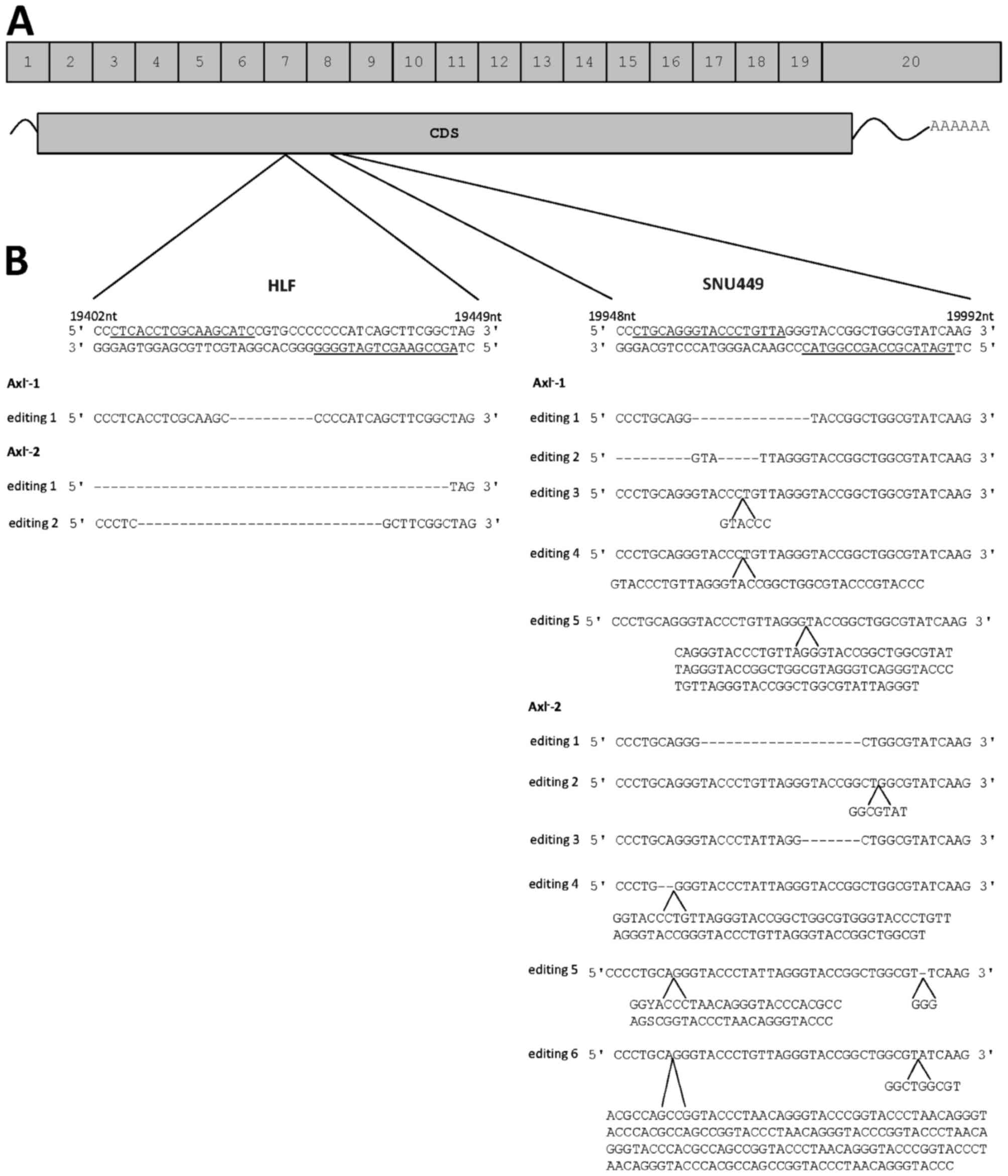
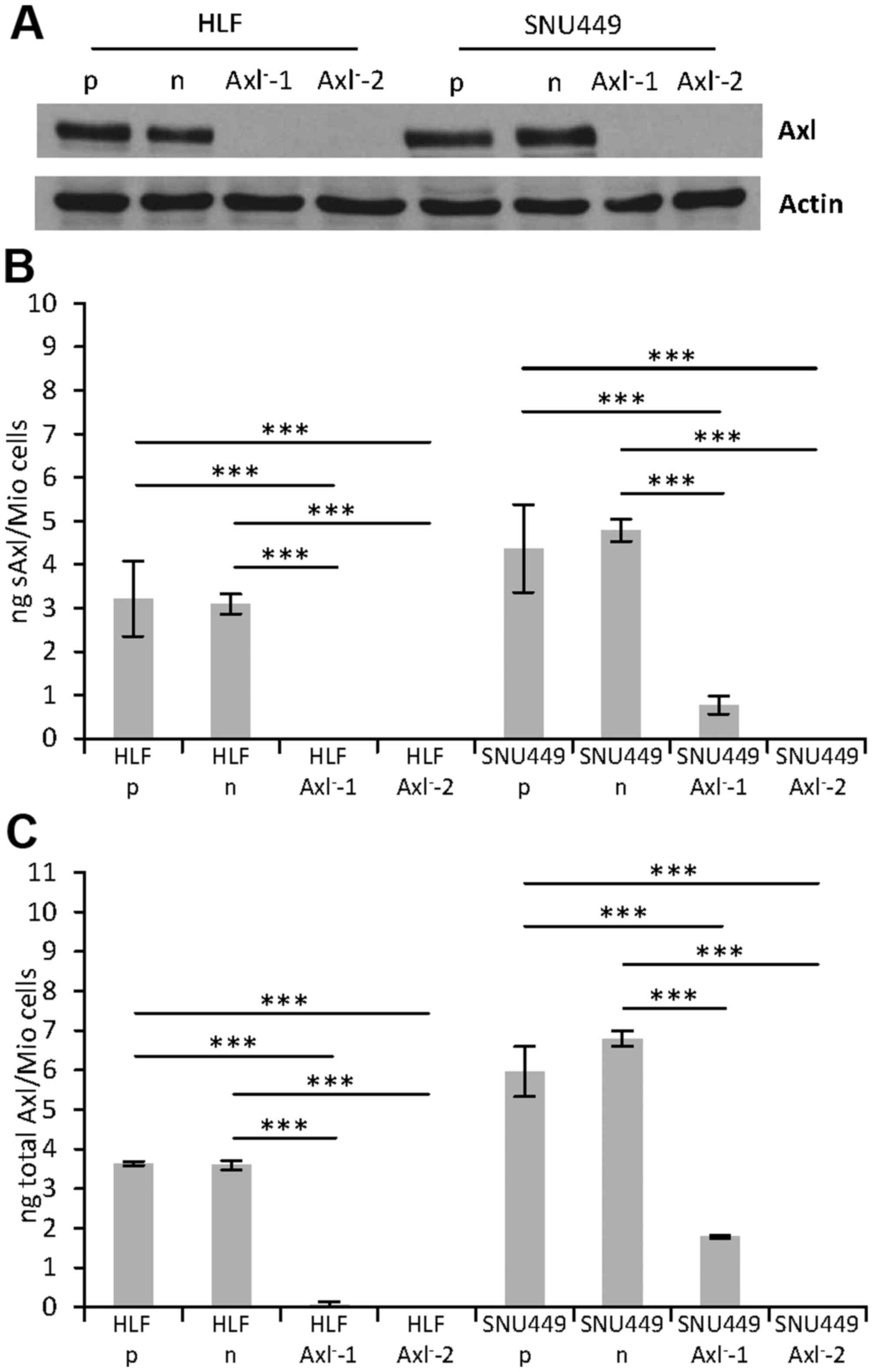
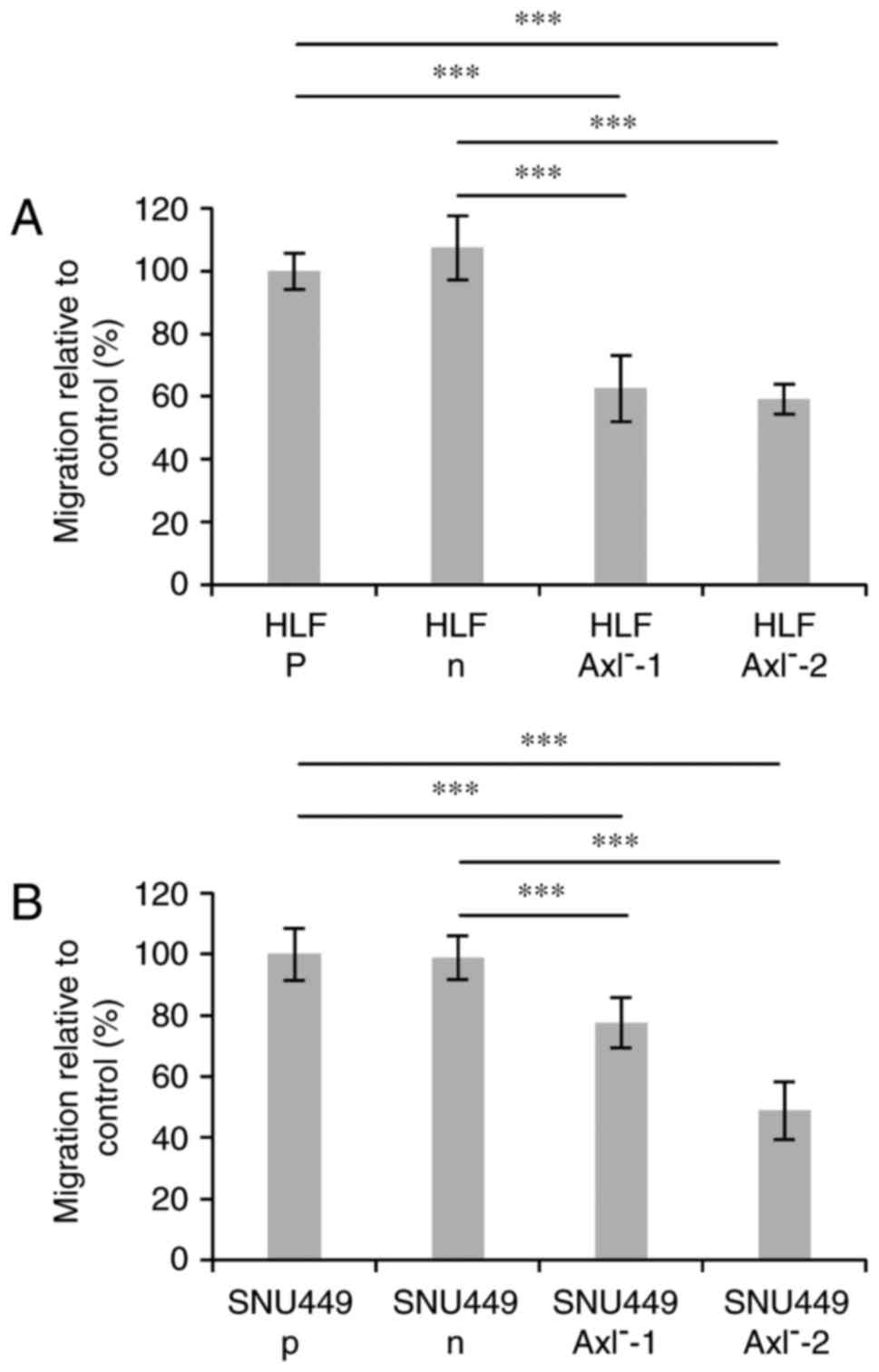
