Introduction
Gastric cancer is a prevalent malignant tumor and
>1,000,000 new cases were diagnosed in 2020. Notably, gastric
cancer ranks as the fifth most common cancer in terms of incidence
and the fourth leading cause of cancer-related mortality worldwide
(1). Despite advancements in
treatment modalities, the overall prognosis remains poor,
highlighting the urgent need for novel therapeutic strategies
(1,2). This type of cancer originates from
the gastric mucosa, and exhibits diverse epidemiological patterns,
risk factor profiles and molecular subtypes influenced by
geographical variability (3–5).
Current therapeutic options, including surgical resection,
chemotherapy, targeted agents and immunotherapy, have demonstrated
limited success in improving the outcome of patients, especially in
cases of metastatic gastric cancer (6–9). The
emergence of chemoresistance, particularly to platinum-based
compounds such as cisplatin (DDP), severely restricts the
effectiveness of standard treatments (10). Consequently, there is a need for
in-depth research into the underlying mechanisms of resistance and
the development of innovative therapeutic strategies. Such
advancements are key for enhancing treatment response, and
tailoring more efficacious and individualized interventions for
patients with gastric cancer.
In the contemporary landscape of oncology, where
targeted therapy and immunotherapy have become the basis of cancer
treatment, research into immune cells within the tumor
microenvironment (TME) is increasingly pivotal (11–13).
Macrophages, as key immune effector cells, have a dual role in the
TME, both influencing and being influenced by tumor dynamics
(14). Macrophages are broadly
classified into two phenotypes: Classically activated macrophages
(M1), which enhance cytokine production and exert antitumor
effects, and alternatively activated macrophages (M2), which are
associated with tumor promotion and immunosuppression (15). In most solid tumors, including
gastric cancer, tumor-associated macrophages (TAMs) are
predominantly skewed towards the M2 phenotype (16). The density and polarization state
of TAMs are intricately associated with essential oncogenic
processes, such as metastasis, invasion and chemoresistance
(16,17).
Exosomes, a subset of extracellular vesicles, have a
key role in intercellular communication by transporting bioactive
molecules, including microRNAs (miRNAs/miRs), lipids and proteins
(18). Previous studies have
suggested that stromal cells in the TME, particularly TAMs, can
modulate tumor cell behavior and facilitate oncogenesis through
exosome secretion (19,20). miRNAs, as a significant component
of exosomes, are encapsulated within a bilipid layer that protects
them from enzymatic degradation, ensuring their stability during
transport to recipient cells (20). Upon delivery, these miRNAs can
regulate gene expression by binding to target mRNAs, resulting in
mRNA degradation or translational repression (19,21,22).
The present study aimed to investigate the role and
underlying mechanisms of exosomes derived from M2-polarized
macrophages in modulating DDP resistance in gastric cancer cells,
using a well-established model of M2-polarized macrophages to mimic
TAM functions in the TME (23,24).
Targeting exosomal miR-3681-3p derived from M2-polarized
macrophages may present a promising therapeutic strategy to
overcome DDP resistance in gastric cancer, offering a potential
pathway for increasing treatment efficacy.
Materials and methods
Cell culture and treatment
AGS gastric cancer cells (cat. no. CRL-1739;
American Type Culture Collection) were cultured in DMEM (cat. no.
11965092, Gibco; Thermo Fisher Scientific, Inc.) supplemented with
10% FBS (cat. no. 16140071; Gibco; Thermo Fisher Scientific, Inc.)
and 1% penicillin-streptomycin (cat. no. 15140122; Gibco; Thermo
Fisher Scientific, Inc.) at 37°C in a humidified atmosphere with 5%
CO2. Primary mouse bone marrow cells (MBMCs; cat. no.
CP-M131; Wuhan Pricella Life Technology Co. Ltd.), were maintained
in DMEM/F-12 (cat. no. 11330032; Gibco; Thermo Fisher Scientific,
Inc.) enriched with 10% FBS and macrophage colony-stimulating
factor (cat. no. HY-P7085A; MedChemExpress) at 37°C and 5%
CO2. For M2 polarization, MBMCs were treated with
interleukin (IL)-4 (30 ng/ml; cat. no. 214-14-1MG) and IL-13 (30
ng/ml; cat. no. 210-13-1MG) (both from Gibco; Thermo Fisher
Scientific, Inc.) for 48 h at 37°C. Co-culture assays were
conducted using a Transwell system, wherein 5×104
M2-polarized macrophages were seeded into 12-well plate inserts
(upper chamber), and 1.5×105 AGS cells were grown in the
lower compartment at 37°C for 3 days.
Immunofluorescence
A total of 2×104 M2-polarized macrophages
were cultured in 35-mm confocal dishes for 48 h, followed by
fixation using 4% paraformaldehyde for 20 min at room temperature.
After fixation, the cells were washed three times for 5 min with
PBS and were then permeabilized with 0.5% Triton X-100 in PBS for
10 min at room temperature (RT). Blocking was achieved by
incubating the cells with 3% BSA (cat. no. 23225; Thermo Fisher
Scientific, Inc.) in PBS for 1 h at RT. Subsequently, the cells
were incubated overnight at 4°C with primary antibodies targeting
CD206 (1:200 dilution; cat. no ab64693; Abcam), CD163 (1:50
dilution; cat. no ab316218; Abcam), CD80 (1:200 dilution; cat. no
ab315832; Abcam) and CD86 (1:100 dilution; cat. no ab239075;
Abcam). This was followed by incubation with fluorophore-conjugated
secondary antibodies (1:200 dilution; cat. no A27039; Thermo Fisher
Scientific, Inc.) for 2 h at room temperature. After three
additional washes with Tris-buffered saline containing 0.1%
Tween-20, the nuclei were stained with DAPI (cat. no. D1306; Thermo
Fisher Scientific, Inc.) for 10 min at 37°C to facilitate nuclear
visualization. Imaging was carried out using a Carl Zeiss LSM 510
META confocal microscope (Zeiss GmbH) equipped with a Plan
Apochromat 63× oil immersion objective featuring a numerical
aperture of 1.4 and differential interference contrast
capability.
Detection of inflammatory factors
Pro-inflammatory cytokine concentrations in the
culture supernatants of M2-polarized macrophages were
quantitatively evaluated using enzyme-linked immunosorbent assay
(ELISA). A total of 1×104 M2 macrophages were plated in
24-well plates and cultured for 48 h. Subsequently, the
supernatants were collected and stored at −80°C until analysis.
Levels of IL-2 (cat. no. JM-02981M1), IL-10 (cat. no. JM-02459M2),
IL-6 (cat. no. JM-02446M1) and tumor necrosis factor-α (TNF-α; cat.
no. JM-02415M2) were determined using specific ELISA kits (Jiangsu
Jingmei Biotechnology Co., Ltd.). All procedures were carried out
according to the manufacturer's instructions.
Cell proliferation and viability
assay
AGS cells were cultured in 96-well plates at a
concentration of 1×103 cells/well overnight, followed by
incubation with 1 µg/ml DDP for 1, 2, 3, 4 and 5 days at 37°C. At
the end of each timepoint, 10 µl Cell Counting Kit-8 (CCK-8)
solution (cat. no. C0039; Beyotime Institute of Biotechnology) was
added to each well, and the plates were incubated at 37°C for 2 h
to allow the reaction to occur. Absorbance at 450 nm was measured
using an Eppendorf BioPhotometer® D30 (Eppendorf SE).
Cell proliferation was quantitatively determined based on the
absorbance readings collected over a 5-day period.
