KT-259; Binzhou, China) according to The
Declaration of Helsinki. Written informed consent was obtained from
all 14 patients included in the present study. The blood specimens
from patients were collected at Binzhou Medical University Hospital
(Shangdong, China) between December 2023 and April 2024. There were
10 male and 4 female patients, with mean ages of 65.30±10.33 and
55.75±21.52, respectively, and an ages range between 34 and 85
years old. The patient cohort consisted of 4 patients with liver
metastases from CRC (stage IV) and 10 patients with CRC without
metastasis (stage I). Blood samples were collected before any
treatment such as surgery and chemotherapy. The patients had no
lung metastasis. Blood samples were centrifuged at 1,500 × g for 20
min at room temperature to collect the plasma. An exosome isolation
kit for plasma (cat. no. MA0403-A; Dalian Meilun Biology Technology
Co., Ltd.) was used to extract plasma exosomes from 200 µl plasma.
An exosome-specific lysis buffer (cat. no. UR33101; Shanghai
Yumeibo Biotechnology Co., Ltd.) was used to extract exosomal
proteins. A Human SPHK1 ELISA Kit [cat. no. ELK4112; Elk (Wuhan)
Biotechnology Co., Ltd.] was used to measure the levels of exosomal
SphK1 on a Multimode Reader (Agilent Technologies, Inc.) at 450 nm.
All aforementioned kits were used according to the manufacturers'
instructions.
Cell culture and treatment
The RKO CRC cell line (Cellverse Co., Ltd.) was
cultured in DMEM high-glucose liquid medium (cat. no. PYG0101;
Wuhan Boster Biological Technology, Ltd.), and the LX-2 human
hepatic stellate cell line (Dalian Meilun Biology Technology Co.,
Ltd.) was cultured in RPMI-1640 liquid medium (cat. no. PYG0049;
Wuhan Boster Biological Technology, Lt.d) at 37°C in 5%
CO2. A total of 1×105 cells were seeded in
6-well cell culture plates. The next day, exosomes
(7×103 particles/cell) were added to co-culture with the
cells for 24 h, and the cells and cell culture supernatant were
collected for subsequent experiments. The PBS group used PBS, with
the same volume of exosomes in the Control group. The AKT agonist
SC79 (cat. no. HY-18749) was purchased from MedChemExpress. LX-2
cells were pre-treated with 8 µg/ml SC79 at 37°C in 5%
CO2 for 2 h, and then the exosomes were added to
co-culture.
Cell transfection and reverse
transcription-quantitative PCR (RT-qPCR)
A total of four short hairpin RNAs (shRNAs)
targeting the SphK1 gene were designed. Each shRNA was used for
cloning into the lentiviral vector [LV3 (H1/GFP&Puro) (10×;
1×108 TU/ml; Shanghai GenePharma Co., Ltd.). The
negative control (NC) was a non-targeting shRNA sequence, which was
a nonsense sequence designed for a mammalian gene and had no
regulatory effect on the expression of the target gene. The
sequences of the shRNAs are presented in Table I. RKO cells were infected with the
lentivirus (packaged by Gemma Pharmaceutical Technology Co., Ltd.).
RKO cells in good condition were digested and resuspended, seeded
into 24-well plates and incubated overnight at 37°C in an incubator
at a density of ~50%. Lentivirus stock (25 µl) was diluted 10-fold
in culture medium containing 10% FBS (according to the guidelines
of Gemma Pharmaceutical Technology Co., Ltd.), and Polybrene was
added to a final concentration of 5 µg/ml. The culture medium in
the 24-well plate was aspirated, and 250 µl diluted virus, as
aforementioned, was added to each well and incubated for 24 h at
37°C in 5% CO2. The diluted virus solution in 24-well
plates was aspirated, and 500 µl medium containing 10% FBS was
added to each well. The RKO cells were cultured at 37°C with 5%
CO2 for 72 h. The efficiency of infecting RKO cells by
microscopy reached >70%. In addition, complete culture medium
with puromycin at final concentrations of 0, 0.5, 1.0, 1.5, 2.0 and
2.5 µg/ml was added to the RKO cells. The concentration of
puromycin was maintained continuously for >120 h to select the
optimal concentration of puromycin that could completely kill the
cells. According to the lethal results of different concentration
gradients of puromycin, 1.0 ug/ml was the best screening
concentration for cells, and this drug concentration was selected
for subsequent screening. Based on the interference effect of the
lentiviruses and the proliferation of the infected RKO cells, RKO
cells containing the K8105 sequence were selected for subsequent
experiments and were referred to as SphK1(−) (Fig. S1A). Although the K9754 sequence
exhibited a higher knockdown efficiency, the RKO cells containing
the K9754 sequence had difficulty in cell expansion. So the K8105
sequence was selected for further experiments. The mRNA level of
SphK1 was detected by RT-qPCR analysis. RNA isolation was performed
using the Total RNA Extraction kit (Gemma Pharmaceutical Technology
Co., Ltd.) and cDNA synthesis was performed using the RT kit (Gemma
Pharmaceutical Technology Co., Ltd.), according to the
manufacturer's protocol. The reverse transcription reaction system
(20 µl) comprised 10 µl reverse transcription buffer (2X), 1.2 µl
reverse transcription primer (1 µM), 0.2 µl MMLV reverse
transcriptase (200 U/µl), total RNA (200 ng/µl) and DEPC water. The
reverse transcription reaction conditions were 42°C for 30 min,
85°C for 10 min and rapid cooling to 4°C. The quantitative PCR
reaction system (20 µl) comprised 10 µl quantitative PCR Master Mix
(2X), 0.08 µl upstream primer (20 µM), 0.08 µl downstream primer
(20 µM), 2 µl cDNA, 0.4 µl Taq DNA polymerase (2.5 U/µl) and
ddH2O. The quantitative PCR reaction conditions were
95°C for 3 min, 95°C for 12 sec and 62°C for 40 sec in 40 cycles.
Gene expression levels were determined using the 2−ΔΔCq
method (18). The primer sequences
(Shanghai Gemma Pharmaceutical Technology Co., Ltd.) were as
follows: GAPDH forward, 5′-GGAAGCTTGTCATCAATGGAAATC-3′ and reverse,
5′-TGATGACCCTTTTGGCTCCC-3′; and SPHK1 forward,
5′-CAGCTCTTCCGGAGTCACGT-3′ and reverse,
5′-CGTCTCCAGACATGACCACCA-3′. GFP expression from the lentivirus was
used for detection. The infecting RKO cells were fixed with 4%
paraformaldehyde. Next, the Antifade mounting medium with DAPI
(4′,6-diamidino-2-phenylindole) (P0131; Shanghai Biyuntian
Biotechnology Co., Ltd.) was used. The cells were observed by
fluorescence microscopy (BX3-CBH; Olympus Corporation). The
efficiency of infecting RKO cells by microscopy reached
>70%.
 | Table I.Short hairpin RNA sequences used for
cloning into the lentiviral vector. |
Table I.
