Introduction
Colorectal cancer (CRC) is a common malignancy with
a high prevalence and associated mortality rate (1). The etiology and pathogenetic changes of
CRC are complex and heterogeneous. The risk factors for CRC include
a diet rich in unsaturated fats, consumption of red meat, excessive
total energy intake, excessive alcohol consumption, and inherited
and somatic mutations (2,3). Significant progress has been made in
identifying genetic changes associated with the pathogenesis of CRC
(3,4).
Although adenomatous polyposis coli (APC) is the most frequently
mutated gene in CRC, mutations in P53, KRAS, NRAS,
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α,
F-box and WD repeat domain-containing protein 7, SMAD2, SMAD 4,
transcription factor 7-like 2 (TCF7L2) and β-catenin (CTNNB1) are
also frequently observed in this cancer type (3,4).
Additionally, BRAF mutations are present in 5–8% of CRC cases, with
a single missense V600E mutation accounting for 80% of these
(5). BRAF mutations are often
associated with aggressiveness, poor differentiation and resistance
to therapy in CRC (4). CRC patients
with BRAF V600E mutations exhibit a poor prognosis and a poor
response to panitumumab and cetuximab, monoclonal antibodies
targeting the epidermal growth factor receptor (EGFR/ERBB1)
(6).
The Wnt/β-catenin pathway serves an important role
in CRC, and numerous pathway components, including APC, axin,
TCF7L2 and β-catenin, are mutated in CRC (3,7).
Additionally, the activated Wnt/β-catenin pathway interacts with a
number of other signaling pathways and regulators in modulating
oncogenic processes (8–12), which further complicates the
regulation and functions of the Wnt/β-catenin pathway in cancer
development.
Until now, human cancer cell lines and cancer cell
xenograft mouse models have been irreplaceable tools used in
preclinical cancer drug development. However, it has been
demonstrated that cancer cell lines often lose the biological
properties of the original cancer, including heterogeneity, genetic
characteristics, migratory and metastatic abilities, the
maintenance of a stem cell population, and dependency on embryonic
signaling pathways (13–17). These deficiencies cause significant
setbacks in cancer therapeutic development and result in financial
and human losses. The emergence of patient-derived cancer xenograft
(PDX) models may provide a reliable alternative to cancer cell
xenografts for the development of cancer drugs (18–20).
However, there have been no previous reports on the establishment
of a CRC PDX model; thus, the present study aimed to establish and
characterize a specific CRC PDX mouse model for drug testing.
Materials and methods
Patients and tissues
The study protocol was approved by the Institutional
Review Board of Capital Medical University (Beijing, China).
Written informed consent was obtained from each participant. Colon
cancer tissues were obtained from 10 patients (7 male, 3 female;
age range, 38–72 years; average age, 57.3 years) who had undergone
surgical resection at Beijing Shijitan Hospital or Beijing
Chao-Yang Hospital (Beijing, China) between February 2015 and June
2015 (Table I). Each primary cancer
tissue was divided into three parts: One part for in vivo
grafting; one part for processing into formalin-fixed,
paraffin-embedded (FFPE) tissue blocks; and one part for genomic
DNA extraction.
 | Table I.Characteristics of primary colon
cancers. |
Table I.
Characteristics of primary colon
cancers.
| Variable | Nο. of cases |
|---|
| Dukes staging |
|
| A | 1 |
| B | 3 |
| C | 6 |
| Differentiation
level |
|
| Poor | 2 |
|
Medium | 5 |
| High | 3 |
| Tumor type |
|
|
Ulcerated | 5 |
|
Infiltrative | 3 |
|
Elevated | 2 |
| Associated
pathological symptoms |
|
|
Intestinal Metaplasia | 3 |
| Mucosal
atrophy | 4 |
|
Neither | 3 |
Establishment of patient-derived colon
cancer xenograft (PDCCX) model
Surgically removed colon cancer tissues (F0) were
immediately placed into 4°C Hank's balanced salt solution (Thermo
Fisher Scientific, Inc., Waltham, MA, USA) with 100 U/ml penicillin
and 100 µg/ml streptomycin and transported to the animal facility
within 2 h. Necrotic tissue and blood were removed from the cancer
tissues prior to the tissue being cut into 1- to 2-mm pieces and
subcutaneously implanted into the right hind flanks of 5
immune-compromised nude mice (6 weeks old; male to female, 1:1;
20±2 g; Changzhou Cavens Laboratory Animal Co., Jiangsu, China) per
patient tumor tissue. Mice were maintained in a germ-free facility
at 22–25°C, 55% humidity, 12 h light/dark cycle and free access to
food and water. The mice were routinely monitored for discomfort,
distress or pain. Once the xenografted tumors reached ~500
mm3, the tumor-bearing mice were sacrificed and the
tumors were resected and separated into three parts as for the
primary tumors. Following 5 generations (F5) of consecutive
xenografts, a PDCCX model was considered to have been established.