In addition, AGS cells were cultured in 96-well
plates at a concentration of 1×103 cells/well overnight,
and were then incubated with 0.5, 1.0, 1.5 and 2.0 µM DDP for 48 h
at 37°C. Then, 10 µl CCK-8 solution was added to each well, and the
plates were incubated at 37°C for 2 h to allow the reaction to
occur. Absorbance at 450 nm was measured using an Eppendorf
BioPhotometer D30. IC50 values were calculated using
non-linear regression in GraphPad Prism (version 8.0;
Dotmatics).
Flow cytometry
The apoptosis of AGS cells was quantified using the
Annexin V-FITC and PI staining kit (cat. no. G1511; Wuhan
Servicebio Technology Co., Ltd.), according to the manufacturer's
protocol. Analysis was carried out using a Beckman DXI800 flow
cytometer (Beckman Coulter, Inc.). The identification and
quantification of apoptotic cells, indicated by PI positivity, were
conducted with FlowJo software (version 10.8; BD Biosciences). The
apoptotic rate was calculated by summing the percentages of cells
in both early and late stages of apoptosis.
Isolation of exosomes and electron
microscopy
M2-polarized macrophage supernatants were collected
and centrifuged at 2,000 × g for 30 min at 4°C. Following
centrifugation, the supernatant was filtered through a 0.22-µm
syringe filter (cat. no. SLGVR13SL; MilliporeSigma) and subjected
to ultracentrifugation at 120,000 × g overnight at 4°C using an
Optima XPN-100 ultracentrifuge (cat. no. CP100MX; Hitachi, Ltd.) to
precipitate exosomes. The exosomes were then resuspended in cold
PBS (pH 7.4) and further purified by ultracentrifugation at 120,000
× g for 90 min at 4°C. The purified exosomes were suspended in cold
PBS or SDS loading buffer (cat. no. P0015F; Beyotime Institute of
Biotechnology; containing 4% SDS and 100 mM Tris-HCl; pH 7.6) and
stored at −80°C for further studies. For the in vitro
experiments, exosomes were used to treat cells at a concentration
of 50 µg protein/105 cells at 37°C for 48 h.
For transmission electron microscopy, freshly
isolated exosome suspensions were fixed in 4% paraformaldehyde for
1 h at 4°C. Subsequently, 20 µl exosome solution was placed onto a
copper grid, excess fluid was removed, and the sample was stained
with 2% phosphotungstic acid for 40 sec at room temperature to
enhance contrast. Visualization of the exosomes was caried out
using a transmission electron microscope (model HT-7700; Hitachi,
Ltd.). In addition, the exosome supernatant was denatured with 5X
SDS buffer and analyzed by western blotting. Protein samples (50
µg/lane) were separated by SDS-PAGE on a 10% SDS-polyacrylamide gel
and were probed with rabbit antibodies targeting calnexin (1:1,000
cat. no. ab133615; Abcam), tumor susceptibility gene 101 (TSG101;
1:1,000; cat. no. 102286-T38; Beijing Sino Technology Co. Ltd.) and
CD9 (1:1,000; cat. no. ab92726; Abcam). The detailed western
blotting procedures are described in the western blotting
subsection.
Nanoparticle tracking analysis
(NTA)
M2-polarized macrophage-extracted exosomes were
diluted to a total volume of 1 ml using Tris-phosphate-modified
saline for subsequent analysis. To assess the size distribution of
the exosomes, NTA was carried out using a NanoSight (model, N30E;
NanoFCM, Inc.) instrument. This analysis adhered to the established
standard operating procedure for NTA (25), enabling precise measurement of
vesicle size based on the Brownian motion of individual
particles.
Western blotting
Proteins were extracted from AGS cells using
radioimmunoprecipitation assay lysis buffer (cat. no. HY-K1001;
MedChemExpress), and their concentration was measured using the BCA
Protein Assay Kit (cat. no. 23225; Thermo Fisher Scientific, Inc.).
For SDS-PAGE, 10 µg each protein lysate was combined with 5X SDS
sample buffer and separated on a 12% SDS-polyacrylamide gel.
Post-electrophoresis, the proteins were transferred to a
polyvinylidene difluoride membrane. To minimize non-specific
interactions, the membrane was blocked with 5% non-fat milk for 2 h
at RT. The membrane was then probed overnight at 4°C with the
following primary antibodies: Rabbit anti-MLH1 (1:2,000; cat. no.
ab92312; Abcam) and mouse anti-GAPDH (1:5,000; cat. no. ab8245;
Abcam). After incubation with primary antibodies, the membrane was
rinsed and exposed to HRP-conjugated secondary antibodies
(1:10,000; cat. nos. ab6721 and ab205719; Abcam) for 2 h at RT.
Protein bands were visualized using enhanced chemiluminescence
western blot detection reagents (Thermo Fisher Scientific, Inc.).
ImageJ software (version 1.53a; National Institutes of Health) was
used for semi-quantification of western blot analysis.
miR-3681-3p mimic and MLH1 small
interfering (si)RNA transfections
The miR-3681-3p mimic (sense:
5′-ACACAGUGCUUCAUCCACUACU-3′; antisense:
5′-AGUAGUGGAUGAAGCACUGUGU-3′) and its negative control (NC: sense:
5′-UUUGUACUACACAAAAGUACUG-3′; antisense:
5′-CAGUACUUUUGUGUAGUACAAA-3′) were synthesized by Shanghai
GenePharma Co., Ltd. Transfection of AGS cells with the miR-3681-3p
mimic was carried out using Oligofectamine™ reagent
(cat. no. 12252-011; Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. This involved 24-h
serum-free pre-treatment of AGS cells, followed by transfection
with a final concentration of 50 nM miR-3681-3p mimic or NC for 48
h. Transfection efficiency was subsequently assessed using reverse
transcription-quantitative PCR (RT-qPCR).
The MLH1 siRNA (sense: 5′-CAGCUAAUGCUAUCAAAGAGA-3′;
antisense: 5′-UCUCUUUGAUAGCAUUAGCUG-3′) and its scrambled NC siRNA
(sense: 5′-UUCUCCGAACGAGUCACGUTT-3′; antisense:
5′-ACGUGACUCGUUCGGAGAA-3′) were also synthesized by Shanghai
GenePharma Co., Ltd. The transfection procedure for MLH1 siRNA
followed the protocol used for the miR-3681-3p mimic.
RT-qPCR
Total RNA was extracted from exosomes derived from
M2 macrophages or AGS cells using the RNAiso Plus reagent (cat. no.
9109; Takara Bio, Inc.). The isolated RNA was then
reverse-transcribed into cDNA using the Bestar™ qPCR RT
kit (cat. no. DBI-2220; DBI Bioscience). RT followed a defined
thermal profile: 5 min at 25°C, 30 min at 42°C and a final step at
85°C for 5 min. qPCR was conducted to assess the expression levels
of target genes using Bestar qPCR MasterMix (cat. no. DBI-2043; DBI
Bioscience) on the Agilent Mx3000P system (cat. no. Mxpro-Mx3000P;
Agilent Technologies, Inc.). The qPCR amplification used a two-step
protocol: 30 sec at 95°C for initial denaturation, followed by 40
cycles of 10 sec at 95°C and 30 sec at 60°C. Relative expression
levels were calculated using the 2−ΔΔCq method (26), with normalization of miR-3681-3p
expression to U6 snRNA levels. The specific primer sequences used
for amplification were as follows: miR-3681-3p forward,
5′-CGCGACACAGTGCTTCATCC-3′, reverse, 5′-AGTAGTGGATGAAGCACTGT-3′;
and U6 snRNA forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse,
5′-AACGCTTCACGAATTTGCGT-3′.