Short hairpin RNA sequences used for
cloning into the lentiviral vector.
| Name | Interference target
position | Target sequence
(5′-3′) |
|---|
| K8104 | SphK1-Homo-837 |
CTGACCAACTGCACGCTATTG |
| K8105 | SphK1-Homo-967 |
ACCTAGAGAGTGAGAAGTATC |
| K8106 |
SphK1-Homo-1349 |
CATGGAGAAGGGCAGGCATAT |
| K9754 |
SphK1-Homo-1277 |
AGCTGGCGTCATGCATCTGTT |
| LV3-NC | - |
TTCTCCGAACGTGTCACGT |
Isolation, characterization and
quantification of exosomes
RKO cells and infected RKO cells were cultured for
48 h in 10% exosome-depleted-FBS (cat. no. UR51101; Shanghai
Yumeibo Biotechnology Co., Ltd.) complete medium. The medium was
collected and processed using the Exosome Extraction and
Purification Kit (cat. no. UR52121; Shanghai Yumeibo Biotechnology
Co., Ltd.) to eliminate cell debris and isolate exosomes. The kit
was used according to the manufacturer's instructions. The pellet
of exosomes was re-suspended in PBS, purified and then stored at
−80°C. Exosomes were identified using a nanoparticle tracking
analyzer (Particle Metrix GmbH) for nanoparticle tracking analysis
(NTA). Exosomes morphology was determined by a transmission
electron microscope (TEM) (HITACHI HT-7700; Hitachi, Ltd.).
Exosome-specific protein markers were detected by western
blotting.
Western blot analysis
Total protein of RKO or LX-2 cells was extracted
using RIPA buffer (cat. no. AR0102; Wuhan Boster Biological
Technology, Ltd.). Total protein from exosomes was extracted as
aforementioned. The bicinchoninic acid method was used to quantify
the total protein obtained from cells and exosomes. A total of 30
µg protein was separated by SDS-PAGE using 10, 12 or 15% gels
according to the molecular weight of the target protein for better
protein separation, and transferred onto PVDF membranes
(MilliporeSigma). The membranes were blocked in 5% milk at room
temperature for 2 h, and subsequently incubated with primary
antibodies at 4°C overnight. The next day, the membranes were
incubated with secondary antibodies at room temperature for 1 h.
The blots were visualized with the ECL substrate (cat. no. AR1191;
Boster Biological Technology). The bands were detected using an
enhanced chemiluminescence gel imaging system (Bio-Rad
Laboratories, Inc.) and blots were semi-quantified using ImageJ
1.51 j8 software (National Institutes of Health), with GAPDH used
as a loading control. The primary antibodies used were as follows:
GAPDH polyclonal antibody (cat. no. 10494-1-AP; 1:4,000), SphK1
polyclonal antibody (cat. no. 10670-1-AP; 1:1,000), TSG101
polyclonal antibody (cat. no. 28283-1-AP; 1:4,000), vimentin
polyclonal antibody (cat. no. 10366-1-AP; 1:2,000), CD81 monoclonal
antibody (cat. no. 66866-1-Ig; 1:1,000), E-cadherin monoclonal
antibody (cat. no. 60335-1-Ig; 1:4,000) and phosphorylated (p-)AKT
(Ser473) monoclonal antibody (cat. no. 66444-1-Ig; 1:2,000) were
purchased from Proteintech Group, Inc. α-SMA (cat. no. BM3902;
1:4,000) and AKT (cat. no. A00024-2; 1:1,000) antibodies were
purchased from Wuhan Boster Biological Technology, Ltd. Calnexin
(cat. no. HY-P80578; 1:1,000), proliferating cell nuclear antigen
(PCNA; cat. no. HY-P80268; 1:2,000), TGF-β1 (cat. no. HY-P80521;
1:500) and TNF-α (cat. no. HY-P80914; 1:1,000) antibodies were
purchased from MedChemExpress. HRP Conjugated AffiniPure Goat
Anti-rabbit lgG (H+L) (cat. no. BA1054; 1:10,000) and HRP
Conjugated AffiniPure Goat Anti-Mouse IgG (H+L) (cat. no. BA1050;
1:10,000) secondary antibodies were purchased from Wuhan Boster
Biological Technology, Ltd.
Transwell assay
RKO cells were suspended in DMEM high-glucose liquid
medium (conventional) (cat. no. PYG0073; Wuhan Boster Biological
Technology, Ltd.) containing 1% FBS (cat. no. PYG0001; Wuhan Boster
Biological Technology, Ltd.). A total of 200 µl of RKO cell
suspension (5×105/ml) was added to the upper chambers of
Transwell plates (cat. no. 3422; Corning, Inc.), and the lower
chamber was filled with 500 µl DMEM high-glucose liquid medium
(conventional) containing 30% FBS in a 12-well plate. After 24 h of
incubation at 37°C, 4% paraformaldehyde was used to fix the cells
for 20 min at room temperature, and then 0.1% crystal violet was
used to stain cells for 8 min at room temperature. Cells that had
migrated to the other side of the membrane were subsequently
counted under a light microscope. The counting was performed using
ImageJ 1.51 j8 software (National Institutes of Health).
Exosome labeling and uptake
PKH67 (cat. no. UR52303; Shanghai Yumeibo
Biotechnology Co., Ltd.) was used to label exosomes with green
fluorescence according to the manufacturers' instructions. PHK67
working solution was added to the RKO exosomes at a concentration
of 5 µM and incubated for 10 min at room temperature. To
investigate whether LX-2 cells could uptake the exosomes,
PKH67-labeled exosomes (7×103 particles/cell) were added
to the 1,000 cells in a 12-well plate. After 24 h of co-culturing
at 37°C, the cells were fixed with 4% paraformaldehyde for 10 min
at room temperature. Subsequently, anti-fade medium with DAPI was
used for mounting (cat. no. P0131; Beyotime Institute of
Biotechnology). The uptake of exosomes was evaluated using
fluorescence microscopy (BX3-CBH; Olympus Corporation).
Cell viability assay
The viability of RKO cells was analyzed using a Cell
Counting Kit-8 (CCK-8) assay kit (cat. no. GK10001; GLPBIO
Technology LLC). A total of 1,000 RKO cells per well were seeded in
a 96-well plate. Over night, the cells fully attached to the
culture plate, and different supernatants of LX-2 cells were added.
The LX-2 cell culture supernatants were extracted from the
coculture system with LX-2 cells and exosomes from RKO cells with
different SphK1 contents for 24 h. After 24 h of incubation at
37°C, the culture medium was removed and 10 µl CCK-8 solution and
90 µl DMEM (cat. no. PYG0073; Wuhan Boster Biological Technology,
Ltd.) were added to the cells. The plate was incubated for 1 h at
37°C. Subsequently, the absorbance of each well was measured using
a microplate reader (BioTek Synergy; Agilent Technologies, Inc.) at
450 nm.
Statistical analysis
Each experiment was repeated at least 3 times. Data
are presented as the means ± SD. SPSS 26.0 (IBM Corp.) and GraphPad
Prism 8.0 (Dotmatics) were used for statistical analysis.
Differences between two groups were compared using unpaired
Student's t test, while one-way ANOVA followed by the Least
Significant Difference or Tukey's post hoc test was used for more
than three groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
Overview
A brief schematic of the experimental processes of
the present study is shown in Fig.
1.
