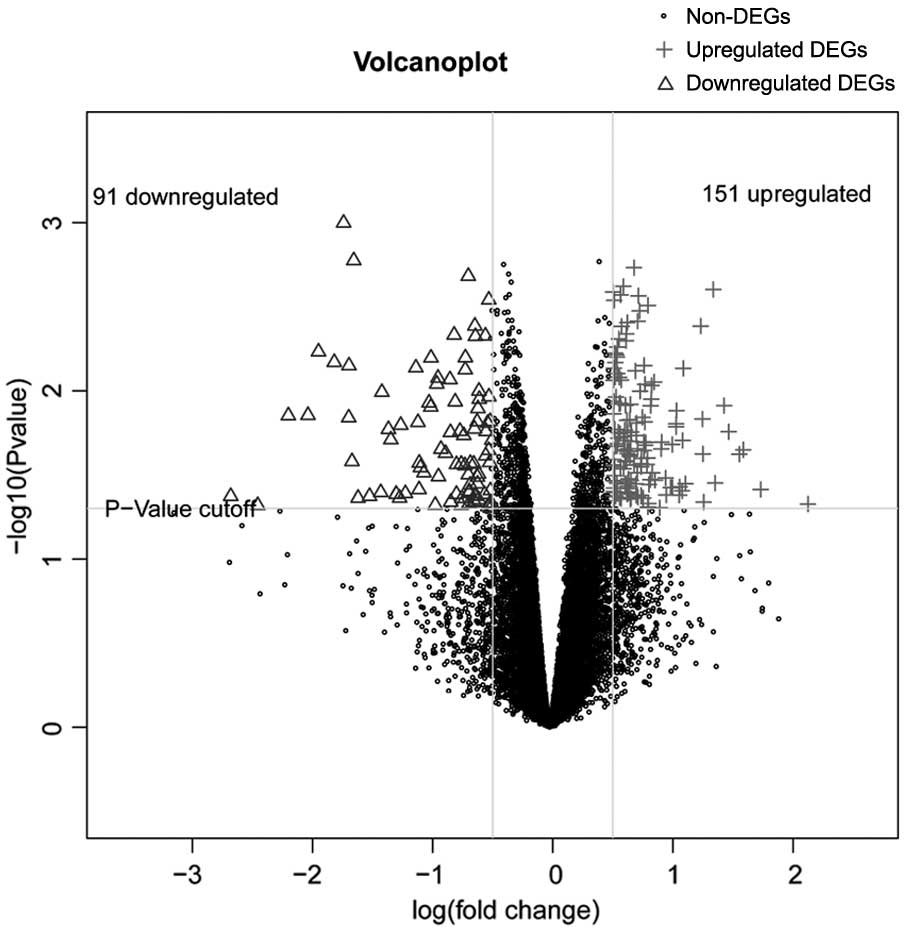
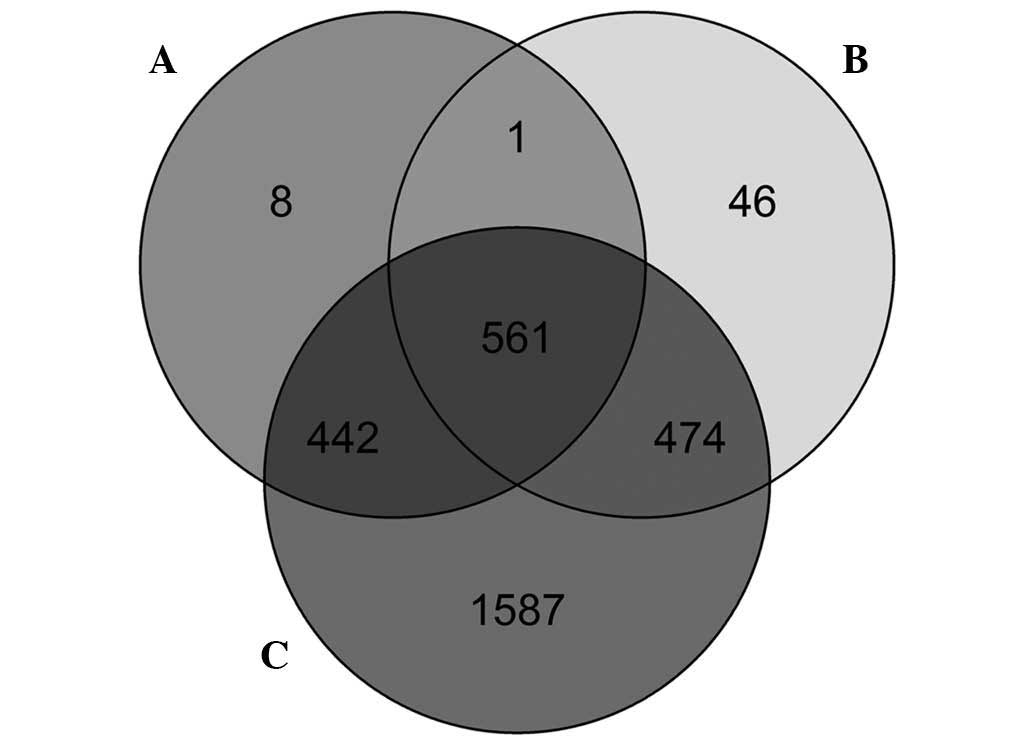
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
van Ham MA, Bakkers JM, Harbers GK, Quint
WG, Massuger LF and Melchers WJ: Comparison of two commercial
assays for detection of human papillomavirus (HPV) in cervical
scrape specimens: Validation of the Roche AMPLICOR HPV test as a
means to screen for HPV genotypes associated with a higher risk of
cervical disorders. J Clin Microbiol. 43:2662–2667. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sood S, Patel FD, Ghosh S, Arora A,
Dhaliwal LK and Srinivasan R: Epigenetic alteration by DNA
methylation of ESR1, MYOD1 and hTERT gene promoters is useful for
prediction of response in patients of locally advanced invasive
cervical carcinoma treated by chemoradiation. Clin Oncol (R Coll
Radiol). 27:720–727. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Clarke MA, Wentzensen N, Mirabello L,
Ghosh A, Wacholder S, Harari A, Lorincz A, Schiffman M and Burk RD:
Human papillomavirus DNA methylation as a potential biomarker for
cervical cancer. Cancer Epidemiol Biomarkers Prev. 21:2125–2137.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kalantari M, Osann K, Calleja-Macias IE,
Kim S, Yan B, Jordan S, Chase DM, Tewari KS and Bernard HU:
Methylation of human papillomavirus 16, 18, 31 and 45 L2 and L1
genes and the cellular DAPK gene: Considerations for use as
biomarkers of the progression of cervical neoplasia. Virology.
448:314–321. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nye MD, Hoyo C, Huang Z, Vidal AC, Wang F,
Overcash F, Smith JS, Vasquez B, Hernandez B, Swai B, et al:
Associations between methylation of paternally expressed gene 3
(PEG3), cervical intraepithelial neoplasia and invasive cervical
cancer. PLoS One. 8:e563252013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen CC, Lee KD, Pai MY, Chu PY, Hsu CC,
Chiu CC, Chen LT, Chang JY, Hsiao SH and Leu YW: Changes in DNA
methylation are associated with the development of drug resistance
