Introduction
Acute myeloid leukemia (AML) is a cancer of the
white blood cells characterized by the clonal proliferation of
myeloid progenitor cells in the bone marrow and peripheral blood
(1). AML accounts for ~80% of all
acute leukemia cases in adults, and the incidence of this disease
has been revealed to increase with age (2,3). At
present, AML is curable in 35–40% of patients <60 years old;
however, among patients >60 years of age, cases of full recovery
are less common (5–15%) (1,4). Due to the fact that survival rates
remain relatively low (overall 5-year survival is <5% in older
patients), novel therapeutics and treatment strategies are required
(3).
The etiology of AML is not yet fully known, but
there are a number of genetic factors that may predispose patients
to this disease, including chromosomal translocations (Breakpoint
cluster region-Abelson murine leukemia viral oncogene homolog 1),
and mutations in FMs-Related Tyrosine Kinase 3 (FLT3), Tumor
Protein 53 and Additional Sex Combs Like 1, Transcriptional
Regulator genes (5). Among the
genetic factors that are involved in the development of AML, are
the runt-related transcription factor 1 (RUNX1) and
runt-related transcription factor 3 (RUNX3) genes. These
genes belong to the runt domain transcription factor family, which
is responsible for encoding the DNA-binding α-subunits of the RUNT
domain transcription factor and serve an important role in the
regulation of transcription (6,7). However,
they may be dysregulated in human cancer cells (as a result of
mutations, translocations or inactivation), and therefore
potentially serve a role in the pathogenesis of cancer (6,8). The
RUNX1 gene, which is located on chromosome 21 (locus
q21.22), serves an important role in hematopoiesis during embryonic
development (9). Furthermore, it is
responsible for the formation of hematopoietic stem cells and
progenitor cells due to its expression in all hematopoietic sites
(10). Previous studies have
demonstrated that the chromosomal translocations and mutations in
the RUNX1 gene may be associated with several types of
leukemia, including AML (11,12).
The RUNX3 gene also encodes transcription
factors and is responsible for the regulation of a number of other
genes, including Transforming growth factor-β and Notch 1 pathways,
Core-Binding Factor β subunit, ETS proto-oncogene 1 and ETS
proto-oncogene 2 transcription factor genes (11). The RUNX3 gene is located on
chromosome 1 (locus p36) (11,13) and is
highly expressed in all hematopoietic stem cells (14). Furthermore, the RUNX3 gene is
hypothesized to act as a tumor suppressor. There is evidence to
suggest that the inactivation of this gene is associated with the
development of various types of cancer, including breast cancer
(15). Additionally, deletion of this
gene is associated with hyperplasia of the gastric mucosa and
gastric cancer development (16,17).
Continuous research regarding the potential functions of
RUNX family genes in tumor development and the influence of
these genes on the expression of other genes may assist in the
early detection of cancer, and the development of more effective
treatment modalities and novel therapeutics for patients with
cancer (8). The role of the
RUNX1 and RUNX3 genes in AML have, thus far, not been
completely elucidated. Therefore, the aim of the present study was
to evaluate the mRNA expression level of the RUNX1 and
RUNX3 genes in patients with AML.
Materials and methods
Sample collection and ethics
statement
The investigated group comprised of 43 (22 female
and 21 male) patients who had been diagnosed with AML at the
Hematology Clinic, Medical University of Lodz (Lodz, Poland). The
mean age at the time of diagnosis was 57.9 years (58.6 for females
and 57.4 for males; age range 17–80 years). Patients were divided
into subgroups according to the French-American-British (FAB)
classification of AML (18).