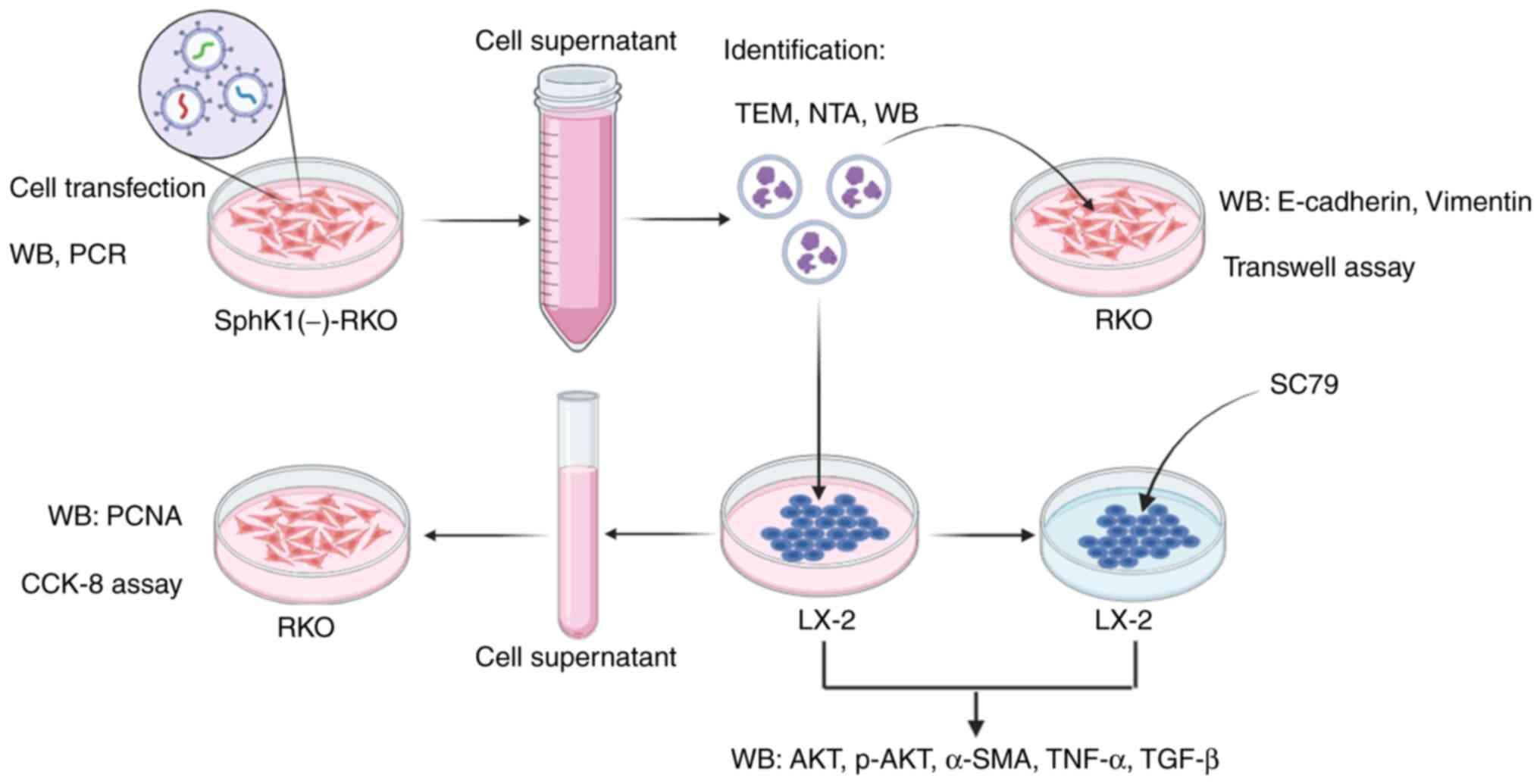 | Figure 1.Brief experimental process of the
present study. Cell transfection technique was used to construct
RKO cells with low expression of SphK1. Western blotting and PCR
were used to verify the expression of SphK1 in RKO cells. The
culture supernatant of SphK1(−)-RKO cells was collected, and the
exosomes were extracted and identified by TEM, NTA and western
blotting. The extracted exosomes were used to co-culture with RKO
cells, and the expression of E-cadherin and vimentin was detected
by western blotting. The migration ability of RKO cells was
detected by Transwell assay. The extracted exosomes were also used
to co-culture with LX-2 cells, and the expression levels of AKT,
p-AKT, α-SMA, TNF-α and TGF-β were detected by western blotting.
The LX-2 cell culture supernatant was extracted and used to culture
RKO cells. CCK-8 was used to detect RKO cell viability, and western
blotting was used to detect PCNA expression in RKO cells. In
addition, LX-2 cells were pretreated with SC79 and treated with RKO
exosomes, and the expression levels of AKT, p-AKT, α-SMA, TNF-α and
TGF-β were detected by western blotting. α-SMA, α-smooth muscle
actin; TEM, transmission electron microscopy; NTA, nanoparticle
tracking analysis; WB, western blotting; SphK1, sphingosine kinase
1; PCNA, proliferating cell nuclear antigen; CCK-8, Cell Counting
Kit-8; p-, phosphorylated. |
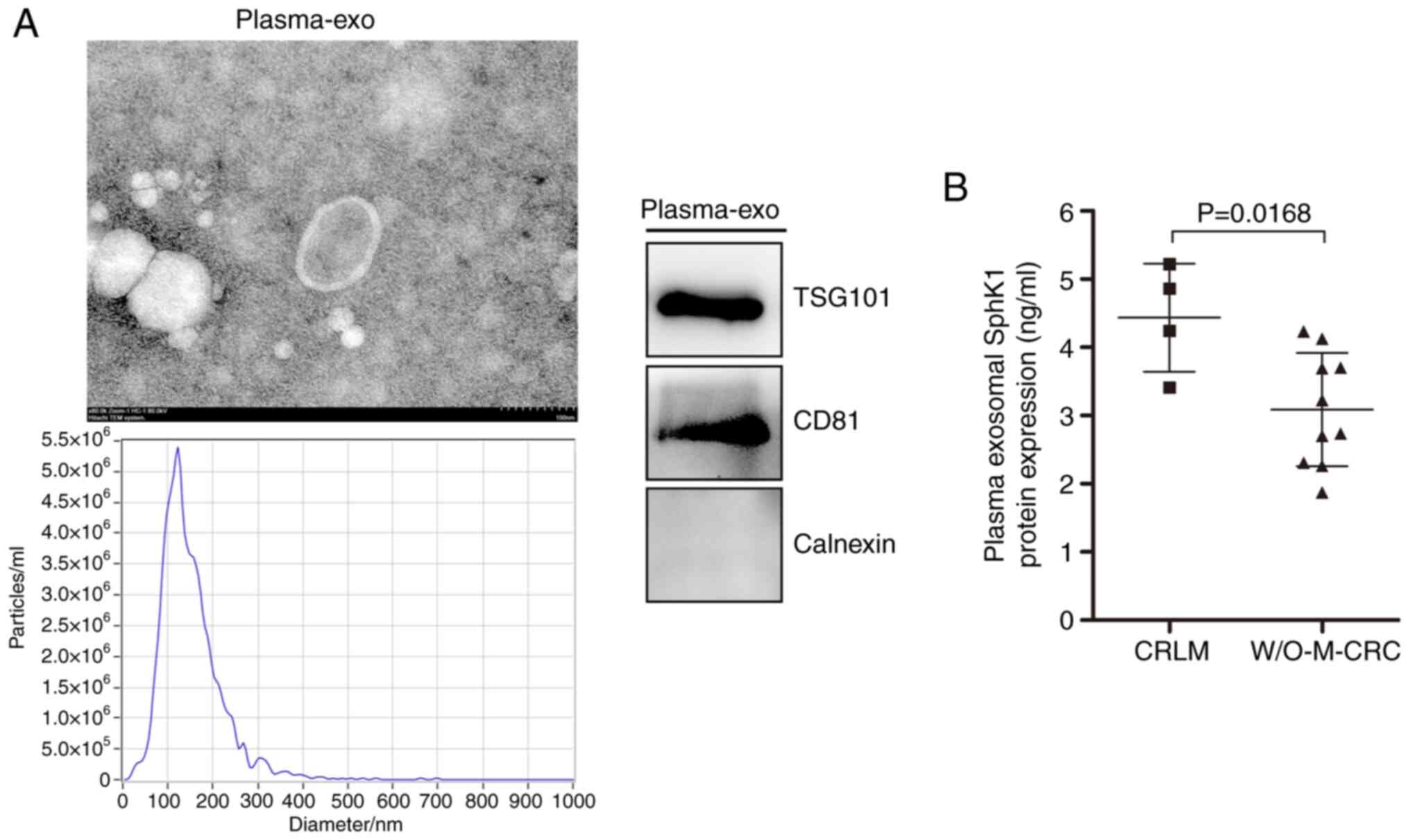
Plasma exosomal SphK1 levels are
significantly upregulated in patients with liver metastasis of
CRC
Plasma exosome identification from a representative
patient using TEM, NTA and western blotting is shown in Fig. 2A. TEM revealed that the morphology
of exosomes was typical with a double-membrane disc structure. NTA
revealed that the isolated exosomes had a mean size of 150.5±66.8
nm. Immunoblotting of exosome-derived proteins revealed the
presence of the typical exosome markers CD81 and TSG101, and the
absence of calnexin. These results revealed that the extracted
plasma exosomes were identified as qualified. An ELISA revealed
that the plasma exosomal SphK1 protein expression was significantly
upregulated in patients with CRC and liver metastasis, compared
with in those with CRC without metastasis (Fig. 2B). These results suggested that
exosomes with increased SphK1 levels may serve a key role in the
liver metastasis associated with CRC.
