Introduction
Acute myeloid leukemia (AML) is a heterogeneous
hematopoietic malignancy characterized by the rapid accumulation
and malignant proliferation of immature myeloid progenitors in the
bone marrow (BM) and peripheral blood (PB) (1). Without treatment, AML quickly becomes
fatal, and historically, it has always been associated with a poor
prognosis. However, AML treatment has markedly improved over the
last few decades, with improvements in risk assessment,
post-remission chemotherapy and hematopoietic stem-cell
transplantation (2,3). However, even though complete remission
(CR) is achieved after chemotherapy by the majority of AML
patients, only ~20% of obtain relatively long-term relapse-free
survival (RFS) (4). Thus, in order to
improve the diagnosis, prevention and treatment of this disease,
detailed knowledge of the mechanisms that form the basis of AML
development and progression must be acquired. Recently, it has been
shown that long non-coding RNAs (lncRNAs) play a crucial role in
hematopoietic differentiation and hematological malignancies,
including AML (5).
Transcriptome analysis by tiling arrays and RNA
sequencing has revealed that only 2% of the human genome is
dedicated to the transcription of protein coding sequences and that
>90% of the genome is transcribed as non-coding RNAs (6,7). lncRNAs
are transcripts of >200 nucleotides and conventionally cannot be
translated into proteins to participate in a large number of
biological processes (8,9). However, recent studies hypothesized that
a number of lncRNAs are key developmental regulators that are
involved in cell homeostasis and proliferation (9,10).
Notably, increasing numbers of studies are indicating that the
abnormal expression of certain lncRNAs is associated with tumor
growth, carcinogenesis or metastasis in a range of malignancies
(11–15).
Hox transcript antisense intergenic RNA (HOTAIR) is
a 2,158-bp lncRNA that is transcribed from the antisense strand of
the homeobox C gene locus of chromosome 12. HOTAIR coordinates with
chromatin-modifying enzymes and regulates gene silencing (16). Several recent studies have identified
the aberrant expression of HOTAIR in a number of cancer types,
including breast, colon, bladder, renal, pancreatic, cervical and
lung cancer, and a high level of HOTAIR expression has been
correlated with enhanced breast, colon and gastric cancer
metastasis. In addition, HOTAIR-knockdown is able to inhibit the
invasion and proliferation of cells, as well as altering cell cycle
progression and inducing cells apoptosis, thus indicating that
HOTAIR may function in the modulation of cancer progression
(17–19).
To the best of our knowledge, no previous studies
exist concerning the expression status, prognostic value and role
of HOTAIR in AML. Thus, the aim of the present study was to
investigate the correlation of HOTAIR expression with
clinicopathological features and the prognosis of the patients with
AML. The findings may improve our understanding of the roles and
the clinic implications of HOTAIR in the development and
progression of AML.
Patients and methods
Patients and specimens
This study was approved by the Research Ethics
Committee of Wenzhou Central Hospital (Wenzhou, Zhejiang, China).
Written informed consent was obtained from all patients according
to the committee's regulations. Between February 2011 and August
2014, 85 patients in the Department of Hematology (Wenzhou Central
Hospital) were diagnosed with AML according to the
French-American-British (FAB) criteria (20). The cohort consisted of 45 males and 40
females, with a medium age of 45.2 years (range, 19.3–72.4 years).
The median leukocyte count at diagnosis was 52,897/µl (range,
793–327,100/µl; normal range, 4,000–11,000/µl). Of the 85 enrolled
patients, 5 patients presented with AML of type M1, 30 of type M2,
12 of type M4, 32 of type M5 and 6 of type M6, according to the
World Health Organization classification system (21). The clinical characteristics of all the
AML patients are summarized in Table
I. A total of 66 patients received standard cytarabine (100
mg/m2 daily, days 1–7) plus daunorubicin (45
mg/m2 daily, days 1–3) 7+3 induction chemotherapy.