Peripheral blood (March 2016-May 2017) was used for research and
the samples obtained were residual material remaining following
routine blood tests. The Ethics Committee of the Medical University
of Lodz approved the present study (RNN/88/16/KE). Written informed
consent was obtained from the patients for participation in the
study.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the blood cells of
participants using the Total RNA Mini kit (A&A Biotechnology,
Gdynia, Poland), according to the manufacturer's protocol. The
purity of obtained RNA was determined by the A260/280 ratio
(DNA/RNA absorbance to protein absorbance). Absorbance at 260 nm
was used to determine the amount of RNA required for reverse
transcription. Isolated RNA samples were stored at −76°C until
further analysis. RNA samples were reverse transcribed into cDNA
using a High Capacity cDNA Reverse Transcription kit (Applied
Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA),
according to the manufacturer's protocol. The thermocycling
parameters were as follows: 25°C for 10 min, then 37°C for 120 min
and 85°C for 5 min. The final concentration of RNA in the reaction
mixture in all samples was 0.02 µg/µl. The presence of cDNA was
verified through PCR amplification normalized to GAPDH
(19). PCR was performed for
qualitative analysis of mRNA expression RUNX1 and
RUNX3. Amplification was performed according to the
manufacturer's protocol for the AccuTaq™LA DNA
Polymerase kit (Sigma Aldrich; Merck KGaA, Darmstadt, Germany). The
reaction mixture consisted of 1 µl cDNA template, 0.7 µl of 10 µM
each primer, 3.5 µl of 1.5 mM 10× PCR buffer without
MgCl2 (Sigma Aldrich; Merck KGaA), 0.7 µl of 25 mM
MgCl2 reagent, 0.4 µl of 0.2 mM dNTP (deoxynucleotides)
mix, 0.2 µl of 0.5 U AccuTaq LA DNA Polymerase and distilled water
to the final volume of 21 µl. Primers used in the present study are
listed in Table I. A negative
control, without cDNA template, was included in every experiment.
Amplification was performed using an MJ Mini Personal Thermal
Cycler (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The
thermocycling conditions were as follows: Initial denaturation at
95°C for 2 min, denaturation at 92°C for 1 min, primer annealing at
58°C for RUNX1 and 56°C for RUNX3 for 30 sec,
elongation at 72°C for 45 sec and final elongation at 72°C for 7
min. Electrophoresis on a 2% agarose gel was used to assess the
products of PCR amplification. The sizes of the reaction products
were as follows: RUNX1, 96 bp and RUNX3, 120 bp. qPCR
was used for quantitative assessment of RUNX1, RUNX3 and
GAPDH mRNA expression, and reactions were performed in a
Rotor-Gene™ 6000 thermocycler (Corbett Life Science;
Qiagen GmbH, Hilden, Germany). GAPDH is a housekeeping gene,
the expression of which is often used to normalize mRNA levels
between samples (19). The reaction
mixture consisted of 5 µl RT HS-PCR Mix Sybr® B (A&A
Biotechnology), 0.7 µl of 10 µM each primer, 1 µl cDNA template and
nuclease-free water to a final volume of 10 µl. Experiments for
investigated and reference genes were performed in triplicate and
reactions were performed in separate tubes. A negative control,
without cDNA template, in triplicate was also included in every
experiment. The reaction parameters were as follows: Initial
denaturation at 95°C for 10 min, denaturation at 95°C for 10 sec,
primer annealing at 55°C for RUNX1 and 58°C for RUNX3
for 15 sec, elongation at 72°C for 20 sec. In order to assess the
specification of products, analysis of melting curves was performed
following amplification. The 2−ΔΔCq method was used to
estimate relative changes in gene expression determined by RT-qPCR
analysis (20). The mean
Cq values of GAPDH, RUNX1 and RUNX3 genes
were used in subsequent calculations.
 | Table I.Primers used for reverse
transcription-quantitative polymerase chain reaction analysis. |
Table I.
Primers used for reverse
transcription-quantitative polymerase chain reaction analysis.
| Gene | Forward primer | Reverse primer |
|---|
| GAPDH |
5′-TGGTATCGTGGAAGGACTCATGAC-3′ |
5′-ATGCCAGTGAGCTTCCCGTTCAGC-3′ |
| RUNX1 |
5′-AGTGGAAGAGGGAAAAGC-3′ |
5′-ATCCACTGTGATTTTGATGG-3′ |
| RUNX3 |
5′-ATGACGAGAACTACTCCG-3′ |
5′-TCAGGGTGAAACTCTTCC-3′ |
Statistical analysis
Statistical analyses were performed using STATISTICA
12.5 (StatSoft Inc., Tulsa, OK, USA). A comparative statistical
analysis was performed using the non-parametric U Mann-Whitney test
in the absence of normality of relative levels of RUNX1 and
RUNX3 gene expression. P<0.05 was considered to indicate
a statistically significant difference.