Exosomes with low SphK1 protein levels
are obtained following transfection of RKO cells with the
interfering lentivirus
Fluorescence microscopy showed the successful
transfection of the RKO cells. The SphK1 mRNA and protein
expression levels were significantly downregulated in the SphK1(−)
RKO group compared with the blank control (Con) group, while there
was no significant difference between the Con group and the
negative control (NC) group (Fig.
3A). The cell culture supernatant of the three groups was
collected, and the exosomes of each group were isolated, purified
and referred to as Con-RKO exosomes (Con-RKOexo), NC-RKO
exosomes (NC-RKOexo) and SphK1(−)-RKO exosomes
(SphK1(−)-RKOexo), accordingly. As shown in Fig. 3B, the exosomes were identified
using TEM, NTA and western blotting. The morphology of the exosomes
was typical with a double-membraned disc structure in TEM. NTA
showed that Con-RKOexo, NC-RKOexo and
SphK1(−)-RKOexo had a mean size of 148.7±54.6,
156.3±56.3 and 158.9±65.6 nm, respectively. Furthermore, the
concentrations of the isolated exosomes were 3.5×1010,
7.0×1010 and 12.0×1011 particles/ml,
respectively (Fig. S1B), as
assessed by the nanoparticle tracking analyzer. Immunoblotting of
exosome-derived proteins revealed the presence of the typical
exosome markers CD81 and TSG101, and the absence of calnexin. In
addition, the SphK1 protein levels were significantly downregulated
in the SphK1(−)-RKOexo group compared with the
Con-RKOexo group, while there was no significant
difference between the Con-RKOexo and
NC-RKOexo groups.
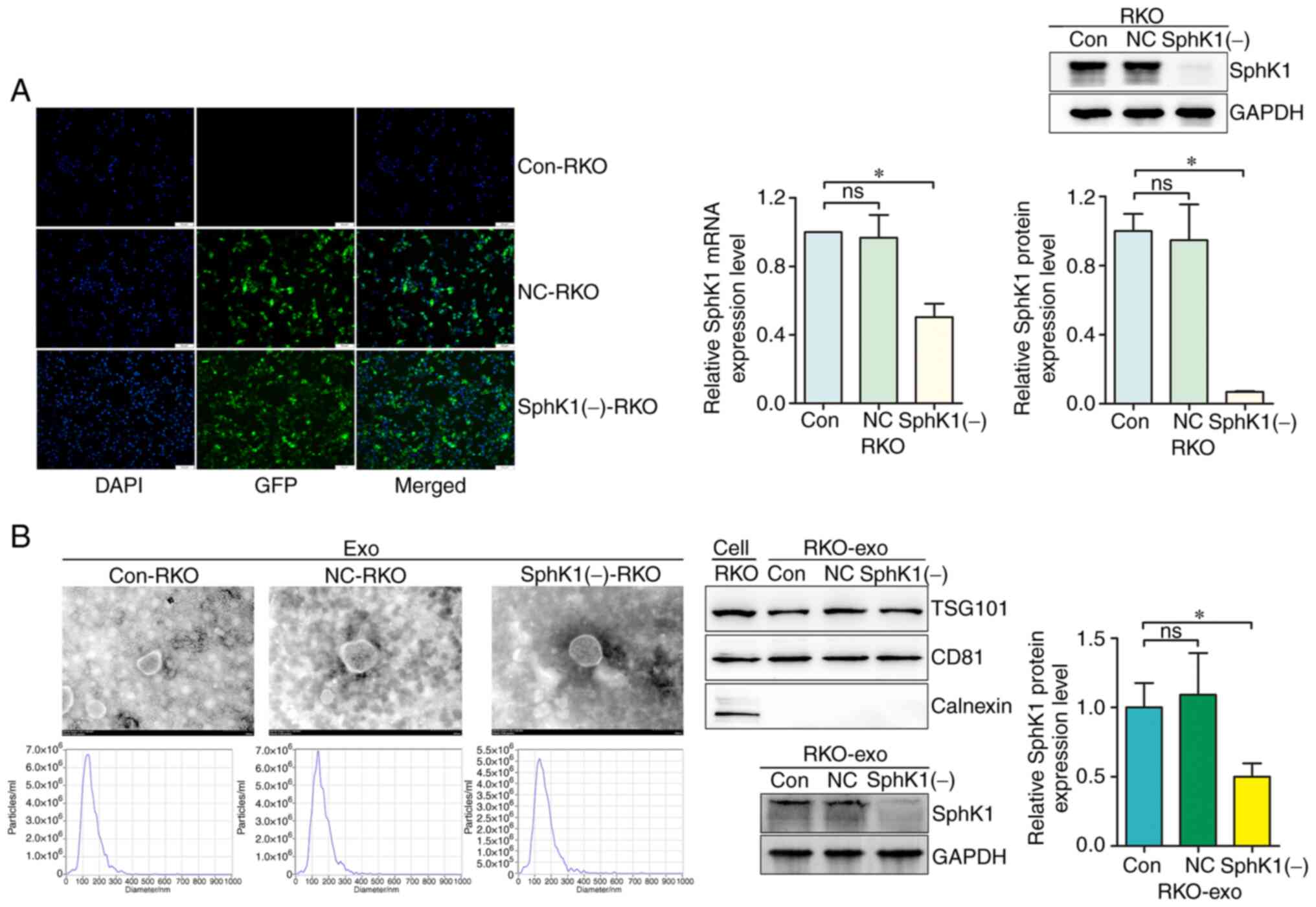 | Figure 3.Exosomes with low levels of SphK1 are
obtained following transfection of RKO cells with the interfering
lentivirus. (A) Transfected RKO cells were observed under a
fluorescence microscope (magnification, ×200; scale bar, 50 µm).
SphK1 mRNA and protein expression of cells was examined by reverse
transcription-quantitative PCR and western blotting, respectively.