Specimens were obtained from the BM or PB (peripheral white blood
cells, >50×109) of the patients at the time of
diagnosis, and from 33 patients who were in CR following two cycles
of chemotherapy. Additionally, 40 PB specimens were obtained from
healthy donors as negative controls. Patients who achieved CR were
then administered high- or medium-dose cytarabine-based
chemotherapy for consolidation according to their physical
condition. The 33 patients with CR were followed up for a median
time of 22 months (range, 9–40 months), and the data were censored
when the patients relapsed or succumbed.
 | Table I.Clinicopathological variables of 85
patients with newly diagnosed AML divided into low (n=52) and high
(n=33) HOTAIR expression groups. |
Table I.
Clinicopathological variables of 85
patients with newly diagnosed AML divided into low (n=52) and high
(n=33) HOTAIR expression groups.
|
|
| HOTAIR
expression |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
variables | No. of patients | High | Low | P-value |
|---|
| Age, years |
|
|
| 0.950 |
| ≤60 | 39 | 15 | 24 |
|
|
>60 | 46 | 18 | 28 |
|
| Gender |
|
|
| 0.259 |
| Male | 45 | 20 | 25 |
|
|
Female | 40 | 13 | 27 |
|
| WBC,
×109/l |
|
|
| 0.000 |
|
<10 | 50 | 9 | 41 |
|
| ≥10 | 35 | 24 | 11 |
|
| HGB, g/dl |
|
|
| 0.007 |
|
<80 | 49 | 25 | 24 |
|
| ≥80 | 36 | 8 | 28 |
|
| PLT,
×109/l |
|
|
| 0.001 |
|
<50 | 46 | 25 | 21 |
|
| ≥50 | 39 | 8 | 32 |
|
| Blasts in BM, % |
|
|
| 0.001 |
|
<50 | 38 | 7 | 31 |
|
| ≥50 | 47 | 26 | 21 |
|
| FAB subtype |
|
|
| 0.347 |
|
M1/M2 | 35 | 17 | 18 |
|
|
M4/M5 | 44 | 13 | 21 |
|
|
Other | 6 | 3 | 3 |
|
| Complete
remission |
|
|
| 0.082 |
| Yes | 33 | 9 | 24 |
|
| No | 52 | 24 | 28 |
|
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Mononuclear cells from PB or BM specimens were
isolated by Ficoll density gradient centrifugation (400 × g, 30
min; Hao Yang Biological Manufacture, Tianjin, China), and then
washed and pelleted. Additionally, PB cluster of differentiation
(CD)34+ cells from healthy donors were obtained using
magnetic bead separation (EasySep Human CD34 Positive Selection
kit; Stem Cell Technologies, Vancouver, BC, Canada). Total mRNA was
extracted using TRIzol reagent (Invitrogen Life Technologies, Grand
Island, NY, USA) according to the manufacturer's instructions. The
quality and concentration of RNA were determined using a Nanodrop
2000 (Thermo Fisher Scientific Inc., Wilmington, DE, USA). Next, 1
µg total RNA was reverse transcribed from each sample to synthesize
cDNA using the RT reagent kit (Fermentas, Glen Burnie, MD, USA)
according to the manufacturer's instructions. qPCR was performed
using the ABI 7300 Sequence Detection System with primer pairs and
SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA,
USA). The primer sequences used were as follows: HOTAIR forward,
5′-CAGTGGGGAACTCTGACTCG-3′ and reverse, 5′-GTGCCTGGTGCTCTCTTACC-3′;
β-actin forward, 5′-CACCATTGGCAATGAGCGGTTCC-3′ and reverse,
5′-GTAGTTTCGTGGATGCCACAGG-3′. The amplification profile was 95°C
for 5 min, followed by 42 cycles of denaturation at 95°C for 15
sec, then annealing and extension at 60°C for 60 sec. The
comparative Ct method (ΔΔCt) was used for the quantification of
gene expression. The relative expression of HOTAIR to β-actin was
calculated using the equation 2−ΔΔCt, where ΔCt =
CtHOTAIR - Ctβ-actin. Each sample was
analyzed in triplicate and the mean expression level was
calculated.