Results
Relative RUNX1 and RUNX3 gene
expression level with sex and age of diagnosis
All 43 samples exhibited GAPDH expression.
The presence of RUNX1 and RUNX3 gene expression was
also identified in all selected samples. Quantitative analyses
revealed that the transcript level of RUNX1 and RUNX3
genes varied among selected cases. It ranged between 0.13 and
18.37, with a median value 0.73 for the RUNX1 gene and
between 0.04 and 8.54 with a median value of 1.28 for the
RUNX3 gene. The investigated group comprised 22 females and
21 males. Statistically significant differences between patient sex
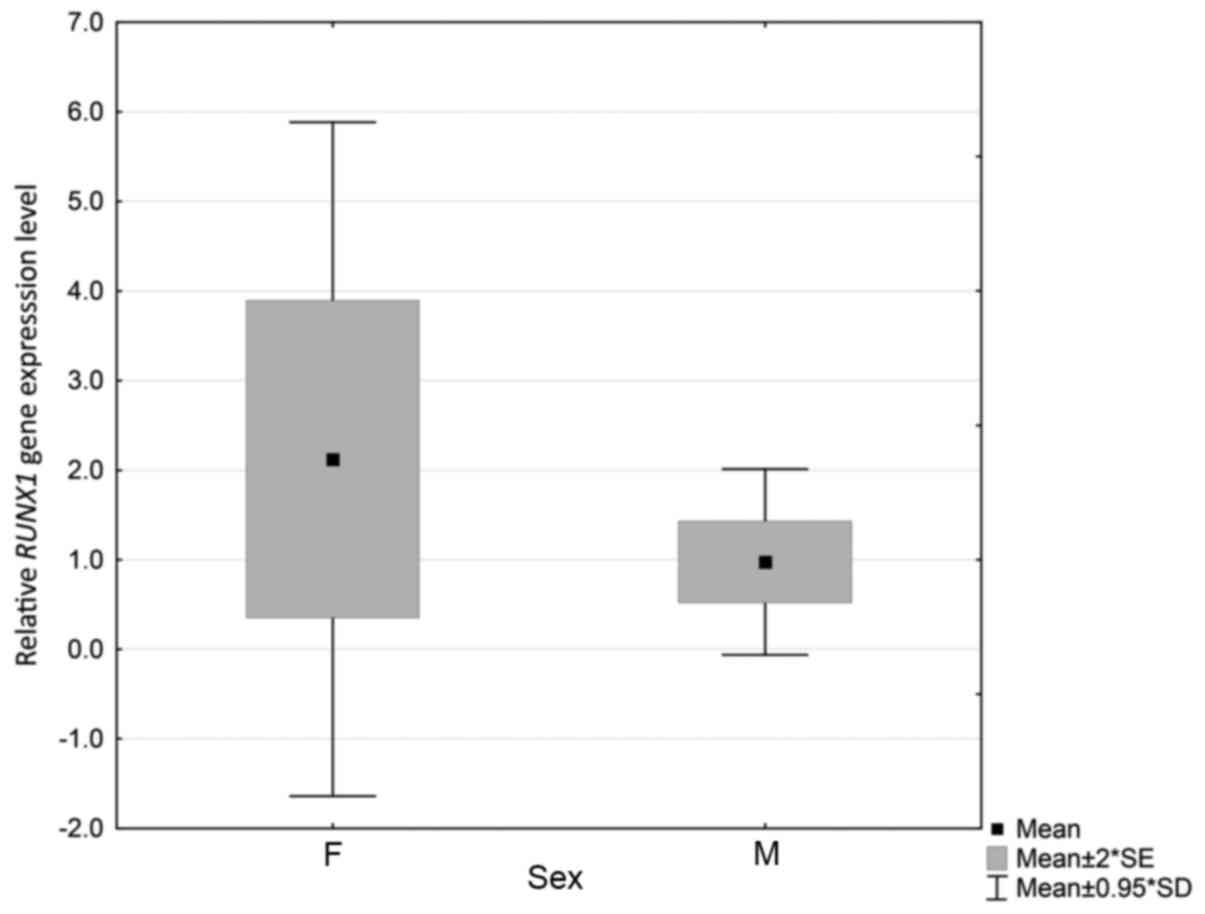
and relative RUNX1 expression were identified (P=0.044).