Target identification and
dual-luciferase reporter assay
RNAhybrid v2.2 online software (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid)
was used for miR-3681-3p target prediction (27). Wild-type and mutant MLH1 gene
3′-untranslated regions (3′-UTRs) were cloned into the pmirGLO
dual-luciferase reporter vector (cat. no. E1910; Promega
Corporation). Briefly, 3×105 293T cells (cat. no.
CL-0005; Wuhan Pricella Life Technology Co. Ltd.) underwent
co-transfection with these vectors alongside either the miR-3681-3p
mimic or NC mimic using the HB-TRLF-1000 LipoFiter transfection kit
(cat. no. HB-TRLF-1000; Hanbio Biotechnology Co., Ltd.). Following
a 48-h incubation period at 37°C, cell lysates were collected, and
luciferase activity was measured according to the instructions of
the Dual-Luciferase Reporter Assay System (cat. no. E1910; Promega
Corporation). Luminescence was quantified using a Lux-T020
microplate reader. The activation of the reporter gene was
evaluated by calculating the normalized luminescence data,
expressed as the ratio of firefly to Renilla luciferase
activities.
Vector construction and lentiviral
transduction
A lentiviral vector engineered for MLH1
overexpression was created using human MLH1 genetic sequences
obtained from the NCBI GenBank (Gene Bank ID: FJ940753.1,
accessible at http://www.ncbi.nlm.nih.gov/nuccore/FJ940753.1) by
Shanghai GeneChem Co., Ltd. The coding sequence of MLH1 was
inserted into the GV358 vector to yield the MLH1 overexpression
vector. An empty GV358 vector was used in the NC group.
AGS cells were cultured in 6-well plates at 37°C for
36 h. Once they had reached ~70% confluence, each well was
uniformly exposed to equivalent volumes (100 µl) of
MLH1-overexpressing lentiviral particles or NC (empty vector)
lentiviral particles at a multiplicity of infection of 10 for 48 h
at 37°C. After 48 h, the AGS cells were harvested for western
blotting and the CCK-8 assay.
Statistical analysis
Quantitative data from the experiments were analyzed
using GraphPad Prism software (version 8.0; Dotmatics). All
experimental protocols were carried out in triplicate to ensure
reproducibility and accuracy. Data are presented as the mean ±
standard deviation. Statistical analysis was performed using the
unpaired two-tailed Student's t-test for comparisons between two
distinct groups. For multiple group comparisons, one-way ANOVA was
applied, followed by Tukey's multiple comparisons test for post hoc
analysis. P<0.05 was considered to indicate a statistically
significant difference.
Results
Co-culturing gastric cancer cells with
M2-polarized macrophages increases resistance to DDP
To investigate how M2-polarized macrophages affect
the resistance of gastric cancer cells to DDP, MBMCs were treated
with IL-13 and IL-4 to promote M2 polarization. These M2
macrophages were characterized by the presence of specific M2
markers, such as CD206 and CD163, and the lack of M1 markers, such
as CD80 and CD86, verified through immunophenotyping (Fig. 1A). Additionally, an increase in
M2-associated cytokines, including IL-2, IL-6, IL-10 and TNF-α was
observed in these cells compared with in resting macrophages
(Fig. 1B). In subsequent
experiments, DDP treatment significantly reduced the proliferation
and increased the apoptosis of AGS gastric cancer cells when
compared with the untreated control. However, co-culture with
M2-polarized macrophages attenuated the cytotoxic effects of DDP,
suggesting that M2 macrophages may confer resistance to DDP in AGS
cells (Fig. 1C and D).
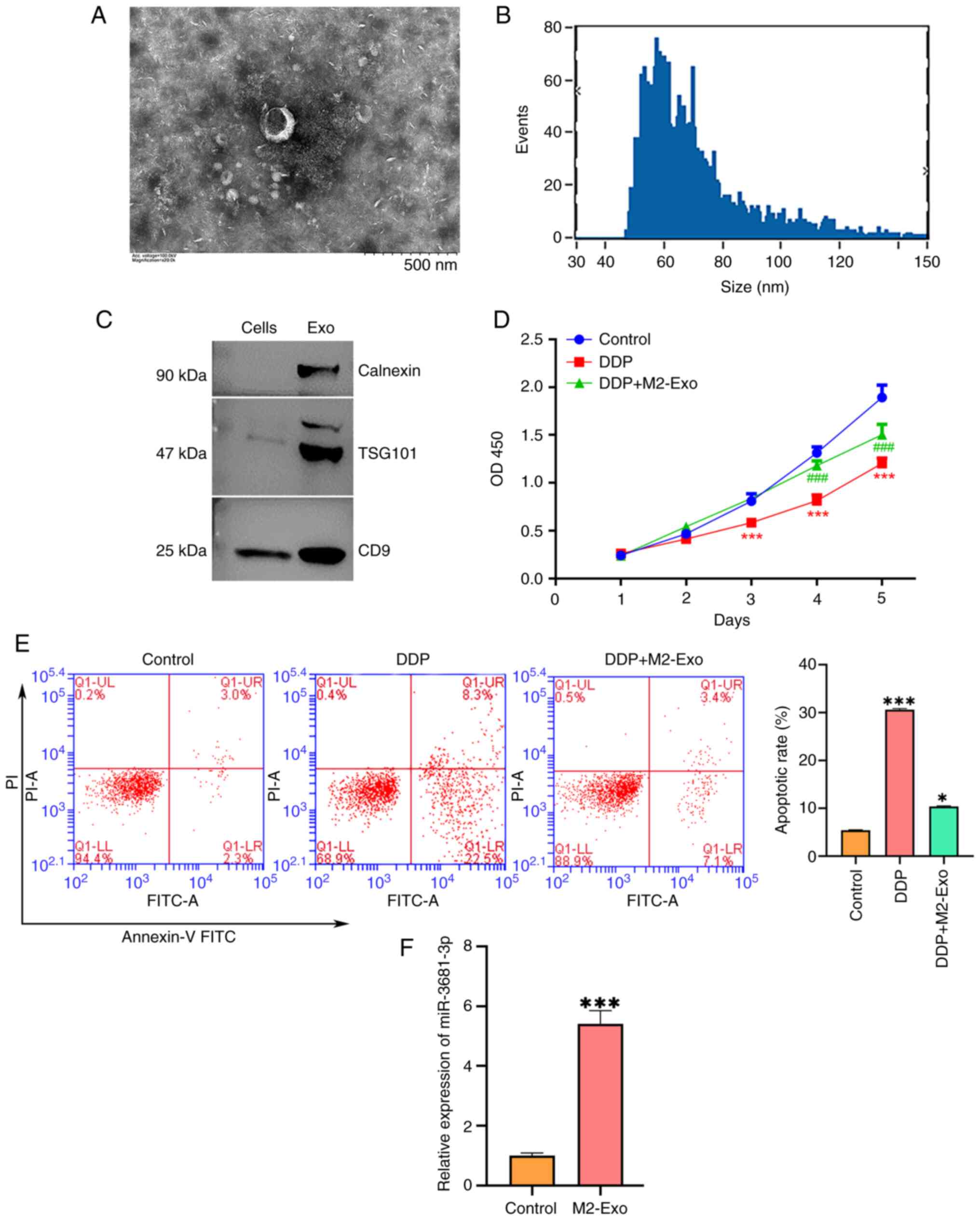
M2-polarized macrophage-derived
exosomes contribute to DDP resistance in gastric cancer cells
Electron microscopy analysis of the M2-polarized
macrophage-derived supernatant revealed the presence of small
vesicles consistent with exosomes, characterized by a 50–100 nm
diameter (Fig. 2A and B). The
exosome fraction was enriched with established markers, including
the endoplasmic reticulum protein calnexin, TSG101 and CD9, as
confirmed by western blotting (Fig.