The protocols for all animal experiments were reviewed and approved
by The Committee for Laboratory Animal Care and Usage of Capital
Medical University.
Immunohistochemistry (IHC)
IHC was used to evaluate six biomarkers at the
protein level. Tumor tissues were fixed in 10% neutral-buffered
formalin (Wuxi Zhanwan Chemicals, Yixing, China; http://www.yxzw.com) at room temperature for 1 week
prior to sectioning. The 4-µm-thick FFPE sections were
deparaffinized in xylene twice (5 min each) and rehydrated in a
descending ethanol series. Antigen retrieval was performed in a
pressure cooker using sodium citrate buffer (10 mM sodium citrate,
0.05% Tween 20, pH 6.0). Endogenous peroxidase activity was blocked
by incubation with 3% H2O2 in PBS at room
temperature for 5–10 min. The sections were incubated with normal
goat serum (Jackson ImmunoResearch Laboratories, Inc., West Grove,
PA) for 30 min at room temperature, followed by incubation with
primary antibodies against β-catenin (1:400; cat. no. 9562), ERBB1
(1:50; cat. no. 4267), c-MET (1:250; cat. no. 8198), caudal type
homeobox 2 (CDX2; 1:1,000; cat. no. 12306), E-cadherin (1:100; cat.
no. 14472) (all from Cell Signaling Technology, Inc., Danvers, MA,
USA) and SMAD3 (1:100; cat. no. ab40854; Abcam, Cambridge, UK)
overnight at 4°C. Following two rinses (5 min each) in PBS, the
sections were incubated with a biotin-conjugated goat anti-rabbit
immunoglobulin (Ig)G (1:200; cat. no. BA1003; Boster Biological
Technology, Pleasanton, CA, USA) following all primary antibody
incubations except those of E-cadherin, for which a goat anti-mouse
IgG secondary antibody was used (1:200; cat. no. BA1001; Boster
Biological Technology) at room temperature for 20 min and rinsed
twice (5 min each) in PBS. Sections were then incubated with
streptavidin-biotin complex reagent (horseradish
peroxidase-conjugated anti-Human IgG SABC kit; cat. no. SA1024;
Boster Biological Technology) at 37°C for 20 min, rinsed four times
for 5 min each, and developed with a Pierce DAB kit (Thermo Fisher
Scientific, Inc.). The sections were counterstained with
hematoxylin, dehydrated in an ethanol gradient, cleared in xylene
and mounted. The slides were observed and imaged (×100,
magnification) using an Olympus IX71 fluorescence microscope
(Olympus Corporation, Tokyo, Japan).
Mutation analyses
Genomic DNA was extracted from primary tumors (F0)
and F5 xenograft tumors using a DNeasy Blood & Tissue kit
(Qiagen China Co., Ltd., Shanghai, China). The genomic DNA was
amplified and sequenced with the primers presented in Table II. Polymerase chain reaction (PCR)
was performed using a Thermal Cycler (Thermo Fisher Scientific,
Inc.) using Phusion High-Fidelity PCR Master Mix (New England
BioLabs, Inc., Ipswich, MA, USA) with the following conditions:
95°C for 5 min followed by 30 cycles of 95°C for 30 sec, 58°C for
30 sec, and 68°C for 30 sec. The PCR products were separated on
agarose gels (0.8%) and sequenced by Sangon Biotech Co., Ltd.
(Shanghai, China). The mutations were analyzed and visualized using
FinchTV (version 1.4.0; Geospiza; PerkinElmer, Waltham, MA,
USA).
 | Table II.Primers sequences used in the present
study. |
Table II.