Levels were higher and varied more among females (Fig. 1). However, no significant differences
were identified between sex and relative RUNX3 gene
expression (P=0.130; data not shown). Another compared parameter
was age at the time of AML diagnosis. The mean age was 57.9, 58.6
years for females and 57.4 years for males; however, no
statistically significant associations were identified between age
at the time of diagnosis and mRNA expression of RUNX1
(P=0.970) or RUNX3 (P=0.469).
Relative RUNX1 and RUNX3 gene
expression level with FAB classification and mortality
Patients were also divided into subgroups according
to the FAB classification of AML (18). Full details are presented in Table II. Statistical analysis revealed no
significant associations between FAB classification subgroups and
relative RUNX1 (P=0.746) and RUNX3 (P=0.771)
expression. Relative expression was also compared with mortality
among the enrolled patients. The results indicated that there is a
statistically significant association between the relative
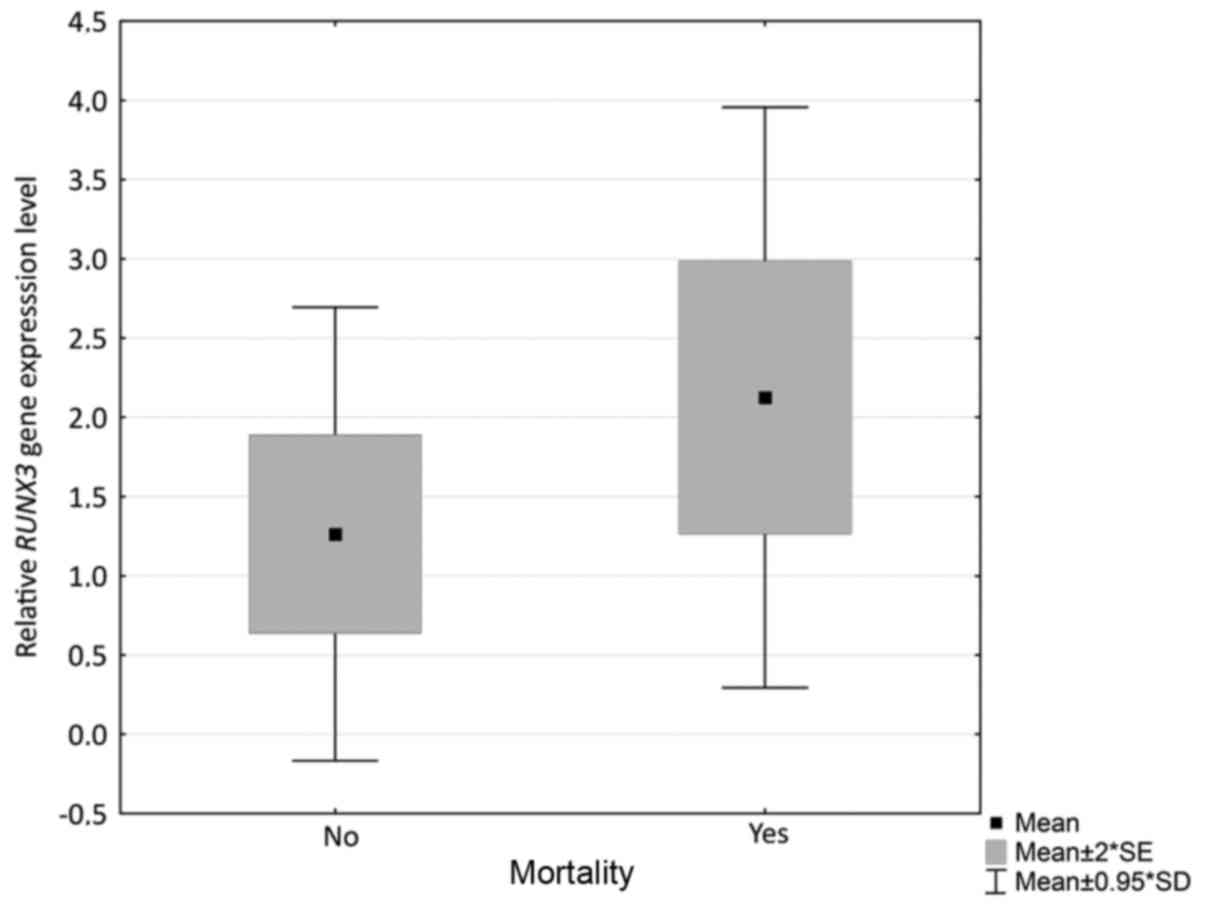
expression of RUNX3 and mortality among patients (P=0.036).