(B) Exosomes were identified by transmission electron microscopy
(magnification, ×80,000; scale bar, 100 nm), nanoparticle tracking
analysis and western blotting for CD81, TSG101 and calnexin.
Exosomal SphK1 was also detected by western blotting. *P<0.05.
ns, not significant; exo, exosomes; SphK1, sphingosine kinase 1;
NC, negative control; Con, control; TSG101, tumor susceptibility
gene 101 protein; GFP, green fluorescent protein. |
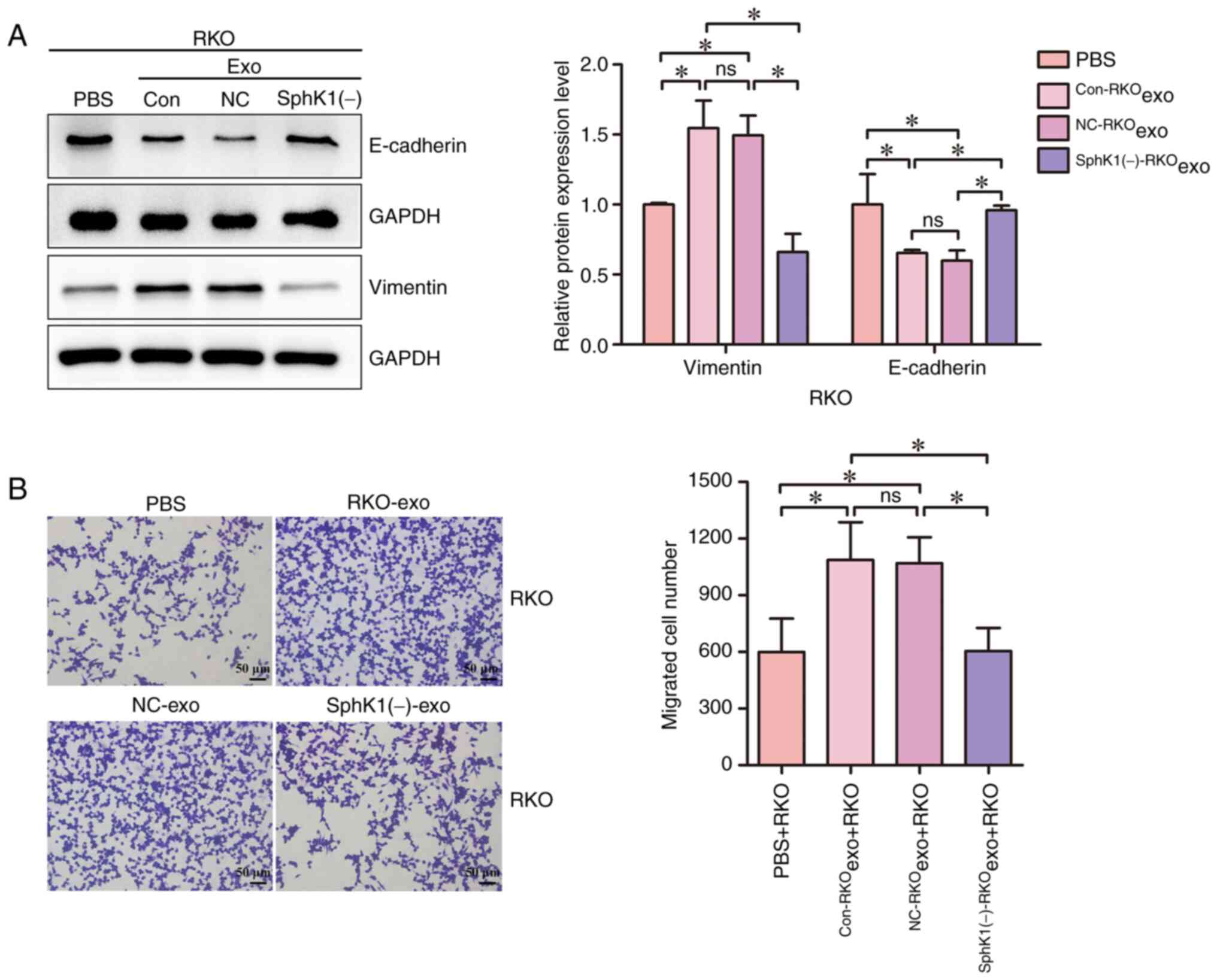
Downregulation of exosomal SphK1
reverses the exosome-mediated migration of CRC cells
RKO cells were treated with equal amounts of
Con-RKOexo, NC-RKOexo and
SphK1(−)-RKOexo. PBS was used as the control group for
exosome treatment. The effect of downregulated exosomal SphK1 on
exosome-mediated CRC cell migration was then analyzed. Compared
with the PBS + RKO group, the Con-RKOexo + RKO and
NC-RKOexo + RKO groups exhibited increased vimentin
expression and decreased E-cadherin expression (Fig. 4A). The number of migrated cells in
the Con-RKOexo + RKO and NC-RKOexo + RKO
groups was significantly increased compared with that in the PBS +
RKO group, indicating that the exosomes derived from CRC cells were
involved in the migration of these cells (Fig. 4B). Compared with the
Con-RKOexo + RKO and NC-RKOexo + RKO groups,
the expression levels of vimentin were decreased, the expression
levels of E-cadherin were increased and the number of migrated
cells was reduced in the SphK1(−)-RKOexo + RKO group
(Fig. 4A and B). These results
indicated that reduced exosomal SphK1 partially reversed the
exosome-induced migration of CRC cells.
CRC-derived exosomes activate hepatic
stellate cells to promote the viability of CRC cells by
upregulating p-AKT, while downregulation of exosomal SphK1
partially reverses this effect
LX-2 cells were treated with PKH67-labeled exosomes
isolated from RKO cells, and fluorescently labelled RKO exosomes
were detected in LX-2 cells (Fig.
5A). This finding suggested that LX-2 cells can uptake RKO
exosomes from the surrounding environment. LX-2 cells were treated
with equal amounts of Con-RKOexo, NC-RKOexo
and SphK1(−)-RKOexo to investigate the potential
mechanism via which CRC-derived exosomes activate hepatic stellate
cells. The levels of AKT, p-AKT, α-SMA, TNF-α and TGF-β were
detected in LX-2 cells. In addition, the LX-2 cell culture
supernatant was collected and co-cultured with RKO cells to detect
changes in the viability of RKO cells. Compared with those in the
PBS + LX-2 group, the expression levels of AKT in the
Con-RKOexo + LX-2 and NC-RKOexo + LX-2 groups
were not significantly changed (Fig.
5B); however, the levels of p-AKT, α-SMA, TNF-α and TGF-β were
increased. The results indicated that CRC-derived exosomes
potentially activated hepatic stellate cells, promoting their
secretion of TNF-α and TGF-β through the AKT pathway. Compared with
those in the Con-RKOexo + LX-2 and NC-RKOexo
+ LX-2 groups, the levels of p-AKT, α-SMA, TNF-α and TGF-β were
decreased in the SphK1(−)-RKOexo + LX-2 group,
indicating that depletion of exosomal SphK1 partially reversed the
activation of hepatic stellate cells induced by CRC-derived
exosomes. The cell supernatant (CS) of the PBS + LX-2,
Con-RKOexo + LX-2, NC-RKOexo + LX-2 and
SphK1(−)-RKOexo + LX-2 groups was co-cultured with RKO
cells. Compared with those in the PBS + LX-2 CS + RKO group, the
PCNA expression and viability (used as indirect measures of cell
proliferation) were significantly increased in the
Con-RKOexo + LX-2 CS + RKO and NC-RKOexo +
LX-2 CS + RKO groups (Fig. 5C and
D). Compared with those in the Con-RKOexo + LX-2 CS
+ RKO and NC-RKOexo + LX-2 CS + RKO groups, the PCNA
expression and viability were significantly decreased in the
SphK1(−)-RKOexo + LX-2 CS + RKO group. The trend of the
cell viability rate was consistent with that of the PCNA expression
levels in RKO cells. Overall, these results suggested that
CRC-derived exosomes activated hepatic stellate cells and promoted
the viability of CRC cells, potentially by activating AKT, and that
reducing exosomal SphK1 expression partially reversed this
effect.