Primers sequences used in the present
study.
| Gene | Forward (5′-3′) | Reverse (5′-3′) | Note |
|---|
| KRAS |
GTGTGACATGTTCTAATATAGTCA |
GAATGGTCCTGCACCAGTAA |
|
| BRAF |
TCATAATGCTTGCTCTGATAGGA |
GGCCAAAAATTTAATCAGTGGA |
|
| CDH1 (NM_004360) |
ACCTCTGTGATGGAGGTC |
CCACATTCGTCACTGCTACG | Nucleotides
961–1498 |
|
|
CTGAAAGTGACTGATGCTG |
TGTGTACGTGCTGTTCTTCAC | Nucleotides
1312–1812 |
| TP53 |
CTTTGCTGCCGTCTTCCAGTTCG |
CTATCTGAGCAGCGCTCATG | Exon 4 |
|
|
GCCATCTACAAGCAGTCA |
AGACCTAAGAGCAATGAGTG | Exon 5 |
|
|
AGGTCTGGCCCCTCCTCAGC |
ACCTCAGGCGGCTCATAGGGCA | Exon 6 |
|
|
TCTCCTAGGTTGGCTCTGAC |
CACAGCAGGCCAGTGTGCAG | Exon 7 |
|
|
TGGGACAGGTAGGACCTGA |
TGAATCTGAGGCATAACTGCACC | Exon 8 |
| APC |
AGGGTTCTAGTTTATCTTCA |
TCTGCTTGGTGGCATGGTTT | Codons
1339–1436 |
|
|
GGCATTATAAGCCCCAGTGA |
AAATGGCTCATCGAGGCTCA | Codons
1417–1516 |
|
|
ACTCCAGATGGATTTTCTTG |
GGCTGGCTTTTTTGCTTTAC | Codons
1497–1596 |
Drug sensitivity assessment
Once the F5 xenograft tumors reached a size of ~150
mm3, tumor-bearing mice were intraperitoneally
administered with saline, cetuximab (1 mg/kg every 3 days, 5 times
in total), 5-fluorouracil (5-FU; 10 mg/kg for 5 days) or
oxaliplatin (3 mg/kg for 5 days). The tumor sizes were measured
every 5 days.
Statistical analysis
The data are expressed as the mean ± standard error
of the mean. Statistical analyses were performed using GraphPad
Prism 6 (GraphPad Software, Inc., La Jolla, CA, USA). The
differences among groups were analyzed using one-way analysis of
variance followed by Tukey's post hoc test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Establishment of colon cancer PDX
models
Between February 2015 and June 2015, 10 colon cancer
specimens were obtained from the Department of General Surgery,
Beijing Shijitan Hospital (Beijing, China) and were grafted into
nude mice (5 mice per patient). Tumors from 3 patients (30%) grew
in nude mice after 2 months. The success rate of second generation
(F2) xenografts was 66.7% (2/3 patients), and these tumors were
passaged through ≥5 generations. The time required for tumor growth
was relatively consistent at 3–4 weeks after F2.
Histological and genetic features were
maintained following xenograft generation
The histological features of all 10 primary tumors
and 2 PDCCX tumors were assessed by pathologists. Compared with the
parental human tumors, PDCCX tumors maintained the original
histological characteristics (Fig.
1).
The expression levels of proteins important for
colon cancer pathogenesis (SMAD3, ERBB1, c-MET, CDX2, E-cadherin
and β-catenin) were consistent in primary tumors (F0), and first
generation (F1) and fifth generation (F5) xenografted tumors
(Fig. 2).