Statistical analysis
Statistical analysis was performed with SPSS 16.0
for Windows (SPSS Inc., Chicago, IL, USA). Data are expressed as
the mean ± standard deviation. The Kruskal-Wallis non-parametric
test was used to evaluate the difference in HOTAIR expression
between the AML patients and the healthy controls. The paired
t-test was used to evaluate the difference in HOTAIR expression
prior to and following chemotherapy. Pearson's χ2 test
was used to evaluate the association between HOTAIR expression and
clinicopathological characteristics. Survival curves were plotted
using the Kaplan-Meier product-limit method, and differences
between survival curves were tested using the log-rank test. RFS
was defined as the time between the achievement of CR and the time
of the hematological relapse or the last follow-up. Overall
survival (OS) was defined as the time between the moment of
diagnosis and mortality or the last follow-up. Cox regression
analysis in a forward stepwise method was used to evaluate the
effect of multiple independent prognostic factors on survival
outcome. Differences were considered to be statistically
significant when P<0.05.
Results
HOTAIR is overexpressed in AML
patients
The HOTAIR expression levels were detected in BM/PB
samples from the patients with AML and the healthy controls by
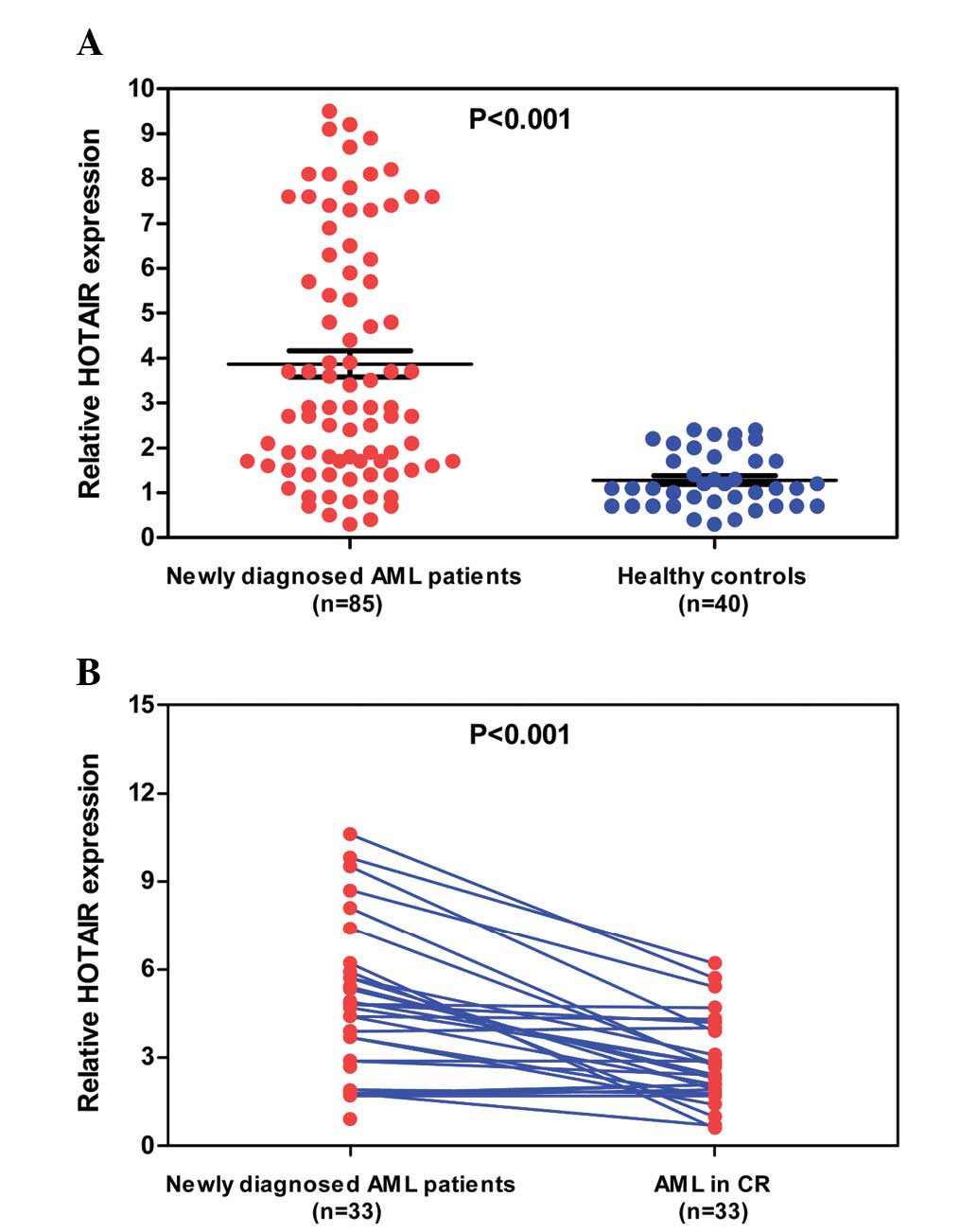
RT-qPCR. As shown in Fig. 1A, the
expression of HOTAIR was significantly upregulated in the AML
patients compared with the healthy controls (mean expression value,
3.87±0.29 vs. 1.28±0.09; P<0.001). Additionally, the 33 AML
patients who achieved CR following one or two cycles of
chemotherapy were monitored for HOTAIR expression during the course
of treatment. The mean expression value of these AML patients
markedly decreased when CR was achieved after chemotherapy (mean
expression value, 4.76±0.47 vs. 2.81±0.27; P<0.001).