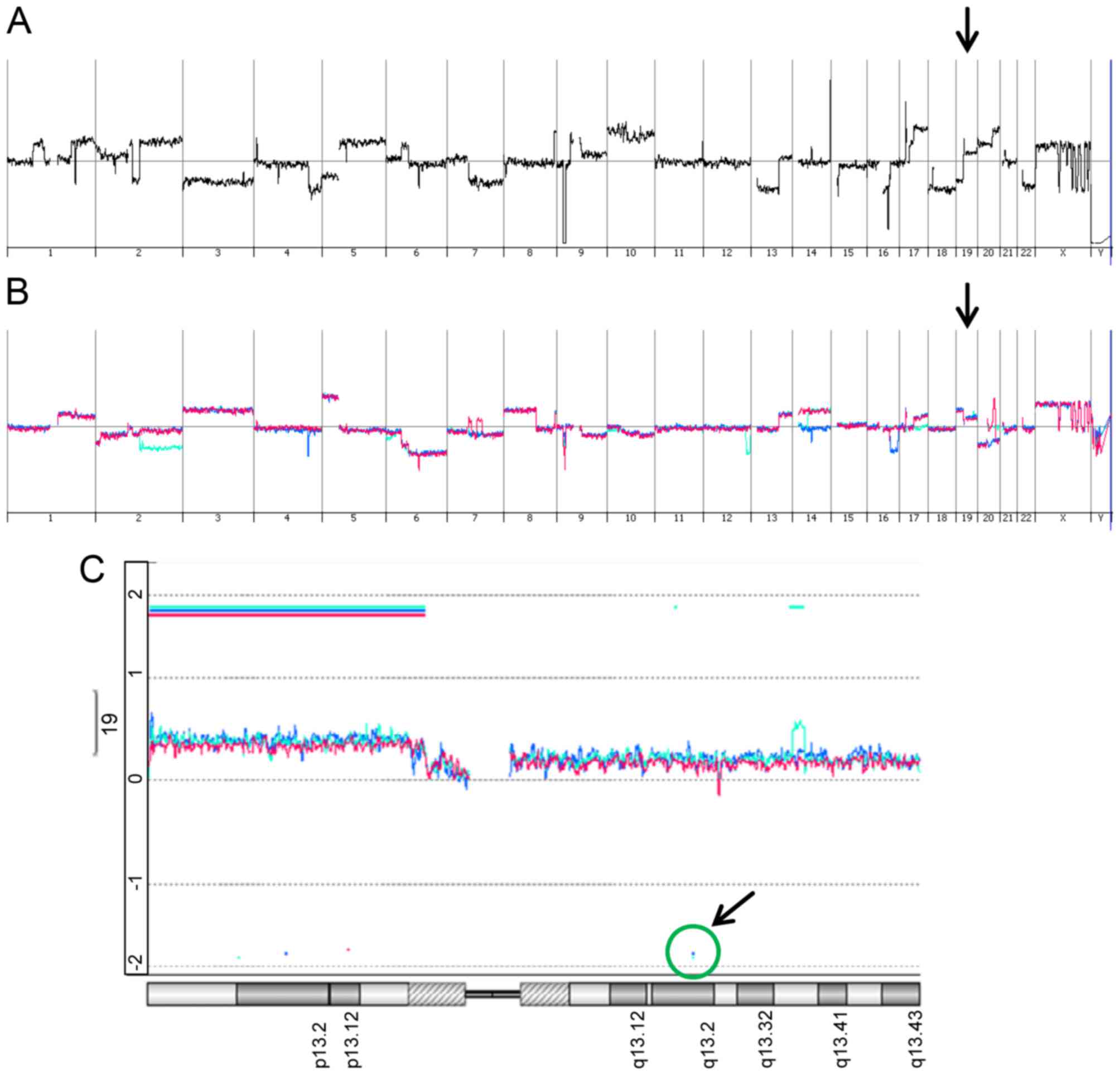
|
1
|
Davidson BL and Paulson HL: Molecular
medicine for the brain: Silencing of disease genes with RNA
interference. Lancet Neurol. 3:145–149. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Xia H, Mao Q, Paulson HL and Davidson BL:
siRNA-mediated gene silencing in vitro and in vivo. Nat Biotechnol.