2C). Notably, exosomes derived from M2 macrophages
significantly promoted the proliferation and inhibited the
apoptosis of DDP-treated AGS cells (Fig. 2D and E). Additionally, the
expression levels of miR-3681-3p were markedly higher in exosomes
from M2 macrophages than those from resting macrophages (Fig. 2F). These findings suggested that
M2-polarized macrophages may mitigate the cytotoxic effects of DDP
on AGS cells through the release of exosomes enriched with
miR-3681-3p.
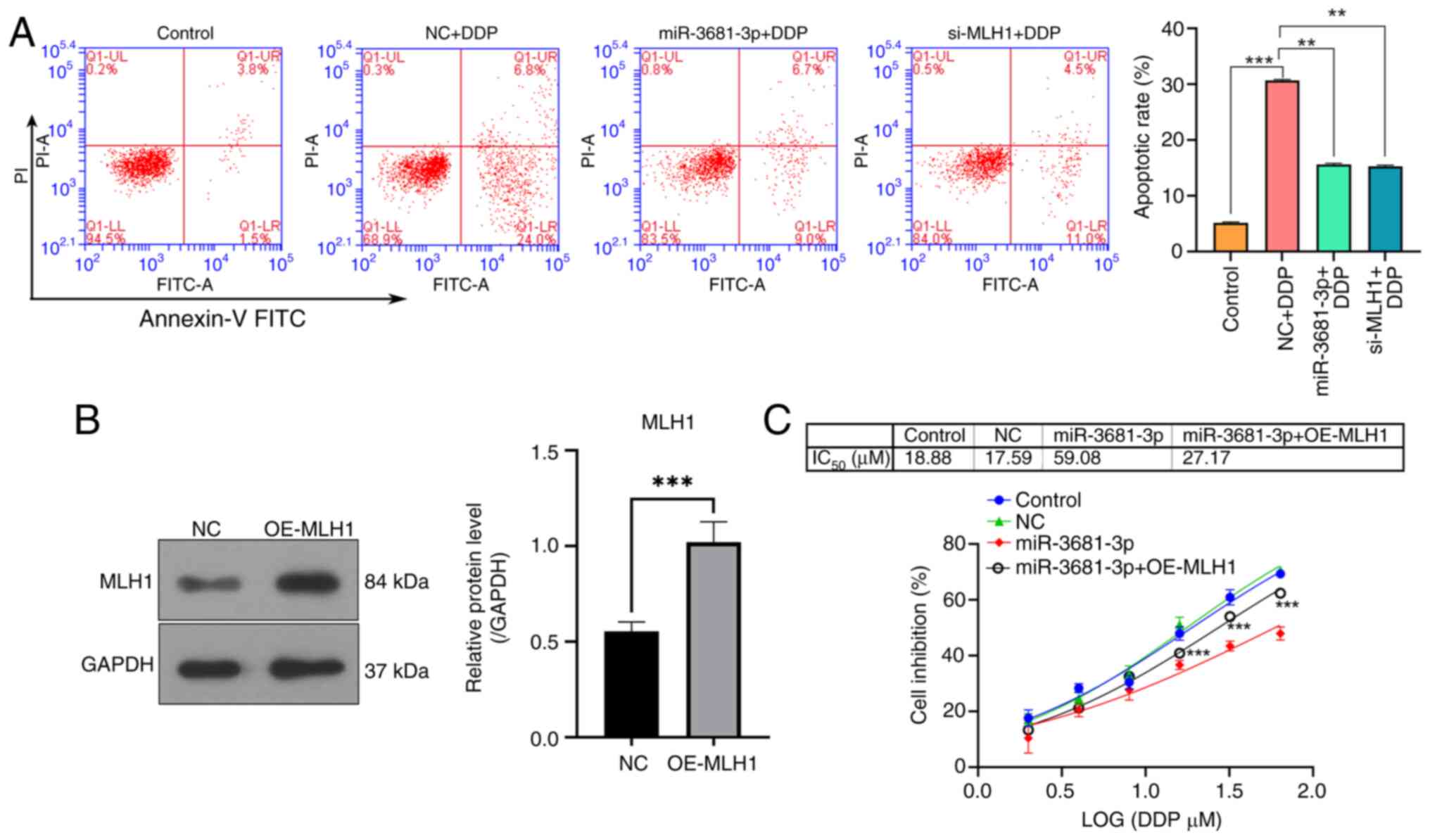
miR-3681-3p increases the DDP
resistance of gastric cancer cells by downregulating MLH1
Potential target genes of miR-3681-3p were
identified using the RNAhybrid v2.2 online software. Analysis
indicated that miR-3681-3p may target the gene encoding MLH1
(Fig. 3A). To validate this
interaction, luciferase reporter vectors were constructed
containing either wild-type or mutant forms of the MLH1 3′-UTR.
Transfection with the miR-3681-3p mimic resulted in a significant
reduction in luciferase activity associated with the wild-type MLH1
3′-UTR compared with the miR-NC. This suppressive effect was absent
in the mutated MLH1 3′-UTR construct, confirming the specificity of
the interaction (Fig. 3B). To
further validate whether miR-3681-3p targets and regulates the
expression of MLH1, the expression of miR-3681-3p was increased in
AGS cells by transfection with a miR-3681-3p mimic (Fig. 3C). Moreover, miR-3681-3p mimic
transfection substantially decreased the expression levels of MLH1
protein (Fig. 3D). Additionally,
knockdown of MLH1 protein expression was achieved by siRNA
transfection (Fig. 3E). Functional
assays demonstrated that both miR-3681-3p mimic transfection and
MLH1 silencing by siRNA rescued the DDP-induced suppression of AGS
cell proliferation (Fig. 3F).
Moreover, both miR-3681-3p mimic transfection and MLH1 silencing
contributed to the drug resistance of AGS cells to DDP, as
evidenced by an increase in the IC50 value from 21.69 to
53.35 and 45.92 µM, respectively (Fig.
3G). Additionally, both miR-3681-3p overexpression and MLH1
knockdown significantly reduced the apoptotic rate of AGS cells
exposed to DDP treatment (Fig.
4A). By contrast, the overexpression of MLH1 significantly
reversed the drug resistance of AGS cells to DDP caused by
miR-3681-3p mimic treatment, as evidenced by a decrease in the
IC50 value from 59.08 to 27.17 µM (Fig. 4B and C).