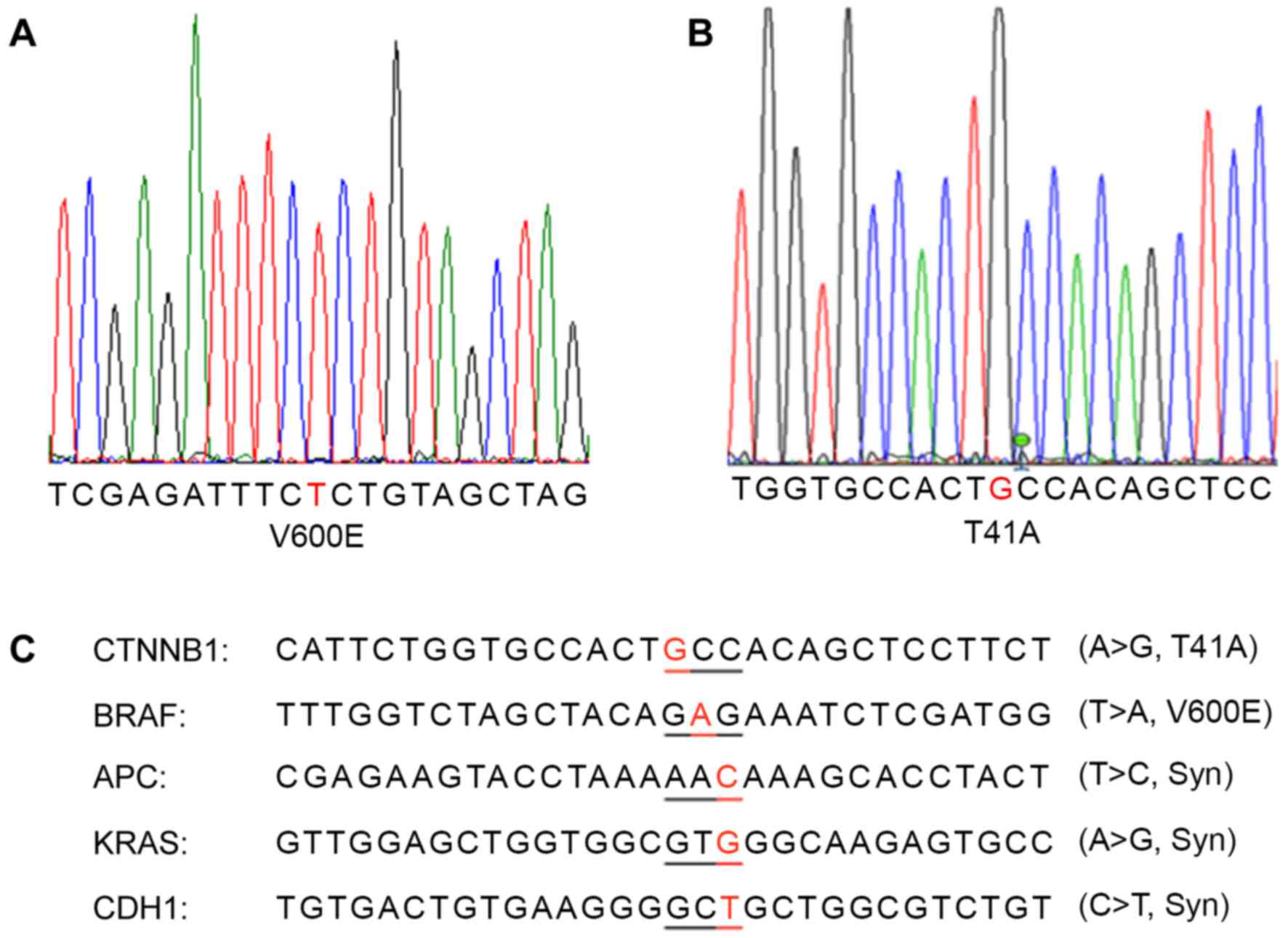
Identification of a colon cancer
carrying a rare BRAF V600E and CTNNB1 T41A double mutation
Through sequencing analysis of the frequently
mutated regions of the most important colon cancer-promoting genes,
a patient with colon cancer exhibiting BRAF V600E (Fig. 3A) and CTNNB1 T41A (Fig. 3B) single-nucleotide mutations was
identified. This cancer also carried synonymous mutations in the
APC, CDH1 and KRAS genes (Fig. 3C).
The same mutations were detected in the primary cancer tissue and
xenografted tumors (F1 and F5).
Drug resistance of colon cancer
carrying BRAF V600E and CTNNB1 T41A double mutation
Three classes of frequently used colon cancer
therapeutics, 5-FU, oxaliplatin and cetuximab, did not
significantly inhibit tumor growth in BRAF V600E/CTNNB1 T41A PDCCX
mice (Fig. 4A). The inhibition rate
of the three drugs never reached 10% (Fig. 4B).
Discussion
In the present study, a PDCCX mouse model was
established and a rare colon cancer carrying BRAF V600E/CTNNB1 T41A
double mutation was identified. The same mutations were identified
in the human primary cancer tissue as well as in the xenografted
tumors (≤5 generations) in the mice. The histology and colon cancer
biomarkers identified in the primary tumors were maintained in F5
engrafted tumors. The BRAF V600E/CTNNB1 T41A PDCCX mouse model was
resistant to inhibition induced by 5-FU, oxaliplatin and
cetuximab.
PDX have emerged as a reliable tool for the
preclinical development of cancer therapeutics, and have been
demonstrated to overcome the limitations of cancer cell lines and
xenograft animal models (19,20). There have been several publications on
the establishment of PDCCX mouse models (21–24).