20:1006–1010. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Miller VM, Xia H, Marrs GL, Gouvion CM,
Lee G, Davidson BL and Paulson HL: Allele-specific silencing of
dominant disease genes. Proc Natl Acad Sci USA. 100:7195–7200.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gaj T, Gersbach CA and Barbas CF III: ZFN,
TALEN, and CRISPR/Cas-based methods for genome engineering. Trends
Biotechnol. 31:397–405. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liberali P, Snijder B and Pelkmans L:
Single-cell and multivariate approaches in genetic perturbation
screens. Nat Rev Genet. 16:18–32. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Joung JK and Sander JD: TALENs: A widely
applicable technology for targeted genome editing. Nat Rev Mol Cell
Biol. 14:49–55. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Miller JC, Tan S, Qiao G, Barlow KA, Wang
J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al: A TALE
nuclease architecture for efficient genome editing. Nat Biotechnol.
29:143–148. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Harrison MM, Jenkins BV, O'Connor-Giles KM
and Wildonger J: A CRISPR view of development. Genes Dev.
28:1859–1872. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jinek M, Chylinski K, Fonfara I, Hauer M,
Doudna JA and Charpentier E: A programmable dual-RNA-guided DNA
endonuclease in adaptive bacterial immunity. Science. 337:816–821.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hryhorowicz M, Lipinski D, Zeyland J and
Slomski R: CRISPR/Cas9 immune system as a tool for genome
engineering. Arch Immunol Ther Exp (Warsz). 65:233–240. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lopes R, Korkmaz G and Agami R: Applying
CRISPR-Cas9 tools to identify and characterize transcriptional
enhancers. Nat Rev Mol Cell Biol. 17:597–604. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jiang W, Bikard D, Cox D, Zhang F and
Marraffini LA: RNA-guided editing of bacterial genomes using
CRISPR-Cas systems. Nat Biotechnol. 31:233–239. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mali P, Yang L, Esvelt KM, Aach J, Guell
M, DiCarlo JE, Norville JE and Church GM: RNA-guided human genome
engineering via Cas9. Science. 339:823–826. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai
SQ, Sander JD, Peterson RT, Yeh JR and Joung JK: Efficient genome
editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol.
31:227–229. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
DiCarlo JE, Norville JE, Mali P, Rios X,
Aach J and Church GM: Genome engineering in Saccharomyces
cerevisiae using CRISPR-Cas systems. Nucleic Acids Res.
41:4336–4343. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bassett AR, Tibbit C, Ponting CP and Liu
JL: Highly efficient targeted mutagenesis of Drosophila with the
CRISPR/Cas9 system. Cell Rep. 4:220–228. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang H, Yang H, Shivalila CS, Dawlaty MM,
Cheng AW, Zhang F and Jaenisch R: One-step generation of mice
carrying mutations in multiple genes by CRISPR/Cas-mediated genome
engineering. Cell. 153:910–918. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li D, Qiu Z, Shao Y, Chen Y, Guan Y, Liu
M, Li Y, Gao N, Wang L, Lu X, et al: Heritable gene targeting in
the mouse and rat using a CRISPR-Cas system. Nat Biotechnol.
31:681–683. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang D, Xu J, Zhu T, Fan J, Lai L, Zhang J
and Chen YE: Effective gene targeting in rabbits using RNA-guided
Cas9 nucleases. J Mol Cell Biol. 6:97–99. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Essletzbichler P, Konopka T, Santoro F,
Chen D, Gapp BV, Kralovics R, Brummelkamp TR, Nijman SM and
Bürckstümmer T: Megabase-scale deletion using CRISPR/Cas9 to
generate a fully haploid human cell line. Genome Res. 24:2059–2065.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ran FA, Hsu PD, Wright J, Agarwala V,
Scott DA and Zhang F: Genome engineering using the CRISPR-Cas9
system. Nat Protoc. 8:2281–2308. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Brinkman EK, Chen T, Amendola M and van
Steensel B: Easy quantitative assessment of genome editing by
sequence trace decomposition. Nucleic Acids Res. 42:e1682014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nemudryi AA, Valetdinova KR, Medvedev SP
and Zakian SM: TALEN and CRISPR/Cas genome editing systems: Tools
of discovery. Acta Naturae. 6:19–40. 2014.PubMed/NCBI
|
|
24
|
Boettcher M and McManus MT: Choosing the
right tool for the job: RNAi, TALEN, or CRISPR. Mol Cell.