Correlations between the expression of
HOTAIR and the clinicopathological factors in AML patients
To identify the clinical relevance of HOTAIR
expression in AML patients, the correlation between HOTAIR
expression and clinicopathological parameters was assessed. Those
AML patients with HOTAIR expression levels at less than the median
value (3.87) were assigned to the low expression group (mean
expression value, 1.98; n=52), and those with expression above the
median value were assigned to the high expression group (mean
expression value, 6.84; n=33). As shown in Table I, high levels of HOTAIR were
associated with higher white blood cell and BM blast counts
(P<0.001 and P=0.001; respectively), and a lower hemoglobin
level and platelet count (P=0.007 and 0.001; respectively).
However, other clinical characteristics, including age, gender and
FAB subtype were not directly associated with the high expression
of HOTAIR.
Association between HOTAIR expression
and clinical outcomes of AML patients
In total, 66 patients received standard induction
chemotherapy, The CR rate following two cycles of chemotherapy was
27.3% (9/33) in the high expression group compared with 46.2%
(24/52) in the low expression group (P=0.082). Despite the high CR
rate in the low expression group, there was no statistically
significant difference between the values of the two groups
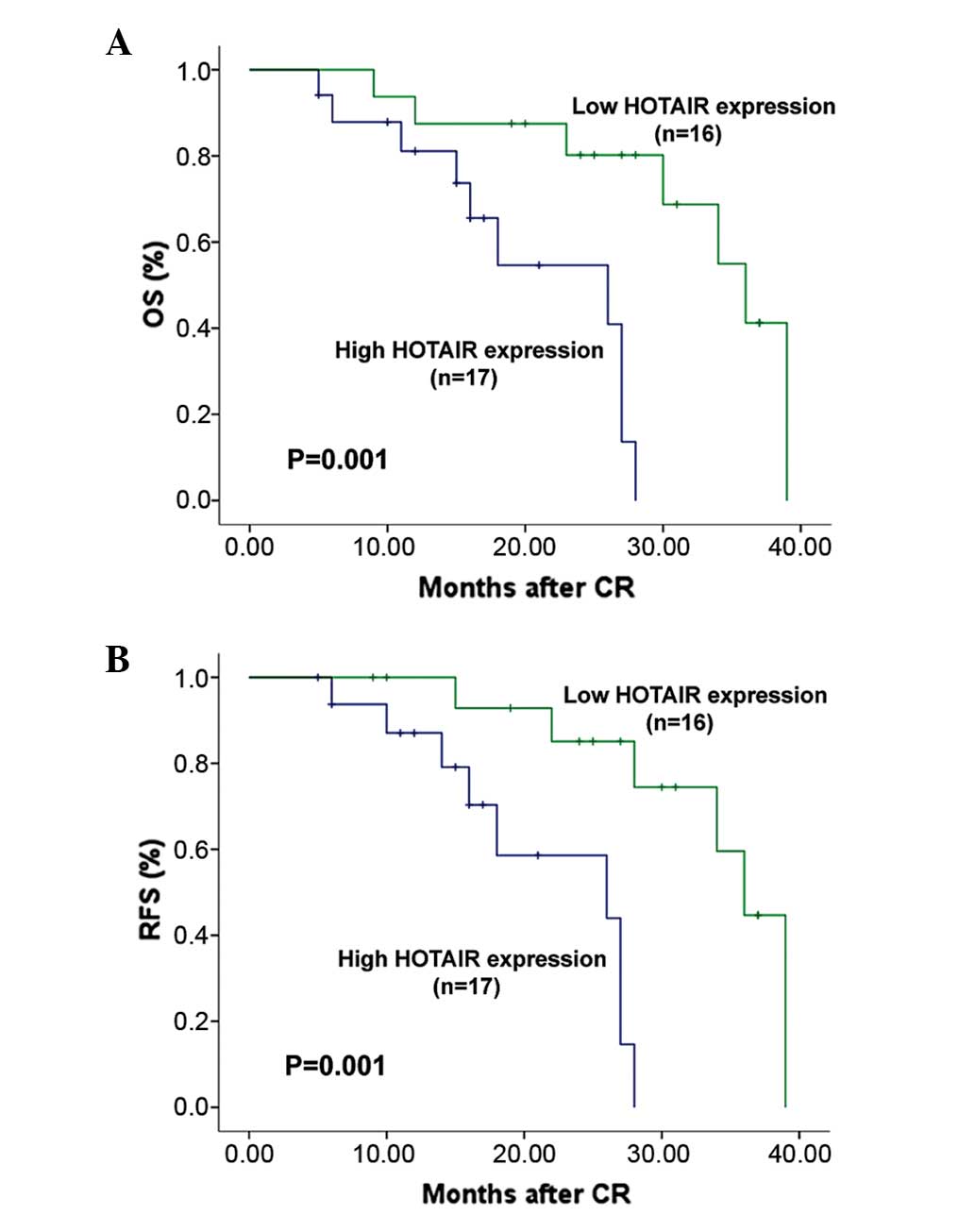
(P=0.082). The 33 patients who achieved a CR were followed up for a
median time of 22 months (range, 9–40 months). OS and RFS survival
curves in high expression and low expression groups are shown in
Fig 2. Patients with high HOTAIR
expression exhibited significantly poorer OS (20.5 vs. 32.1 months;
P=0.001) and RFS (21.5 vs. 33.6 months; P=0.001) times compared
with those with low HOTAIR expression. In multivariate analysis,
the Cox regression analysis revealed that HOTAIR overexpression was
an independent prognostic factor for OS (hazard ratio, 3.37; 95%
confidence interval, 0.99–8.31; P=0.008). Statistical values for
HOTAIR expression and other clinical parameters derived from the
Cox stepwise proportional hazards model are presented in Table II.
 | Table II.Cox multivariate analysis of factors
for overall survival in AML patients. |
Table II.
Cox multivariate analysis of factors