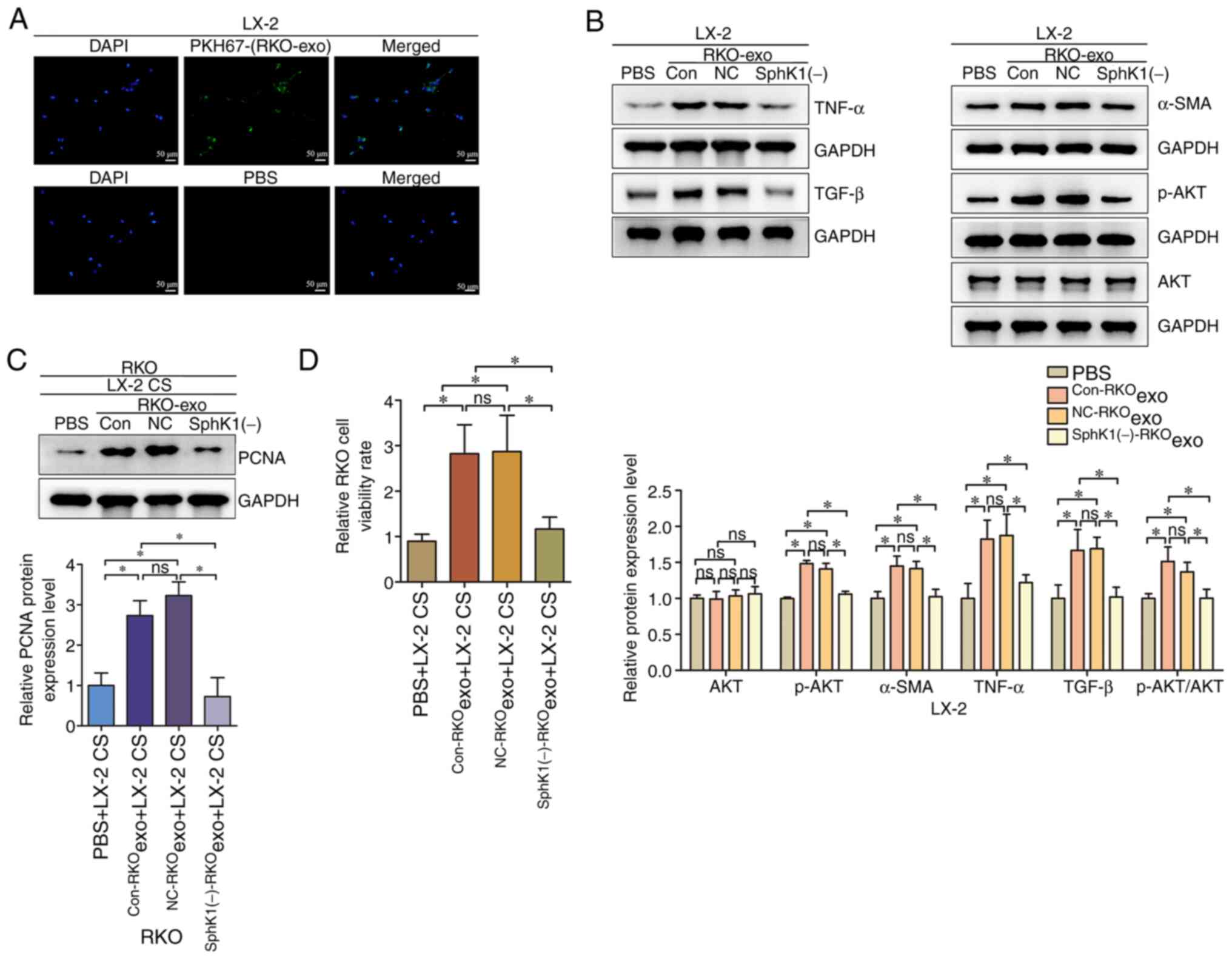 | Figure 5.CRC-derived exosomes regulate p-AKT to
activate hepatic stellate cells and promote the proliferation of
CRC cells via exosomal SphK1. (A) PKH67-labeled RKO-exosomes were
detected in LX-2 cells (magnification, ×200; scale bar, 50 µm). (B)
Western blotting for AKT, p-AKT, α-SMA, TNF-α and TGF-β in LX-2
cells. (C) Western blotting for PCNA in RKO cells. (D) The
viability rate of RKO cells was analyzed using a Cell Counting
Kit-8 assay. *P<0.05. ns, not significant; exo, exosomes; CS,
cell supernatant; α-SMA, α-smooth muscle actin; SphK1, sphingosine
kinase 1; NC, negative control; Con, control; PCNA, proliferating
cell nuclear antigen; p-, phosphorylated; CRC, colorectal
cancer. |
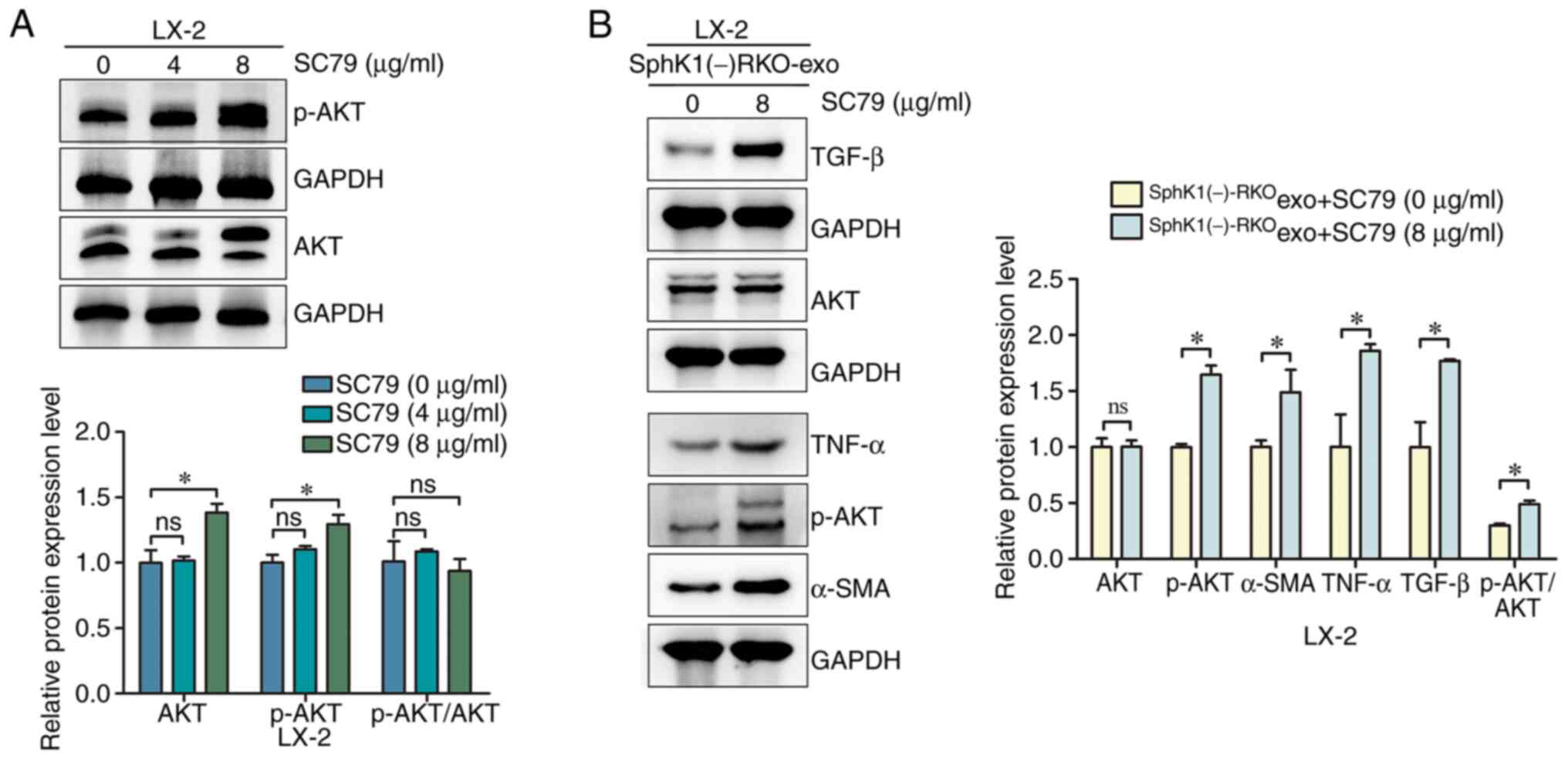
AKT agonist SC79 reverses the
inhibition of hepatic stellate cell activation induced by the
reduction of CRC-exosomal SphK1
LX-2 cells were treated with 0, 4 and 8 µg/ml SC79.