in cervical cancer cells. Cancer Cell Int. 15:982015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lando M, Fjeldbo CS, Wilting SM, Snoek CB,
Aarnes EK, Forsberg MF, Kristensen GB, Steenbergen RD and Lyng H:
Interplay between promoter methylation and chromosomal loss in gene
silencing at 3p11-p14 in cervical cancer. Epigenetics. 10:970–980.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sun NX, Ye C, Zhao Q, Zhang Q, Xu C, Wang
SB, Jin ZJ, Sun SH, Wang F and Li W: Long noncoding RNA-EBIC
promotes tumor cell invasion by binding to EZH2 and repressing
E-cadherin in cervical cancer. PLoS One. 9:e1003402014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ye C, Sun NX, Ma Y, Zhao Q, Zhang Q, Xu C,
Wang SB, Sun SH, Wang F and Li W: MicroRNA-145 contributes to
enhancing radiosensitivity of cervical cancer cells. FEBS Lett.
589:702–709. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Burris HH, Baccarelli AA, Motta V, Byun
HM, Just AC, Mercado-Garcia A, Schwartz J, Svensson K, Téllez-Rojo
MM and Wright RO: Association between length of gestation and
cervical DNA methylation of PTGER2 and LINE 1-HS. Epigenetics.
9:1083–1091. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang D, Yan L, Hu Q, Sucheston LE, Higgins
MJ, Ambrosone CB, Johnson CS, Smiraglia DJ and Liu S: IMA: An R
package for high-throughput analysis of Illumina's 450K Infinium
methylation data. Bioinformatics. 28:729–730. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Diboun I, Wernisch L, Orengo CA and
Koltzenburg M: Microarray analysis after RNA amplification can
detect pronounced differences in gene expression using limma. BMC
Genomics. 7:2522006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Almouzni G and Cedar H: Maintenance of
Epigenetic Information. Cold Spring Harb Perspect Biol. 8:pii:
a019372. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yin FF, Wang N, Bi XN, Yu X, Xu XH, Wang
YL, Zhao CQ, Luo B and Wang YK: Serine/threonine kinases 31(STK31)
may be a novel cellular target gene for the HPV16 oncogene E7 with
potential as a DNA hypomethylation biomarker in cervical cancer.