for overall survival in AML patients.
| Factors | Hazard ratio | 95% confidence
interval | P-value |
|---|
| WBC | 0.87 | 0.44–1.39 | 0.047 |
| HGB | 0.93 | 0.74–1.79 | 0.088 |
| PLT | 2.07 | 1.13–3.62 | 0.034 |
| Blasts in BM | 1.15 | 0.41–1.77 | 0.078 |
| Complete
remission | 1.78 | 1.14–2.55 | 0.031 |
| HOTAIR
expression | 3.37 | 0.99–8.31 | 0.008 |
Discussion
It is becoming evident that mammalian genomes encode
thousands of lncRNAs, and multiple lines of evidence increasingly
support the idea that certain lncRNAs could be used as biomarkers
that predict the prognosis of tumor targets of human cancer
(22,23). HOTAIR was first identified as one of
231 lncRNAs that are associated with the human HOX loci, which
binds to polycomb repressive complex 2 (PRC2), a transcriptional
co-repressor, and recruits it to silence the target genes (18). HOTAIR is also able to interact with a
second histone modification complex, the LSD1/CoREST/REST complex,
which functions by coordinating the targeting of PRC2 and LSD1 to
chromatin for methylation of coupled histone H3K27 and
demethylation of K4 (24). HOTAIR is
believed to be an oncogene due to its elevated expression levels in
a number of cancer types, and due to its ability to mediate the
invasion and metastasis of cancer cells. For example, Huang et
al revealed that HOTAIR expression in cervical cancer tissues
was significantly upregulated compared with the matched
non-tumorous tissues, and increased HOTAIR expression was
significantly correlated with the International Federation of
Gynecology and Obstetrics stage, lymph node metastasis, depth of
cervical invasion and tumor size (25). HOTAIR was associated with the
carcinogenesis and invasion of gastric adenocarcinoma,
HOTAIR-targeted RNA interference can reduce the proliferation,
invasion and migration abilities of gastric cancer cell lines
(26). Similarly, dysregulation of
HOXA5 expression has also been reported in association with
tumorigenesis and progression in lung cancer (27–29). These
observations suggest that HOTAIR has a direct role in the
modulation of cancer progression and may be useful in patients with
cancer as a novel prognostic or progression marker. However, in
AML, the HOTAIR expression status and its prognostic roles are
unclear.