Compared with the 0 µg/ml SC79 group, the levels of AKT and p-AKT
did not significantly change in the 4 µg/ml SC79 group, while the
levels of AKT and p-AKT were significantly increased in the 8 µg/ml
SC79 group (Fig. 6A). While the
expression of p-AKT/AKT did not change in either 4 of 8 µg/ml, this
may be due to the fact that SC79 was an AKT and p-AKT agonist,
which could increase AKT and p-AKT simultaneously. The effect of
the 8 µg/ml SC79 treatment in SphK1(−)-RKOexo + LX-2
cells was subsequently investigated. Compared with the
SphK1(−)-RKOexo + SC79 (0 µg/ml) + LX-2 group, the
SphK1(−)-RKOexo + SC79 (8 µg/ml) + LX-2 group exhibited
increased levels of p-AKT, α-SMA, TNF-α and TGF-β (Fig. 6B), indicating that the AKT agonist
SC79 reversed the inhibition of hepatic stellate cell activation
induced by the reduced levels of SphK1 in CRC-derived exosomes.
Discussion
SphK1 is a lipid kinase and a key regulatory enzyme
maintaining the level of sphingosine-1-phosphate. It is involved in
the development of several diseases and pathological conditions,
including cancer, infectious diseases and inflammation (11,19).
Several studies have found that SphK1 expression is upregulated in
various types of cancer, such as colorectal cancer (11). Furthermore, SphK1 was shown to
regulate AKT, FAK, ERK, NF-κB and other signaling pathways to
promote cancer cell proliferation, induce EMT and enhance stem
cell-like properties of cancer cells, reshape the cancer
microenvironment, modulate cancer immunity, and promote distant
metastasis of cancer cells (20–22). SphK1 also regulates the AKT,
FAK and ERK pathways to induce EMT and autophagy in CRC cells
(12,13,23).
Therefore, SphK1 may constitute an important target for the
treatment of cancer, which was further emphasized by the results of
the present study. The present study demonstrated that plasma
exosomal SphK1 was significantly upregulated in the plasma of
patients with liver metastasis of CRC compared with patients
without metastasis. Furthermore, SphK1 was present in CRC-derived
exosomes. Downregulation of exosomal SphK1 partially reversed the
exosome-induced migration of CRC cells. Similarly, a study
indicated that ovarian cancer exosomes released SphK1 in the tumor
microenvironment to promote ovarian cancer progression (24). These results collectively revealed
that cancer cell-derived exosomal SphK1 may promote tumor
progression.
The liver is the main site of metastasis for CRC
cells (25). Hepatic stellate
cells are important components of the liver microenvironment and
serve a key role in the liver metastasis of CRC cells (26,27).
The expression levels of TNF-α, TGF-β and α-SMA in hepatic stellate
cells increase upon activation of these cells (28,29).
Exosomes from metastatic CRC cells have been shown to activate
hepatic stellate cells, reshaping the liver microenvironment, and
forming a ‘soil’ that supports cancer cell colonization (14). The present study revealed that
hepatic stellate cells could uptake CRC-derived exosomes from the
surrounding environment. Further investigation demonstrated that
CRC-derived exosomes activated hepatic stellate cells, as evidenced
by increased TNF-α and TGF-β expression, potentially by
upregulating p-AKT. TNF-α and TGF-β are conducive to the
colonization and proliferation of cancer cells in the liver
(16,17). Consistently, the culture
supernatant of activated hepatic stellate cells promoted CRC cell
viability in the present study. These results suggested that
CRC-derived exosomes activated hepatic stellate cells to promote
the viability of CRC cells, potentially through the AKT pathway.
The present study revealed that the downregulation of exosomal
SphK1 partially reversed this effect. Additionally, the AKT agonist
SC79 promoted hepatic stellate cell activation, which was inhibited
by reduced levels of exosomal SphK1. These results suggested that
CRC-derived exosomal SphK1 regulated p-AKT to activate hepatic
stellate cells and promote the viability of CRC cells.
In summary, the present study demonstrated that
CRC-derived exosomal SphK1 promoted hepatic stellate cell
activation and CRC cell migration in vitro. Furthermore, in
vivo experiments are required to confirm the role of
CRC-derived exosomal SphK1 in the liver metastasis of CRC.
In the future, the application of exosome
engineering in cancer treatment will become increasingly prominent.
The versatility of exosomes allows the modification of their
original configuration and expands their applications in the
biomedical field using various methods, including genetic
engineering, chemical procedures, physical techniques and
microfluidic techniques. Further studies may help improve the
understanding of the functions and components of CRC-derived
exosomes, contributing to the development of novel genetically
engineered exosomes to inhibit CRC metastasis.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Medical and Health
Science and Technology Development Program of Shandong Province
(grant no. 2019WS331).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
WZ conducted the experiments and statistical
analyses, and drafted the manuscript. CX contributed to study
design and helped draft the manuscript. Both authors have read and
approved the final manuscript. WZ and CX confirm the authenticity
of all the raw data.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Ethics Committee of Binzhou Medical University (approval no.
KT-259; Binzhou, China). Written informed consent was obtained from
all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Viale PH: The American cancer society's
facts & figures: 2020 edition. J Adv Pract Oncol. 11:135–136.