However, these studies did not systemically characterize the
genetic alterations of the established PDCCX lines, thereby
limiting the applicability of the lines, and also lacked the
clarity required for defining the targeted patient population for
the therapeutics developed using those lines. The PDCCX line
established here had a defined cancer genetic profile and verified
oncogenic drivers.
The new PDCCX line reported in the present study
carried two well-defined point mutations, BRAF V600E and CTNNB1
T41A, which result in activation of growth factor receptor/Ras/MEK
pathways (25) and the Wnt/β-catenin
pathway (26), respectively, thereby
leading to widespread drug resistance. BRAF V600E has been revealed
to be associated with resistance to cancer treatments and a poor
prognosis (27–29). Wild-type BRAF was shown to be required
for response to the EGFR-targeting monoclonal antibodies cetuximab
and panitumumab in CRC patients, as BRAF V600E carriers did not
respond to these two drugs in a previous study (27). A large-scale phase III trial (PETACC-8
trial) demonstrated that the BRAF V600E mutation led to shorter
disease-free survival and overall survival times in patients with
microsatellite-stable colon cancers treated with a combination of
leucovorin, fluorouracil and oxaliplatin, with or without cetuximab
(29). By contrast, activation of the
Wnt/β-catenin pathway led to increased stemness of cancer cells and
drug resistance (30–33). Wnt/β-catenin signaling was
constitutively activated in breast cancer stem cells, and blockade
of the Wnt/β-catenin pathway inhibited cancer metastasis (30). Elevated β-catenin activity promoted
carboplatin resistance in the ovarian cancer A2780 cell line
(31). Increased nuclear
translocation and activation of β-catenin converted CRC cells into
cancer stem cells (32). In a
non-small-cell lung cancer cell line carrying the EGFR T790M
mutation, inhibition of β-catenin activity resulted in inhibition
of cancer cell stemness and sensitivity to an irreversible tyrosine
kinase inhibitor (33). Taken
together, these results indicate that activating BRAF and β-catenin
mutations are individual promoters of tumorigenesis and cancer drug
resistance, and that the BRAF V600E and CTNNB1 T41A double mutation
expands the range of therapeutics that the cancer is resistant to,
as well as the complexity and difficulty of developing effective
treatments. The PDCCX line established in the present study may
provide a reliable tool for developing novel therapeutics that
target both the BRAF V600E and CTNNB1 T41A mutations.
Genetic stability of PDX models is critical for
their application in the preclinical evaluation of drug efficacy.
It was previously reported that the positivity of biomarkers was
consistent between parental human gastric cancer tissues and
corresponding PDX models (34).
Similarly, the biomarker expression was consistent between primary
cancer tissues and third-generation PDX of lymphatic and hepatic
metastatic colon tumors (35). In the
current study, all mutations identified in the primary cancer
tissues were present, with similar expression levels, in the F5
PDCCX tumors, indicating that this rare PDCCX model may be
beneficial in screening and evaluating therapeutics targeting BRAF
and CTNNB1 activating mutations.
In summary, the present study successfully
established and characterized a rare PDCCX mouse model carrying
BRAF V600E and CTNNB1 T41A activating mutations. Histology, genetic
alterations and biomarkers were well-maintained in the PDCCX tumors
(F1 and F5). This PDCCX model demonstrated strong resistance to
several classes of frequently used colon cancer therapeutics.
However, due to the limited number of available cancer samples, we
were unable to obtain a sufficient number of PDCCX models carrying
either a BRAF V600E or CTNNB1 activating mutation to conclusively
determine the interactions between these two colon cancer
promoters, which requires consideration in future studies.
Acknowledgements
The present study was partly supported by the
Beijing Municipal Hospital Authority Key Medical Professional
Development Program for Minimally Invasive Cancer Therapy (grant
no. ZYLX201512).
References
|
1
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Huxley RR, Ansary-Moghaddam A, Clifton P,
Czernichow S, Parr CL and Woodward M: The impact of dietary and
lifestyle risk factors on risk of colorectal cancer: A quantitative
overview of the epidemiological evidence. Int J Cancer.
125:171–180. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fearon ER: Molecular genetics of
colorectal cancer. Annu Rev Pathol. 6:479–507. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cancer Genome Atlas Network, .