Mortality was more frequent among patients with higher RUNX3
expression levels (Fig. 2); however,
no such association was observed between mortality and RUNX1
expression (P=0.445, data not shown).
 | Table II.French-American-British
classification of investigated patients. |
Table II.
French-American-British
classification of investigated patients.
| Diagnosis | Number of
cases | RUNX1 P-value | RUNX3 P-value |
|---|
| AML undefined | 21 (10F, 11M) |
|
|
| AML 0 | 1F |
|
|
| AML1 | 3 (2F, 1M) |
|
|
| AML2 | 8 (5F, 3M) | 0.746 | 0.771 |
| AML3 | 2 (1F, 1M) |
|
|
| AML4 | 4 (2F, 2M) |
|
|
| AML5 | 3 (1F, 2M) |
|
|
| AML6 | 1M |
|
|
Discussion
Due to the presence of various mutations in the
RUNX1 and RUNX3 genes in patients with AML, we
hypothesized that these genes may influence mRNA formation and may
contribute to the different levels of expression among the
investigated cases. To the best of our knowledge, the present study
is the first to present the RUNX1 and RUNX3 gene
expression levels in patients with AML determined by RT-qPCR
analysis in a Polish population, as previous studies have only been
conducted in Chinese populations thus far.
The RUNX1 gene serves an important role in
hematopoiesis (9). Its abnormal
expression is present in various malignancies, including ovarian
cancer, cytogenetically normal AML (CN-AML) and breast cancer
(21–24). However, the significance of the
RUNX1 gene in cancer development is not fully known.
Previous studies have suggested that the RUNX1 gene
functions as a tumor suppressor in AML (25), and that loss of the RUNX1 gene
may lead to weak differentiation and leukemia development (26). One previous study has reported that a
normal expression level of RUNX1 gene inhibits cell
proliferation and promotes differentiation of hematopoietic
progenitor cells (21). By contrast,
deactivating the RUNX1 gene may cause amplification of
myeloid progenitors and the subsequent development of AML. A
previous study undertaken by Silva et al (25) suggested that the RUNX1 gene
acts as a classical tumor suppressor gene; however, other studies
have suggested that RUNX1 functions as an oncogene and that
it may cause AML development due to its pro-survival role in
leukemia cell proliferation (27–30). The
results of these studies also suggested that the prognostic impact
in CN-AML depends on the RUNX1 expression level. A study
undertaken by Goyama et al (28) reported that overexpression of the
RUNX1 gene inhibited the growth of regular cord blood cells
by inducing myeloid differentiation. It was suggested that the
RUNX1 gene may be a valuable novel marker for risk
stratification in patients with AML and that it is an excellent
candidate for anticancer-targeted therapy due to the modulation of
its post-translational modifications (29).
It is now hypothesized that, due to its expression
level, the RUNX1 gene may serve a role as a tumor promoter
or tumor suppressor in different types of cancer and hematological
malignancies including AML (21). A
study undertaken by Fu et al (21) estimated RUNX1 expression using
microarrays and revealed that a high level of RUNX1 mRNA
expression in CN-AML was associated with a poorer overall survival
(OS) and event-free survival (EFS) than low RUNX1 mRNA
expression. The median OS and EFS times in patients with a higher
RUNX1 expression level were poorer than that of the low
RUNX1 expression group (P=0.009 and P=0.011, respectively).
Among 157 patients with CN-AML with a higher RUNX1 gene
expression, significantly more patients exhibited the FAB M2
subtype than in the group with lower RUNX1 gene expression.
Furthermore, the RUNX1 high expression group included
significantly more patients with the FAB M1 subtype than the
RUNX1 low expression group (P=0.0014), suggesting that the
leukemia cells from patients with a high expression of RUNX1
are derive from relatively less mature cells. According to this
aforementioned study, RUNX1 gene expression may have
prognostic significance in AML and it may be a biomarker of an
unfavorable outcome in CN-AML, where overexpression of the
RUNX1 gene is widespread among patients (high expression of
RUNX1 is associated with poorer disease outcomes) (21).