58:575–585. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Roschke AV and Rozenblum E: Multi-layered
cancer chromosomal instability phenotype. Front Oncol. 3:3022013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Teoh NC, Dan YY, Swisshelm K, Lehman S,
Wright JH, Haque J, Gu Y and Fausto N: Defective DNA strand break
repair causes chromosomal instability and accelerates liver
carcinogenesis in mice. Hepatology. 47:2078–2088. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Reichl P, Dengler M, van Zijl F, Huber H,
Führlinger G, Reichel C, Sieghart W, Peck-Radosavljevic M,
Grubinger M, Mikulits W, et al: Axl activates autocrine
transforming growth factor-β signaling in hepatocellular carcinoma.
Hepatology. 61:930–941. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Reichl P, Fang M, Starlinger P, Staufer K,
Nenutil R, Muller P, Greplova K, Valik D, Dooley S, Brostjan C, et
al: Multicenter analysis of soluble Axl reveals diagnostic value
for very early stage hepatocellular carcinoma. Int J Cancer.
137:385–394. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Petz M, Them N, Huber H, Beug H and
Mikulits W: La enhances IRES-mediated translation of laminin B1
during malignant epithelial to mesenchymal transition. Nucleic
Acids Res. 40:290–302. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hwang HJ, Kim GJ, Lee GB, Oh JT, Chun YH
and Park SH: A comprehensive karyotypic analysis on Korean
hepatocellular carcinoma cell lines by cross-species color banding
and comparative genomic hybridization. Cancer Genet Cytogenet.
141:128–137. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dor I, Namba M and Sato J: Establishment
and some biological characteristics of human hepatoma cell lines.
Gan. 66:385–392. 1975.PubMed/NCBI
|
|
32
|
Aguirre AJ, Meyers RM, Weir BA, Vazquez F,
Zhang CZ, Ben-David U, Cook A, Ha G, Harrington WF, Doshi MB, et
al: Genomic copy number dictates a gene-independent cell response
to CRISPR/Cas9 targeting. Cancer Discov. 6:914–929. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Munoz DM, Cassiani PJ, Li L, Billy E, Korn
JM, Jones MD, Golji J, Ruddy DA, Yu K, McAllister G, et al: CRISPR
screens provide a comprehensive assessment of cancer
vulnerabilities but generate false-positive hits for highly
amplified genomic regions. Cancer Discov. 6:900–913. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hastings RJ and Franks LM: Cellular
heterogeneity in a tissue culture cell line derived from a human
bladder carcinoma. Br J Cancer. 47:233–244. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Rauch BJ, Silvis MR, Hultquist JF, Waters
CS, McGregor MJ, Krogan NJ and Bondy-Denomy J: Inhibition of
CRISPR-Cas9 with bacteriophage proteins. Cell. 168(150–158):
e1102017.
|
|
36
|
Pawluk A, Amrani N, Zhang Y, Garcia B,
Hidalgo-Reyes Y, Lee J, Edraki A, Shah M, Sontheimer EJ, Maxwell KL
and Davidson AR: Naturally occurring off-switches for CRISPR-Cas9.
Cell. 167:1829–1838.e9. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liang X, Potter J, Kumar S, Zou Y,
Quintanilla R, Sridharan M, Carte J, Chen W, Roark N and
Ranganathan S: Rapid and highly efficient mammalian cell
engineering via Cas9 protein transfection. J Biotechnol. 208:44–53.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kim S, Kim D, Cho SW, Kim J and Kim JS:
Highly efficient RNA-guided genome editing in human cells via
delivery of purified Cas9 ribonucleoproteins. Genome Res.
24:1012–1019. 2014. View Article : Google Scholar : PubMed/NCBI
|