Comprehensive molecular characterization of human colon and rectal
cancer. Nature. 487:330–337. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Davies H, Bignell GR, Cox C, Stephens P,
Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W,
et al: Mutations of the BRAF gene in human cancer. Nature.
417:949–954. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yuan ZX, Wang XY, Qin QY, Chen DF, Zhong
QH, Wang L and Wang JP: The prognostic role of BRAF mutation in
metastatic colorectal cancer receiving anti-EGFR monoclonal
antibodies: A meta-analysis. PLoS One. 8:e659952013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kolligs FT, Bommer G and Göke B:
Wnt/beta-catenin/tcf signaling: A critical pathway in
gastrointestinal tumorigenesis. Digestion. 66:131–144. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lei S, Dubeykovskiy A, Chakladar A,
Wojtukiewicz L and Wang TC: The murine gastrin promoter is
synergistically activated by transforming growth factor-beta/Smad
and Wnt signaling pathways. J Biol Chem. 279:42492–42502. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Guo RJ, Huang E, Ezaki T, Patel N,
Sinclair K, Wu J, Klein P, Suh ER and Lynch JP: Cdx1 inhibits human
colon cancer cell proliferation by reducing beta-catenin/T-cell
factor transcriptional activity. J Biol Chem. 279:36865–36875.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ripple MJ, Parker Struckhoff A,
Trillo-Tinoco J, Li L, Margolin DA, McGoey R and Del Valle L:
Activation of c-Myc and Cyclin D1 by JCV T-Antigen and β-catenin in
colon cancer. PLoS One. 9:e1062572014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lian J, Tang J, Shi H, Li H, Zhen T, Xie
W, Zhang F, Yang Y and Han A: Positive feedback loop of
hepatoma-derived growth factor and β-catenin promotes
carcinogenesis of colorectal cancer. Oncotarget. 6:29357–29374.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang R, Sun Q, Wang P, Liu M, Xiong S, Luo
J, Huang H, Du Q, Geller DA and Cheng B: Notch and Wnt/β-catenin
signaling pathway play important roles in activating liver cancer
stem cells. Oncotarget. 7:5754–5768. 2016.PubMed/NCBI
|
|
13
|
Pandita A, Aldape KD, Zadeh G, Guha A and
James CD: Contrasting in vivo and in vitro fates of glioblastoma
cell subpopulations with amplified EGFR. Genes Chromosomes Cancer.
39:29–36. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Vescovi AL, Galli R and Reynolds BA: Brain
tumour stem cells. Nat Rev Cancer. 6:425–436. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sasai K, Romer JT, Lee Y, Finkelstein D,
Fuller C, McKinnon PJ and Curran T: Shh pathway activity is
down-regulated in cultured medulloblastoma cells: Implications for
preclinical studies. Cancer Res. 66:4215–4222. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
De Witt Hamer PC, VanTilborg AA, Eijk PP,
Sminia P, Troost D, Van Noorden CJ, Ylstra B and Leenstra S: The
genomic profile of human malignant glioma is altered early in
primary cell culture and preserved in spheroids. Oncogene.
27:2091–2096. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Clement V, Sanchez P, de Tribolet N,
Radovanovic I and Ruiz i Altaba A: HEDGEHOG-GLI1 signaling
regulates human glioma growth, cancer stem cell self-renewal, and
tumorigenicity. Curr Biol. 17:165–172. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Daniel VC, Marchionni L, Hierman JS,
Rhodes JT, Devereux WL, Rudin CM, Yung R, Parmigiani G, Dorsch M,
Peacock CD and Watkins DN: A primary xenograft model of small-cell
lung cancer reveals irreversible changes in gene expression imposed
by culture in vitro. Cancer Res. 69:3364–3373. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tentler JJ, Tan AC, Weekes CD, Jimeno A,
Leong S, Pitts TM, Arcaroli JJ, Messersmith WA and Eckhardt SG:
Patient-derived tumour xenografts as models for oncology drug
development. Nat Rev Clin Oncol. 9:338–350. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bousquet G and Janin A: Patient-derived
xenograft: An adjuvant technology for the treatment of metastatic
disease. Pathobiology. 83:170–176. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fichtner I, Slisow W, Gill J, Becker M,
Elbe B, Hillebrand T and Bibby M: Anticancer drug response and
expression of molecular markers in early-passage xenotransplanted
colon carcinomas. Eur J Cancer. 40:298–307. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Guenot D, Guérin E, Aguillon-Romain S,
Pencreach E, Schneider A, Neuville A, Chenard MP, Duluc I, Du
Manoir S, Brigand C, et al: Primary tumour genetic alterations and
intra-tumoral heterogeneity are maintained in xenografts of human
colon cancers showing chromosome instability. J Pathol.