In the present study, through the use of qPCR, it
was confirmed for the first time that the expression of lncRNA
HOTAIR was markedly unregulated in patients with newly diagnosed
AML compared with healthy controls; these results were consistent
with other studies regarding solid tumors. Moreover, the level of
HOTAIR expression was significantly decreased following
chemotherapy when patients achieved CR, indicating that HOTAIR
expression is consistent with tumor burden, and that HOTAIR
expression can be used as a prognostic marker of relapse. In
addition, the present results indicated that the upregulation of
HOTAIR in AML patients was significantly correlated with higher
white blood cell and BM blast counts, and a lower hemoglobin level
and platelet count, which represented more aggressive
clinicopathological features. Finally, AML patients with high
HOTAIR expression tended to have poorer OS and RFS times compared
with those with low HOTAIR expression, indicating that the
expression of HOTAIR is significant in the classification of AML
prognosis. Taken together, these data suggest that HOTAIR may
function as an oncogene in the development of AML, and may
represent a candidate prognostic biomarker for AML patients.
The aforementioned findings that HOTAIR
overexpression was associated with aggressive tumor progression
indicated that its possible prognostic value in AML patients should
be investigated in the present study. According to the univariate
and multivariate analyses, HOTAIR overexpression was identified as
an independent predictor for the OS of AML patients, which was in
agreement with recent findings in NSCLC and cervical cancer
(25,26), suggesting that the detection of
increased HOTAIR expression may aid in the identification of AML
patients with a poor prognosis, and could therefore be a novel
prognostic marker for AML patients.
In summary, the present study provides evidence for
the first time that HOTAIR may act as an oncogenic gene in AML, and
that it may represent a potential biomarker of poor prognosis and a
potential therapeutic target for AML intervention. However, the
precise molecular mechanisms behind the involvement of HOTAIR in
AML require further investigation.
References
|
1
|
Troy JD, Atallah E, Geyer JT and Saber W:
Myelodysplastic syndromes in the united states: An update for
clinicians. Ann Med. 46:283–289. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Daver N and Cortes J: Molecular targeted
therapy in acute myeloid leukemia. Hematology. 1 (17
Suppl):S59–S62. 2012.
|
|
3
|
Stone R, Sekeres M and Garcia-Manero G:
Evolving strategies in the treatment of MDS and AML. Clin Adv
Hematol Oncol. 7:1–14; quiz 12 p following 14. 2009.PubMed/NCBI
|
|
4
|
Tallman MS, Gilliland DG and Rowe JM: Drug
therapy for acute myeloid leukemia. Blood. 106:1154–1163. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fatica A: Noncoding RNAs in acute myeloid
leukemia: From key regulators to clinical players. Scientifica
(Cairo). 2012:9257582012.PubMed/NCBI
|
|
6
|
ENCODE Project Consortium, . Birney E,
Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH,
Weng Z, Snyder M, Dermitzakis ET, et al: Identification and
analysis of functional elements in 1% of the human genome by the
ENCODE pilot project. Nature. 447:799–816. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mattick JS: Non-coding RNAs: The
architects of eukaryotic complexity. EMBO Rep. 2:986–991. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wilusz JE, Sunwoo H and Spector DL: Long
noncoding RNAs: Functional surprises from the RNA world. Genes Dev.
23:1494–1504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Johnsson P and Morris KV: Expanding the
functional role of long noncoding RNAs. Cell Res. 24:1284–1285.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bhan A and Mandal SS: Long noncoding RNAs:
Emerging stars in gene regulation, epigenetics and human disease.