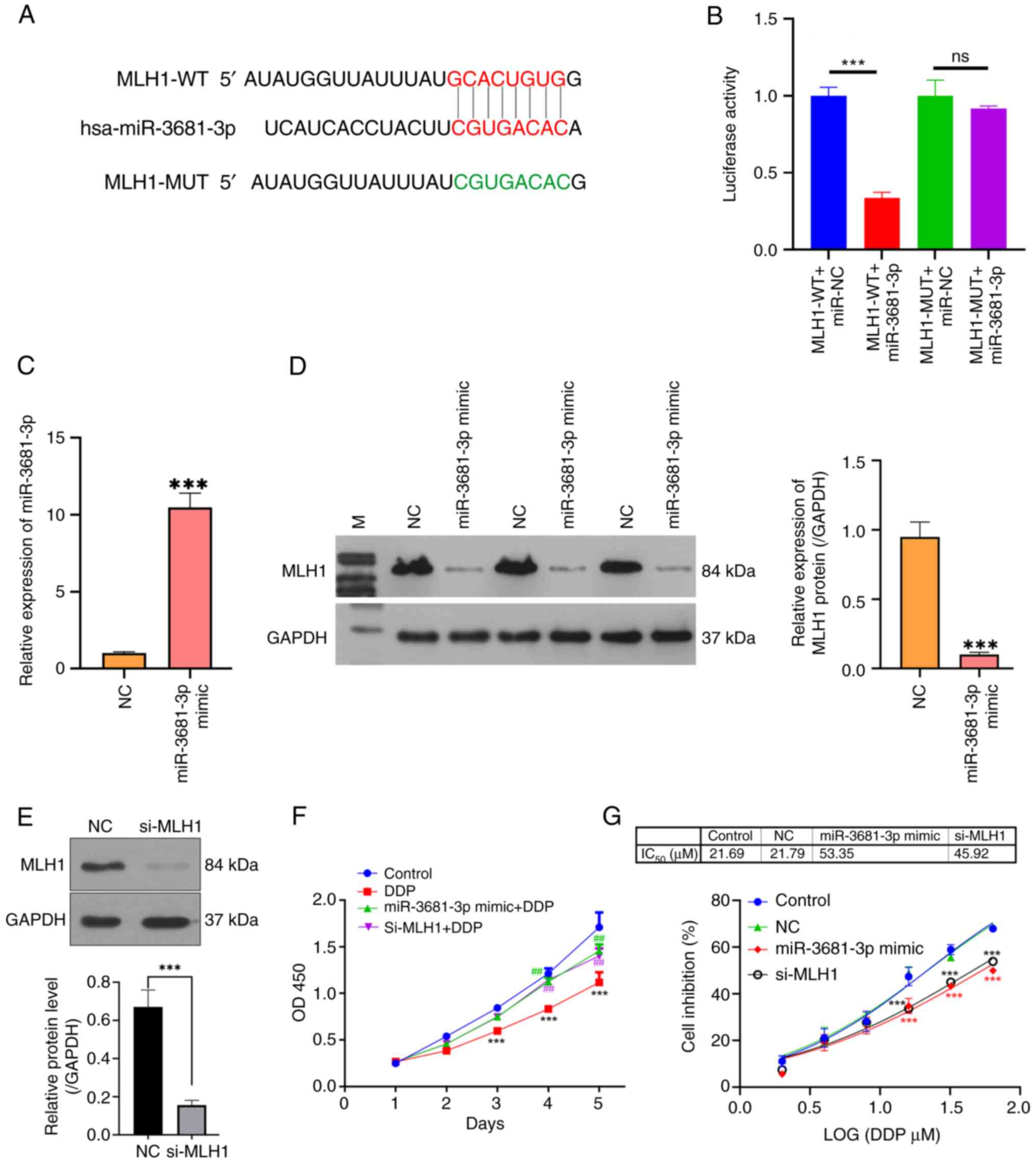 | Figure 3.miR-3681-3p increases DDP resistance
in gastric cancer cells by downregulating MLH1. (A) Bioinformatics
analysis was utilized to predict the binding site of miR-3681-3p to
MLH1. (B) Results of the dual-luciferase reporter assay assessing
the interaction between miR-3681-3p and MLH1. ***P<0.001. (C)
Reverse transcription-quantitative PCR was used to measure the
expression levels of miR-3681-3p in AGS cells after miR-3681-3p
mimic transfection. (D) Western blot analysis was carried out to
determine the protein levels of MLH1 in AGS cells transfected with
the miR-3681-3p mimic. (E) Protein levels of MLH1 were detected by
western blotting after si-MLH1 transfection. Grayscale analysis was
conducted using ImageJ software. ***P<0.001 vs. NC group. (F)
Cell proliferation of miR-3681-3p mimic-transfected and
si-MLH1-transfected AGS cells was assessed by CCK-8 assay. (G) The
IC50 value was calculated using GraphPad software based
on CCK-8 assay results. Quantitative data are presented as the mean
± SD (n=3). ***P<0.001 vs. control group; ##P<0.01
vs. DDP group. CCK-8, Cell Counting Kit-8; DDP, cisplatin; miR,
microRNA; MLH1, MutL protein homolog 1; MUT, mutant; NC, negative
control; ns, not significant; OD, optical density; si, small
interfering; WT, wild-type. |
Discussion
Gastric cancer, characterized by unchecked
proliferation of neoplastic cells in the gastric mucosa, is a
leading cause of cancer-related mortality worldwide (2,9). Its
high mortality rate is largely attributed to late-stage diagnosis
and high metastatic potential (7,9).
DDP, the first platinum analog approved for clinical use in 1971,
is extensively used as a first-line treatment for gastric cancer,
either alone or in combination with other chemotherapeutic agents
(28–30). However, DDP-based therapies face
notable limitations in certain patients, primarily due to adverse
effects, such as nephrotoxicity, insufficient therapeutic efficacy
and the development of tumor resistance (31,32).
In gastric cancer, considerable knowledge gaps exist
regarding DDP resistance, particularly in understanding the
specific mechanisms underlying this resistance in gastric cancer
cells. Factors such as genetic mutations, epigenetic alterations
and the role of extracellular vesicles remain inadequately
investigated (33–35). Additionally, there is a notable
absence of reliable biomarkers to predict which patients may
develop resistance to DDP, impacting the implementation of
personalized therapy (36). While
some new drugs and treatments are under investigation, effective
options for managing drug-resistant gastric cancer are limited
(37). Researchers are actively
working to elucidate the molecular mechanisms of DDP resistance
using advanced technologies, including genomics, transcriptomics
and proteomics (38,39). Clinical trials are being conducted
to assess new treatment combinations and alternative therapies
aimed at overcoming DDP resistance (40,41).
The identification of biomarkers of resistance through genomic
analysis is key for developing personalized treatment plans and
advancing precision medicine (42,43).
In the near future, a wealth of new research findings is
anticipated that will enhance the understanding of DDP resistance
mechanisms, thereby laying the groundwork for innovative
therapeutic strategies (44,45).
Ongoing research holds promise for identifying effective biomarkers
that can aid in selecting suitable patients for DDP therapy
(46). Additionally, novel drugs
and treatment combinations are likely to emerge in clinical
practice, potentially improving outcomes for patients with
DDP-resistant gastric cancer (47). Researchers are expected to
strengthen interdisciplinary collaboration, merging basic and
clinical research efforts to expedite the development and
application of new therapies (48,49).
Through these collective efforts, the aim is to effectively tackle
the challenges posed by DDP resistance, and enhance treatment
efficacy and survival rates for patients with gastric cancer.
Investigating the molecular mechanisms underlying
DDP resistance is key for enhancing therapeutic efficacy. In the
present study, the role of exosomal miR-3681-3p, derived from
M2-polarized macrophages, in fostering DDP resistance in gastric
cancer cells was elucidated. This was achieved through the
downregulation of MLH1 expression levels, providing novel
mechanistic insights into the modulation of chemoresistance. The
role of the miR-3681-3p/MLH1 axis indicates a promising therapeutic
approach to overcome DDP resistance in gastric cancer.