208:643–652. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dangles-Marie V, Pocard M, Richon S,
Weiswald LB, Assayag F, Saulnier P, Judde JG, Janneau JL, Auger N,
Validire P, et al: Establishment of human colon cancer cell lines
from fresh tumors versus xenografts: Comparison of success rate and
cell line features. Cancer Res. 67:398–407. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Linnebacher M, Maletzki C, Ostwald C,
Klier U, Krohn M, Klar E and Prall F: Cryopreservation of human
colorectal carcinomas prior to xenografting. BMC Cancer.
10:3622010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Swaika A, Crozier JA and Joseph RW:
Vemurafenib: An evidence-based review of its clinical utility in
the treatment of metastatic melanoma. Drug Des Devel Ther.
8:775–787. 2014.PubMed/NCBI
|
|
26
|
Lasota J, Felisiak-Golabek A, Aly FZ, Wang
ZF, Thompson LD and Miettinen M: Nuclear expression and
gain-of-function β-catenin mutation in glomangiopericytoma
(sinonasal-type hemangiopericytoma): Insight into pathogenesis and
a diagnostic marker. Mod Pathol. 28:715–720. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Di Nicolantonio F, Martini M, Molinari F,
Sartore-Bianchi A, Arena S, Saletti P, De Dosso S, Mazzucchelli L,
Frattini M, Siena S and Bardelli A: Wild-type BRAF is required for
response to panitumumab or cetuximab in metastatic colorectal
cancer. J Clin Oncol. 26:5705–5712. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mori Y, Nagasaka T, Mishima H, Umeda Y,
Inada R, Kishimoto H, Goel A and Fujiwara T: The rare BRAF
VK600-601E mutation as a possible indicator of poor prognosis in
rectal carcinoma-a report of a case. BMC Med Genet. 16:12015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Taieb J, Zaanan A, Le Malicot K, Julié C,
Blons H, Mineur L, Bennouna J, Tabernero J, Mini E, Folprecht G, et
al: Prognostic effect of BRAF and KRAS mutations in patients with
stage III colon cancer treated with leucovorin, fluorouracil, and
oxaliplatin with or without cetuximab: A post hoc analysis of the
PETACC-8 trial. JAMA Oncol. 14:1–11. 2016.
|
|
30
|
Jang GB, Kim JY, Cho SD, Park KS, Jung JY,
Lee HY, Hong IS and Nam JS: Blockade of Wnt/β-catenin signaling
suppresses breast cancer metastasis by inhibiting CSC-like
phenotype. Sci Rep. 5:124652015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Barghout SH, Zepeda N, Xu Z, Steed H, Lee
CH and Fu Y: Elevated β-catenin activity contributes to carboplatin
resistance in A2780cp ovarian cancer cells. Biochem Biophys Res
Commun. 468:173–178. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wangpu X, Yang X, Zhao J, Lu J, Guan S, Lu
J, Kovacevic Z, Liu W, Mi L, Jin R, et al: The metastasis
suppressor, NDRG1, inhibits ‘stemness’ of colorectal cancer via
down-regulation of nuclear β-catenin and CD44. Oncotarget.
6:33893–33911. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Togashi Y, Hayashi H, Terashima M, de
Velasco MA, Sakai K, Fujita Y, Tomida S, Nakagawa K and Nishio K:
Inhibition of β-Catenin enhances the anticancer effect of
irreversible EGFR-TKI in EGFR-mutated non-small-cell lung cancer
with a T790M mutation. J Thorac Oncol. 10:93–101. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang T, Zhang L, Fan S, Zhang M, Fu H,
Liu Y, Yin X, Chen H, Xie L, Zhang J, et al: Patient-derived
gastric carcinoma xenograft mouse models faithfully represent human
tumor molecular diversity. PLoS One. 10:e01344932015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jin K, Li G, Cui B, Zhang J, Lan H, Han N,
Xie B, Cao F, He K, Wang H, et al: Assessment of a novel VEGF
targeted agent using patient-derived tumor tissue xenograft models
of colon carcinoma with lymphatic and hepatic metastases. PLoS One.
6:e283842011. View Article : Google Scholar : PubMed/NCBI
|