Chem Med Chem. 9:1932–1956. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cheng W, Zhang Z and Wang J: Long
noncoding RNAs: New players in prostate cancer. Cancer Lett.
339:8–14. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xia T, Liao Q, Jiang X, Shao Y, Xiao B, Xi
Y and Guo J: Long noncoding RNA associated-competing endogenous
RNAs in gastric cancer. Sci Rep. 4:60882014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhou S, Wang J and Zhang Z: An emerging
understanding of long noncoding RNAs in kidney cancer. J Cancer Res
Clin Oncol. 140:1989–1995. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao J and Lawless MW: Long noncoding RNAs
and their role in the liver cancer axis. Nat Rev Gastroenterol
Hepatol. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tsai MC, Spitale RC and Chang HY: Long
intergenic noncoding RNAs: New links in cancer progression. Cancer
Res. 71:3–7. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Woo CJ and Kingston RE: HOTAIR lifts
noncoding RNAs to new levels. Cell. 129:1257–1259. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wu Y, Zhang L, Wang Y, Li H, Ren X, Wei F,
Yu W, Wang X, Zhang L, Yu J, et al: Long noncoding RNA HOTAIR
involvement in cancer. Tumour Biol. 35:9531–9538. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cai B, Song XQ, Cai JP and Zhang S:
HOTAIR: A cancer-related long non-coding RNA. Neoplasma.
61:379–391. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang J, Zhang P, Wang L, Piao HL and Ma
L: Long non-coding RNA HOTAIR in carcinogenesis and metastasis.
Acta Biochim Biophys Sin (Shanghai). 46:1–5. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bennett JM, Catovsky D, Daniel MT,
Flandrin G, Galton DA, Gralnick HR and Sultan C: Proposed revised
criteria for the classification of acute myeloid leukemia. A report
of the French-American-British Cooperative Group. Ann Intern Med.
103:620–625. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Arber DA, Brunning RD, Le Beau MM, Falini
B, Vardiman JW, Porwit A, Thiele J and Bloomfield CD: Acute myeloid
leukaemia with recurrent genetic abnormalitiesWHO classification of
tumours of haematopoietic and lymphoid tissues. Swerdlow S, Campo E
and Harris NL: 4th edition. IARC Press; Lyon, France: pp. 110–123.
2008
|
|
22
|
Guttman M, Amit I, Garber M, French C, Lin
MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al:
Chromatin signature reveals over a thousand highly conserved large
non-coding RNAs in mammals. Nature. 458:223–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Maruyama R and Suzuki H: Long noncoding
RNA involvement in cancer. BMB Rep. 45:604–611. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tsai MC, Manor O, Wan Y, Mosammaparast N,
Wang JK, Lan F, Shi Y, Segal E and Chang HY: Long noncoding RNA as
modular scaffold of histone modification complexes. Science.
329:689–693. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang L, Liao LM, Liu AW, Wu JB, Cheng XL,
Lin JX and Zheng M: Overexpression of long noncoding RNA HOTAIR
predicts a poor prognosis in patients with cervical cancer. Arch
Gynecol Obstet. 290:717–723. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lee NK, Lee JH, Park CH, Yu D, Lee YC,
Cheong JH, Noh SH and Lee SK: Long non-coding RNA HOTAIR promotes
carcinogenesis and invasion of gastric adenocarcinoma. Biochem
Biophys Res Commun. 451:171–178. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu XH, Liu ZL, Sun M, Liu J, Wang ZX and
De W: The long non-coding RNA HOTAIR indicates a poor prognosis and
promotes metastasis in non-small cell lung cancer. BMC Cancer.
13:4642013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ono H, Motoi N, Nagano H, Miyauchi E,
Ushijima M, Matsuura M, Okumura S, Nishio M, Hirose T, Inase N, et
al: Long noncoding RNA HOTAIR is relevant to cellular
proliferation, invasiveness and clinical relapse in small-cell lung
cancer. Cancer Med. 3:632–642. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhao W, An Y, Liang Y and Xie XW: Role of
HOTAIR long noncoding RNA in metastatic progression of lung cancer.
Eur Rev Med Pharmacol Sci. 18:1930–1936. 2014.PubMed/NCBI
|