Virol J. 13:602016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Choi CH, Chung JY, Kim JH, Kim BG and
Hewitt SM: Expression of fibroblast growth factor receptor family
members is associated with prognosis in early stage cervical cancer
patients. J Transl Med. 14:1242016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kotacková L, Baláziová E and Sedo A:
Expression pattern of dipeptidyl peptidase IV activity and/or
structure homologues in cancer. Folia Biol (Praha). 55:77–84.
2009.PubMed/NCBI
|
|
18
|
Havre PA, Abe M, Urasaki Y, Ohnuma K,
Morimoto C and Dang NH: The role of CD26/dipeptidyl peptidase IV in
cancer. Front Biosci. 13:1634–1645. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Busek P, Krepela E, Mares V, Vlasicova K,
Sevcik J and Sedo A: Expression and function of dipeptidyl
peptidase IV and related enzymes in cancer. Adv Exp Med Biol.
575:55–62. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Buffon A, Beckenkamp A, Santana DB,
Nascimento J, Paccez J, Zerbini LF, Wink MR and Bruno AN: Abstract
A43: Investigation of dipeptidyl peptidase IV/CD26 role in cervical
cancer cell lines. Mol Cancer Ther. 12:A432013. View Article : Google Scholar
|
|
21
|
Tang L, Su M, Zhang Y, Ip W, Martinka M,
Huang C and Zhou Y: Endothelin-3 is produced by metastatic melanoma
cells and promotes melanoma cell survival. J Cutan Med Surg.
12:64–70. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Benaduce AP, Batista D, Grilo G, Jorge K,
Cardero D, Milikowski C and Kos L: Novel UV-induced melanoma mouse
model dependent on Endothelin3 signaling. Pigment Cell Melanoma
Res. 27:839–842. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Garcia RJ, Ittah A, Mirabal S, Figueroa J,
Lopez L, Glick AB and Kos L: Endothelin 3 induces skin pigmentation
in a keratin-driven inducible mouse model. J Invest Dermatol.
128:131–142. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wiesmann F, Veeck J, Galm O, Hartmann A,
Esteller M, Knüchel R and Dahl E: Frequent loss of endothelin-3
(EDN3) expression due to epigenetic inactivation in human breast
cancer. Breast Cancer Res. 11:R342009. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Espinosa AM, Alfaro A, Roman-Basaure E,
Guardado-Estrada M, Palma Í, Serralde C, Medina I, Juárez E,
Bermúdez M, Márquez E, et al: Mitosis is a source of potential
markers for screening and survival and therapeutic targets in
cervical cancer. PLoS One. 8:e559752013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen YC, Huang RL, Huang YK, Liao YP, Su
PH, Wang HC, Chang CC, Lin YW, Yu MH, Chu TY and Lai HC:
Methylomics analysis identifies epigenetically silenced genes and
implies an activation of β-catenin signaling in cervical cancer.
Int J Cancer. 135:117–127. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kelleher FC, O'Sullivan H, Smyth E,
McDermott R and Viterbo A: Fibroblast growth factor receptors,
developmental corruption and malignant disease. Carcinogenesis.
34:2198–2205. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Okada T, Murata K, Hirose R, Matsuda C,
Komatsu T, Ikekita M, Nakawatari M, Nakayama F, Wakatsuki M, Ohno
T, et al: Upregulated expression of FGF13/FHF2 mediates resistance
to platinum drugs in cervical cancer cells. Sci Rep. 3:28992013.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang SS, Smiraglia DJ, Wu YZ, Ghosh S,
Rader JS, Cho KR, Bonfiglio TA, Nayar R, Plass C and Sherman ME:
Identification of novel methylation markers in cervical cancer
using restriction landmark genomic scanning. Cancer Res.
68:2489–2249. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Anastas JN and Moon RT: WNT signalling
pathways as therapeutic targets in cancer. Nat Rev Cancer.
13:11–26. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ruterbusch JJ, Levin AM, Kittles R,
Rybicki BA and Bock CH: Abstract A66: Admixture and fine mapping in
African Americans identifies a susceptibility locus for prostate
cancer on chromosome 7. Cancer Epidemiol Biomarkers Prev.
21:A662012. View Article : Google Scholar
|