M2 macrophages, which are involved in tissue repair
and immunoregulation, significantly influence tumor progression by
creating a supportive environment for cancer growth, angiogenesis
and metastasis (50–52). These macrophages undergo metabolic
reprogramming in the TME, supporting their survival and enhancing
their immunosuppressive functions (24,53).
M2 macrophages have been shown to mediate tumor resistance to
various chemotherapeutic agents. For instance, M2 macrophages
activate signaling pathways such as CXCR2, promoting resistance to
sorafenib in hepatocellular carcinoma (54). In glioblastoma, hypoxia-driven M2
polarization facilitates cancer aggressiveness and resistance to
temozolomide (55). Additionally,
M2 macrophages modulate cholesterol homeostasis, suppress
ferroptosis and support their own polarization, exacerbating
hepatocellular carcinoma progression and contributing to drug
resistance (56). In gastric
adenocarcinoma, M2 macrophages have been reported to mediate
resistance to 5-fluorouracil through the regulation of integrin β3,
focal adhesion kinase and cofilin expression (57). In the present study, M2
polarization of MBMCs was induced through the administration of
IL-13 and IL-4. This induction resulted in the expression of
M2-specific markers (CD206 and CD163) and the upregulation of
associated cytokines (IL-2, IL-6, IL-10 and TNF-α). Notably, the
results of the present study indicated that co-culturing gastric
cancer cells with M2-polarized macrophages increased resistance to
DDP, suggesting an association between M2 polarization and DDP
resistance.
M2 macrophages affect tumor cells either directly or
by secreting exosomes containing specific miRNAs that modulate
tumor behavior and immune responses (58–60).
For example, exosomal miR-23a-3p from M2 macrophages has been shown
to promote oral squamous cell carcinoma progression by inhibiting
PTEN (58). Similarly, exosomes
carrying miR-660-5p from M2 macrophages facilitate hepatocellular
carcinoma development by downregulating KLF3 (59). Notably, exosomal miR-588 from
M2-polarized macrophages has been implicated in DDP resistance in
gastric cancer cells (60). These
findings highlight the complex roles of M2 macrophages and their
miRNA contents in driving tumor drug resistance, emphasizing the
potential for therapeutic strategies targeting these mechanisms to
enhance cancer treatment efficacy.
Previous studies have reported that miR-3681-3p is
downregulated in lung adenocarcinoma and is associated with
aggressive behaviors (61,62). However, the role and mechanism of
miR-3681-3p in gastric cancer remain poorly understood. The present
study revealed that exosomes derived from M2-polarized macrophages
contributed to DDP resistance in gastric cancer cells, with a
significant elevation of miR-3681-3p in these exosomes.
Furthermore, miR-3681-3p mimic transfection significantly reduced
the sensitivity of gastric cancer cells to DDP, resulting in an
increased IC50 value. These findings offer new insights
into the key role of exosomes from M2-polarized macrophages in
regulating cancer progression and highlight the novel function of
miR-3681-3p in gastric cancer. The association between exosomal
miR-3681-3p from M2 macrophages and its role in mediating drug
resistance is an important scientific question that warrants
further investigation. Understanding the mechanisms by which
elevated miR-3681-3p contributes to DDP resistance in gastric
cancer cells is essential for developing effective therapeutic
interventions.
The MLH1 gene has a key role in maintaining genomic
integrity through the mismatch repair mechanism, correcting errors
during DNA replication (63).
Deficiency or loss of MLH1 is associated with the initiation and
progression of various types of cancer (64–66).
Specifically, in colorectal carcinoma, MLH1-proficient cells were
revealed to be less sensitive to 5-FU-induced cytotoxic effects
(64). Moreover, MLH1 deficiency
is associated with cetuximab resistance in colon cancer due to
activation of the Her-2/PI3K/AKT signaling pathway (65). Notably, negative MLH1 expression
was previously detected in 9.8% (28/285) of patients with gastric
cancer, and was associated with chemoresistance and resulted in no
improvement in survival after neoadjuvant chemotherapy (67). The present study revealed that
miR-3681-3p directly targets MLH1. MLH1 downregulation
significantly diminished the sensitivity of gastric cancer cells to
DDP, as evidenced by an increased IC50 value. By
contrast, the overexpression of MLH1 effectively reversed the DDP
resistance of AGS cells caused by miR-3681-3p mimic treatment, as
indicated by the decrease in the IC50 value. These
results confirmed that miR-3681-3p may directly target MLH1.
Mechanistically, high levels of miR-3681-3p in exosomes from
M2-polarized macrophages may promote DDP resistance by
downregulating MLH1 expression. However, MLH1 is likely one of
multiple targets of miR-3681-3p, suggesting the need for further
investigation into additional regulatory factors involved in this
pathway.
While the findings of the present study provided
insights into the miR-3681-3p/MLH1 axis as a potential therapeutic
target in gastric cancer, this study has some limitations. The
interactions between miR-3681-3p and MLH1 require further
validation through additional experiments. Moreover, the findings
were derived exclusively from the AGS cell line, necessitating
corroboration across diverse cancer cell lines to enhance their
applicability. Future studies should also include animal models to
comprehensively elucidate the role of the miR-3681-3p/MLH1 axis in
gastric cancer treatment.
In conclusion, exosomal miR-3681-3p from M2
macrophages may contribute to DDP resistance in gastric cancer
cells by suppressing MLH1 expression. The miR-3681-3p/MLH1 axis
could be a promising molecular target for developing new
therapeutic strategies for gastric cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Scientific Research
Project of High-Level Talents of Affiliated Hospital of Youjiang
Medical University for Nationalities (grant no. R202011710), the
Scientific Research Project of Young and Middle-Aged Key Talents of
Affiliated Hospital of Youjiang Medical University for
Nationalities (grant no. Y20212603), the Project of Open Subject of
Guangxi Key Laboratory of Molecular Pathology of Hepatobiliary
Diseases of Affiliated Hospital of Youjiang Medical University for
Nationalities (grant no. GXZDSYS-009), the Project of Scientific
Research and Technology Development Plan of Baise City (grant nos.
Encyclopedia 20213301, Encyclopedia 20213242, Encyclopedia
20232080), Self-Funded Scientific Research Projects of Guangxi
Health and Health Commission (grant nos. Z20211114 and Z20190202),
Self-Funded Scientific Research Projects of the Administration of
Traditional Chinese Medicine of the Guangxi Zhuang Autonomous
Region (grant no. GXZYL20220304) and Scientific Research Basic
Ability Enhancement Project for Young and Middle-Aged Teachers of
Guangxi Colleges And Universities (grant no. 2021KY0538).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
WJW and ZYC conceptualized and designed the study.
WJW, JXL, CL and RTH engaged in the acquisition, analysis and
interpretation of the data. JJH, QJ and JZW contributed to the
interpretation of the data, along with manuscript drafting and
finalization. QL and GDX performed the formal analysis. ZYC secured
funding. WJW and ZYC confirm the authenticity of all the raw data.
All authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
The protocol of the present study was approved by
the Medical Ethics Committee of Affiliated Hospital of Youjiang
Medical University for Nationalities (approval no.
YYFY-LL-2024-283).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ricci AD, Rizzo A and Brandi G: DNA damage
response alterations in gastric cancer: Knocking down a new wall.