2022.PubMed/NCBI
|
|
2
|
Pallares-Rusiñol A, Bernuz M, Moura SL,
Fernández-Senac C, Rossi R, Martí M and Pividori MI: Advances in
exosome analysis. Adv Clin Chem. 112:69–117. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rahnama M, Heidari M, Poursalehi Z and
Golchin A: Global trends of exosomes application in clinical
trials: A scoping review. Stem Cell Rev Rep. 20:2165–2193. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yáñez-Mó M, Siljander PR, Andreu Z, Zavec
AB, Borràs FE, Buzas EI, Buzas K, Casal E, Cappello F, Carvalho J,
et al: Biological properties of extracellular vesicles and their
physiological functions. J Extracell Vesicles. 4:270662015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Huang C, Li J, Xie Z, Hu X and Huang Y:
Relationship between exosomes and cancer: Formation, diagnosis, and
treatment. Int J Biol Sci. 21:40–62. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ye Z, Yi J, Jiang X, Shi W, Xu H, Cao H,
Qin L, Liu L, Wang T, Ma Z and Jiao Z: Gastric cancer-derived
exosomal let-7 g-5p mediated by SERPINE1 promotes macrophage M2
polarization and gastric cancer progression. J Exp Clin Cancer Res.
44:22025. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xie H, Wu Y, Huang J, Shen Q, Li X, Wang
L, Lin J, Chi Z, Ke K, Lin X, et al: NK cell exosomes alleviate
PD-l1 expression and facilitate tumor immunity by repressing
PI3K-AKT-mTOR signaling. Immunol Invest. 2:1–14. 2025. View Article : Google Scholar
|
|
8
|
Yin J, Zhu W, Feng S, Yan P and Qin S: The
role of cancer-associated fibroblasts in the invasion and
metastasis of colorectal cancer. Front Cell Dev Biol.
12:13755432024. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rom AD, Dragicevic S, Jankovic R, Skodric
SR, Sabljak P, Markovic V, Stojkovic JR, Barisic G and Nikolic A:
Markers of epithelial-mesenchymal transition and mucinous histology
are significant predictors of disease severity and tumor
characteristics in early-onset colorectal cancer. Diagnostics
(Basel). 14:15122024. View Article : Google Scholar
|
|
10
|
Ramos R, Vinyals A, Campos-Martin R, Cabré
E, Bech JJ, Vaquero J, Gonzalez-Sanchez E, Bertran E, Ferreres JR,
Lorenzo D, et al: New insights into the exosome-induced migration
of uveal melanoma cells and the pre-metastatic niche formation in
the liver. Cancers (Basel). 16:29772024. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Alkafaas SS, Elsalahaty MI, Ismail DF,
Radwan MA, Elkafas SS, Loutfy SA, Elshazli RM, Baazaoui N, Ahmed
AE, Hafez W, et al: The emerging roles of sphingosine 1-phosphate
and SphK1 in cancer resistance: A promising therapeutic target.
Cancer Cell Int. 24:892024. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu SQ, Xu CY, Wu WH, Fu ZH, He SW, Qin MB
and Huang JA: Sphingosine kinase 1 promotes the metastasis of
colorectal cancer by inducing the epithelial-mesenchymal transition
mediated by the FAK/AKT/MMPs axis. Int J Oncol. 54:41–52.
2019.PubMed/NCBI
|
|
13
|
Xu CY, Liu SQ, Qin MB, Zhuge CF, Qin L,
Qin N, Lai MY and Huang JA: SphK1 modulates cell migration and
EMT-related marker expression by regulating the expression of p-FAK
in colorectal cancer cells. Int J Mol Med. 39:1277–1284. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao S, Mi Y, Zheng B, Wei P, Gu Y, Zhang
Z, Xu Y, Cai S, Li X and Li D: Highly-metastatic colorectal cancer
cell released miR-181a-5p-rich extracellular vesicles promote liver
metastasis by activating hepatic stellate cells and remodelling the
tumour microenvironment. J Extracell Vesicles. 1:e121862022.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang W, Gao K, Bai Y, Xu D, Zhao M, Tao X
and Wang J: Wedelolactone attenuates liver fibrosis and hepatic
stellate cell activation by suppressing the hippo pathway.
Rejuvenation Res. 27:207–219. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Landskron G, De la Fuente M, Thuwajit P,
Thuwajit C and Hermoso MA: Chronic inflammation and cytokines in
the tumor microenvironment. J Immunol Res. 2014:1491852014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Paschos KA and Bird NC: Liver regeneration
and its impact on post-hepatectomy metastatic tumour recurrence.
Anticancer Res. 30:2161–2170. 2010.PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zou F, Wang S, Xu M, Wu Z and Deng F: The
role of sphingosine-1-phosphate in the gut mucosal microenvironment
and inflammatory bowel diseases. Front Physiol. 14:12356562023.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kao WH, Liao LZ, Chen YA, Lo UG, Pong RC,
Hernandez E, Chen MC, Teng CJ, Wang HY, Tsai SC, et al: SPHK1
promotes bladder cancer metastasis via PD-L2/c-Src/FAK signaling
cascade. Cell Death Dis. 15:6782024. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yu M, Wang S, Zeng Y, Liu P and Li H:
SPHK1 promotes pancreatic cancer lymphangiogenesis through the
activation of ERK in LECs. Mol Biotechnol.
11:10.1007/s12033–024-01192-9. 2024.
|
|
22
|
Zhang C, Zhou C, Xu J and Xue S: Increased
sphingosine kinase 1 expression is associated with poor prognosis
in human solid tumors: A meta-analysis. Dis Markers.
2022:84439322022.PubMed/NCBI
|
|
23
|
Xu C, Zhang W, Liu S, Wu W, Qin M and
Huang J: Activation of the SphK1/ERK/p-ERK pathway promotes
autophagy in colon cancer cells. Oncol Lett. 15:9719–9724.
2018.PubMed/NCBI
|
|
24
|
Gupta P, Kadamberi IP, Mittal S, Tsaih SW,
George J, Kumar S, Vijayan DK, Geethadevi A, Parashar D, Topchyan
P, et al: Tumor derived extracellular vesicles drive T cell
exhaustion in tumor microenvironment through sphingosine mediated
signaling and impacting immunotherapy outcomes in ovarian cancer.
Adv Sci (Weinh). 9:e21044522022. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Niu Y, Yang W, Qian H and Sun Y:
Intracellular and extracellular factors of colorectal cancer liver
metastasis: A pivotal perplex to be fully elucidated. Cancer Cell
Int. 22:3412022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu B, Wu T, Lin B, Liu X, Liu Y, Song G,
Fan C and Ouyang G: Periostin-TGF-β feedforward loop contributes to
tumour-stroma crosstalk in liver metastatic outgrowth of colorectal
cancer. Br J Cancer. 130:358–368. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Huang WH, Zhou MW, Zhu YF, Xiang JB, Li
ZY, Wang ZH, Zhou YM, Yang Y, Chen ZY and Gu XD: The role of
hepatic stellate cells in promoting liver metastasis of colorectal
carcinoma. Onco Targets Ther. 12:7573–7580. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang Y, Stoess C, Holzmann G, Mogler C,
Stupakov P, Altmayr F, Schulze S, Wang B, Steffani M, Friess H, et
al: Signalling of the neuropeptide calcitonin gene-related peptide
(CGRP) through RAMP1 promotes liver fibrosis via TGFβ1/Smad2 and
YAP pathways. Exp Cell Res. 442:1141932024. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Watakabe K, Miyoshi M, Kakinuma S, Sato A,
Tsuchiya J, Shimizu T, Mochida T, Inada K, Kaneko S, Kawai-Kitahata
F, et al: A20 in hepatic stellate cells suppresses chronic
hepatitis by inhibiting DCLK1-JNK pathway-dependent chemokines.
FASEB J. 38:e237572024. View Article : Google Scholar : PubMed/NCBI
|