Future Oncol. 17:865–868. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
López MJ, Carbajal J, Alfaro AL, Saravia
LG, Zanabria D, Araujo JM, Quispe L, Zevallos A, Buleje JL, Cho CE,
et al: Characteristics of gastric cancer around the world. Crit Rev
Oncol Hematol. 181:1038412023. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Guggenheim DE and Shah MA: Gastric cancer
epidemiology and risk factors. J Surg Oncol. 107:230–236. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang K, Lu L, Liu H, Wang X, Gao Y, Yang
L, Li Y, Su M, Jin M and Khan S: A comprehensive update on early
gastric cancer: Defining terms, etiology, and alarming risk
factors. Expert Rev Gastroenterol Hepatol. 15:255–273. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Smyth EC, Nilsson M, Grabsch HI, van
Grieken NC and Lordick F: Gastric cancer. Lancet. 396:635–648.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Song Z, Wu Y, Yang J, Yang D and Fang X:
Progress in the treatment of advanced gastric cancer. Tumour Biol.
39:10104283177146262017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Patel TH and Cecchini M: Targeted
therapies in advanced gastric cancer. Curr Treat Options Oncol.
21:702020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ricci AD, Rizzo A, Rojas Llimpe FLR, Di
Fabio F, De Biase D and Rihawi K: Novel HER2-directed treatments in
advanced gastric carcinoma: AnotHER paradigm shift? Cancers
(Basel). 13:16642021. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rao X, Zhang C, Luo H, Zhang J, Zhuang Z,
Liang Z and Wu X: Targeting gastric cancer stem cells to enhance
treatment response. Cells. 11:28282022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bilotta MT, Antignani A and Fitzgerald DJ:
Managing the TME to improve the efficacy of cancer therapy. Front
Immunol. 13:9549922022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bejarano L, Jordāo MJC and Joyce JA:
Therapeutic targeting of the tumor microenvironment. Cancer Discov.
11:933–959. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rizzo A, Santoni M, Mollica V, Logullo F,
Rosellini M, Marchetti A, Faloppi L, Battelli N and Massari F:
Peripheral neuropathy and headache in cancer patients treated with
immunotherapy and immuno-oncology combinations: The MOUSEION-02
study. Expert Opin Drug Metab Toxicol. 17:1455–1466. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xia Y, Rao L, Yao H, Wang Z, Ning P and
Chen X: Engineering macrophages for cancer immunotherapy and drug
delivery. Adv Mater. 32:e20020542020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mantovani A, Sozzani S, Locati M, Allavena
P and Sica A: Macrophage polarization: Tumor-associated macrophages
as a paradigm for polarized M2 mononuclear phagocytes. Trends
Immunol. 23:549–555. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hu X, Ma Z, Xu B, Li S, Yao Z, Liang B,
Wang J, Liao W, Lin L, Wang C, et al: Glutamine metabolic
microenvironment drives M2 macrophage polarization to mediate
trastuzumab resistance in HER2-positive gastric cancer. Cancer
Commun (Lond). 43:909–937. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shi Q, Shen Q, Liu Y, Shi Y, Huang W, Wang
X, Li Z, Chai Y, Wang H, Hu X, et al: Increased glucose metabolism
in TAMs fuels O-GlcNAcylation of lysosomal Cathepsin B to promote
cancer metastasis and chemoresistance. Cancer Cell.
40:1207–1222.e10. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kalluri R and LeBleu VS: The biology,
function, and biomedical applications of exosomes. Science.
367:eaau69772020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang Y, Guo Z, Chen W, Wang X, Cao M, Han
X, Zhang K, Teng B, Cao J, Wu W, et al: M2 macrophage-derived
exosomes promote angiogenesis and growth of pancreatic ductal
adenocarcinoma by targeting E2F2. Mol Ther. 29:1226–1238. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu M, Zhou C, Weng J, Chen Z, Zhou Q, Gao
J, Shi G, Ke A, Ren N, Sun H and Shen Y: Tumor associated
macrophages-derived exosomes facilitate hepatocellular carcinoma
malignance by transferring lncMMPA to tumor cells and activating
glycolysis pathway. J Exp Clin Cancer Res. 41:2532022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
O'Brien K, Breyne K, Ughetto S, Laurent LC
and Breakefield XO: RNA delivery by extracellular vesicles in
mammalian cells and its applications. Nat Rev Mol Cell Biol.
21:585–606. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhao S, Mi Y, Guan B, Zheng B, Wei P, Gu
Y, Zhang Z, Cai S, Xu Y, Li X, et al: Tumor-derived exosomal
miR-934 induces macrophage M2 polarization to promote liver
metastasis of colorectal cancer. J Hematol Oncol. 13:1562020.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Han S, Bao X, Zou Y, Wang L, Li Y, Yang L,
Liao A, Zhang X, Jiang X, Liang D, et al: d-lactate modulates M2
tumor-associated macrophages and remodels immunosuppressive tumor
microenvironment for hepatocellular carcinoma. Sci Adv.
9:eadg26972023. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gao J, Liang Y and Wang L: Shaping
polarization of tumor-associated macrophages in cancer
immunotherapy. Front Immunol. 13:8887132022. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hu Q, Yao J, Wu X, Li J, Li G, Tang W, Liu
J and Wan M: Emodin attenuates severe acute pancreatitis-associated
acute lung injury by suppressing pancreatic exosome-mediated
alveolar macrophage activation. Acta Pharm Sin B. 12:3986–4003.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Rehmsmeier M, Steffen P, Hochsmann M and
Giegerich R: Fast and effective prediction of microRNA/target
duplexes. RNA. 10:1507–1517. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu C, Li S and Tang Y: Mechanism of
cisplatin resistance in gastric cancer and associated microRNAs.
Cancer Chemother Pharmacol. 92:329–340. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang Q, Huang C, Wang D, Tao Z, Zhang H,
Zhao Y, Wang M, Zhou C, Xu J, Shen B and Zhu W: Gastric cancer
derived mesenchymal stem cells promoted DNA repair and cisplatin
resistance through up-regulating PD-L1/Rad51 in gastric cancer.
Cell Signal. 106:1106392023. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hou G, Bai Y, Jia A, Ren Y, Wang Y, Lu J,
Wang P, Zhang J and Lu Z: Inhibition of autophagy improves
resistance and enhances sensitivity of gastric cancer cells to
cisplatin. Can J Physiol Pharmacol. 98:449–458. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Peng L, Sang H, Wei S, Li Y, Jin D, Zhu X,
Li X, Dang Y and Zhang G: circCUL2 regulates gastric cancer
malignant transformation and cisplatin resistance by modulating
autophagy activation via miR-142-3p/ROCK2. Mol Cancer. 19:1562020.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Baba H, Takeuchi H, Inutsuka S, Yamamoto
M, Endo K, Ohno S, Maehara Y and Sugimachi K: Clinical value of SDI
test for predicting effect of postoperative chemotherapy for
patients with gastric cancer. Semin Surg Oncol. 10:140–144. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen SH and Chang JY: New insights into
mechanisms of cisplatin resistance: From tumor cell to
microenvironment. Int J Mol Sci. 20:41362019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yan XY, Qu XZ, Xu L, Yu SH, Tian R, Zhong
XR, Sun LK and Su J: Insight into the role of p62 in the cisplatin
resistant mechanisms of ovarian cancer. Cancer Cell Int.
20:1282020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang S, Li MY, Liu Y, Vlantis AC, Chan JY,
Xue L, Hu BG, Yang S, Chen MX, Zhou S, et al: The role of microRNA
in cisplatin resistance or sensitivity. Expert Opin Ther Targets.
24:885–897. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bellmunt J, Pons F and Orsola A: Molecular
determinants of response to cisplatin-based neoadjuvant
chemotherapy. Curr Opin Urol. 23:466–471. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Misumi T, Tanabe K, Fujikuni N and Ohdan
H: Stimulation of natural killer cells with rhCD137 ligand enhances
tumor-targeting antibody efficacy in gastric cancer. PLoS One.
13:e02048802018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Olsen LC and Færgeman NJ: Chemical
genomics and emerging DNA technologies in the identification of
drug mechanisms and drug targets. Curr Top Med Chem. 12:1331–1345.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lill JR, Mathews WR, Rose CM and Schirle
M: Proteomics in the pharmaceutical and biotechnology industry: A
look to the next decade. Expert Rev Proteomics. 18:503–526. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Dinić J, Efferth T, Garcia-Sosa AT,
Grahovac J, Padrón JM, Pajeva I, Rizzolio F, Saponara S, Spengler G
and Tsakovska I: Repurposing old drugs to fight multidrug resistant
cancers. Drug Resist Updat. 52:1007132020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Brewer JR, Chang E, Agrawal S, Singh H,
Suzman DL, Xu J, Weinstock C, Fernandes LL, Cheng J, Zhang L, et
al: Regulatory considerations for contribution of effect of drugs
used in combination regimens: Renal cell cancer case studies. Clin
Cancer Res. 26:6406–6411. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Biankin AV, Piantadosi S and Hollingsworth
SJ: Patient-centric trials for therapeutic development in precision
oncology. Nature. 526:361–370. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Mahomed S, Padayatchi N, Singh J and
Naidoo K: Precision medicine in resistant Tuberculosis: Treat the
correct patient, at the correct time, with the correct drug. J
Infect. 78:261–268. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yang C, Deng X, Tang Y, Tang H and Xia C:
Natural products reverse cisplatin resistance in the hypoxic tumor
microenvironment. Cancer Lett. 598:2171162024. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Otvos L: The latest trends in peptide drug
discovery and future challenges. Expert Opin Drug Discov.
19:869–872. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Asaoka Y, Ikenoue T and Koike K: New
targeted therapies for gastric cancer. Expert Opin Investig Drugs.
20:595–604. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Liu L, Wu N and Li J: Novel targeted
agents for gastric cancer. J Hematol Oncol. 5:312012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Portilla LM, Evans G, Eng B and Fadem TJ:
Advancing translational research collaborations. Sci Transl Med.
2:63cm302010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Munoz-Sanjuan I and Bates GP: The
importance of integrating basic and clinical research toward the
development of new therapies for Huntington disease. J Clin Invest.
121:476–483. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Li M, Yang Y, Xiong L, Jiang P, Wang J and
Li C: Metabolism, metabolites, and macrophages in cancer. J Hematol
Oncol. 16:802023. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Chen S, Saeed AFUH, Liu Q, Jiang Q, Xu H,
Xiao GG, Rao L and Duo Y: Macrophages in immunoregulation and
therapeutics. Signal Transduct Target Ther. 8:2072023. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Locati M, Curtale G and Mantovani A:
Diversity, mechanisms, and significance of macrophage plasticity.
Annu Rev Pathol. 15:123–147. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wang S, Liu R, Yu Q, Dong L, Bi Y and Liu
G: Metabolic reprogramming of macrophages during infections and
cancer. Cancer Lett. 452:14–22. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wang HC, Haung LY, Wang CJ, Chao YJ, Hou
YC, Yen CJ and Shan YS: Tumor-associated macrophages promote
resistance of hepatocellular carcinoma cells against sorafenib by
activating CXCR2 signaling. J Biomed Sci. 29:992022. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhang G, Tao X, Ji B and Gong J:
Hypoxia-driven M2-polarized macrophages facilitate cancer
aggressiveness and temozolomide resistance in glioblastoma. Oxid
Med Cell Longev. 2022:16143362022.PubMed/NCBI
|
|
56
|
Huang J, Pan H, Sun J, Wu J, Xuan Q, Wang
J, Ke S, Lu S, Li Z, Feng Z, et al: TMEM147 aggravates the
progression of HCC by modulating cholesterol homeostasis,
suppressing ferroptosis, and promoting the M2 polarization of
tumor-associated macrophages. J Exp Clin Cancer Res. 42:2862023.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Ngabire D, Niyonizigiye I, Patil MP, Seong
YA, Seo YB and Kim GD: M2 macrophages mediate the resistance of
gastric adenocarcinoma cells to 5-fluorouracil through the
expression of integrin β 3, focal adhesion kinase, and cofilin. J
Immunol Res. 2020:17314572020. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Li J, Bao Y, Peng S, Jiang C, Zhu L, Zou
S, Xu J and Li Y: M2 macrophages-derived exosomal miRNA-23a-3p
promotes the progression of oral squamous cell carcinoma by
targeting PTEN. Curr Issues Mol Biol. 45:4936–4947. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Tian B, Zhou L, Wang J and Yang P:
miR-660-5p-loaded M2 macrophages-derived exosomes augment
hepatocellular carcinoma development through regulating KLF3. Int
Immunopharmacol. 101:1081572021. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Cui HY, Rong JS, Chen J, Guo J, Zhu JQ,
Ruan M, Zuo RR, Zhang SS, Qi JM and Zhang BH: Exosomal microRNA-588
from M2 polarized macrophages contributes to cisplatin resistance
of gastric cancer cells. World J Gastroenterol. 27:6079–6092. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wang B, Cai Y, Li X, Kong Y, Fu H and Zhou
J: ETV4 mediated lncRNA C2CD4D-AS1 overexpression contributes to
the malignant phenotype of lung adenocarcinoma cells via
miR-3681-3p/NEK2 axis. Cell Cycle. 20:2607–2618. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zhu MC, Zhang YH, Xiong P, Fan XW, Li GL
and Zhu M: Circ-GSK3B up-regulates GSK3B to suppress the
progression of lung adenocarcinoma. Cancer Gene Ther. 29:1761–1772.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
DuPrie ML, Palacio T, Calil FA, Kolodner
RD and Putnam CD: Mlh1 interacts with both Msh2 and Msh6 for
recruitment during mismatch repair. DNA Repair (Amst).
119:1034052022. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Manzoor S, Saber-Ayad M, Maghazachi AA,
Hamid Q and Muhammad JS: MLH1 mediates cytoprotective nucleophagy
to resist 5-fluorouracil-induced cell death in colorectal
carcinoma. Neoplasia. 24:76–85. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Han Y, Peng Y, Fu Y, Cai C, Guo C, Liu S,
Li Y, Chen Y, Shen E, Long K, et al: MLH1 deficiency induces
cetuximab resistance in colon cancer via her-2/PI3K/AKT signaling.
Adv Sci (Weinh). 7:20001122020. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Scarpa M, Ruffolo C, Kotsafti A, Canal F,
Erroi F, Basato S, DallAgnese L, Fiorot A, Pozza A, Brun P, et al:
MLH1 deficiency down-regulates TLR4 expression in sporadic
colorectal cancer. Front Mol Biosci. 8:6248732021. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Hashimoto T, Kurokawa Y, Takahashi T,
Miyazaki Y, Tanaka K, Makino T, Yamasaki M, Nakajima K, Ikeda JI,
Mori M and Doki Y: Predictive value of MLH1 and PD-L1 expression
for prognosis and response to preoperative chemotherapy in gastric
cancer. Gastric Cancer. 22:785–792. 2019. View Article : Google Scholar : PubMed/NCBI
|