These results differed from those obtained in the
present study, where there were no associations among mortality,
FAB classification of AML and the expression level of RUNX1.
Furthermore, the present study revealed statistically significant
differences in RUNX1 gene expression levels between females
and males; as females tended to exhibit a higher and more variable
expression level. This suggested that sex may affect RUNX1
expression, thereby influencing the process of leukemia
development.
The RUNX3 gene is involved in neurogenesis
and thymopoiesis, and serves a role as a tumor suppressor in
gastric cancer (7,31–33). A
study undertaken by Jiang et al reported that the
RUNX3 expression level is associated with breast cancer
development and that it is decreased in this type of cancer. The
principal cause for this inactivation mechanism may be
hypermethylation in the promoter region (34). A study undertaken by Cheng et
al (7) demonstrated that
RUNX3 gene expression was an independent prognostic factor
in childhood AML, and that a higher RUNX3 gene expression
level was associated with a shorter EFS and OS time (7). The results of a study undertaken by
Lacayo et al (35) also
demonstrated that a higher level of RUNX3 gene expression
was associated with a shortened EFS rate in childhood AML. However,
this study was conducted on patients belonging to an FLT3
mutant group, which may have also affected EFS (35). Based on these aforementioned studies,
it is possible that the RUNX3 gene expression level is
associated with a shorter survival time in childhood AML (7,35).
Furthermore, according to the study undertaken by Cheng et
al (7), the RUNX3 gene
expression level was not associated with age or sex. However, in a
group of patients with a lower RUNX3 gene expression level,
this was significantly associated with the presence of t(8;21) or
inv(16) translocations (7). Lower
RUNX3 gene expression levels were frequently identified in
patients with FAB M2 and M4 AML subtypes. Furthermore, RUNX3
was significantly underexpressed in the prognostically favorable
subgroup of AML with the t(8;21) and inv(16) translocations
(7).
The RUNX3 expression level differed among the
patients enrolled in the present study, and the study undertaken by
Cheng et al (7) obtained
similar levels of RUNX3 expression in patients with
childhood AML, although the results of the present study were more
varied and slightly higher. This may be due to differences in age
between the investigated groups. Cheng et al (7) identified no statistically significant
associations between clinicopathological features (sex, age or FAB
classification) and relative RUNX3 expression level. The
results obtained in the present study are comparable, as no
associations between sex or age at the time of diagnosis or FAB
classification and RUNX3 expression were identified.
Statistically significant differences were identified between the
expression level and the incidence of mortality among patients, as
mortality occurred more frequently in the group with a higher
RUNX3 expression level. These observations are also similar
to those reported by Cheng et al (7) which leads to the conclusion that
RUNX3 may serve as a potential prognostic factor in AML.
The lack of an association between the selected
clinicopathological features and relative RUNX1 and
RUNX3 expression may be a limitation of the present study,
particularly due to the relatively small group of investigated
patients. Future studies would benefit from an increased number of
patients and the collection of more detailed clinical information,
including the results of peripheral blood morphology, previously
applied treatment, percent of blasts in bone marrow.
The results of the present study suggested that sex
may be associated with the expression level of the RUNX1
gene and may influence the difference in the process of AML
development between females and males. Based on the results of
earlier studies (7,35) and those of the present study,
RUNX3 may serve as a potential novel prognostic factor.
Patients with a higher RUNX3 expression level generally have
poorer outcomes. However, the obtained results must be confirmed in
a larger cohort.
Acknowledgements
Not applicable.
Funding
The present study was supported by statutory funds
of the Department of Pharmaceutical Biochemistry and Molecular
Diagnostics, Medical University of Lodz (grant no.
503/3-015-02/503-31-001) and funds of the Faculty of Pharmacy,
Medical University of Lodz (grant nos. 502-03/3-015-02/502-34-089
and 502-03/3-015-02/502-34-088).
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Author's contributions
AK and DS planned and conducted experiments, and
assisted in the preparation of the manuscript for publication. MŻ
and AJ conducted experiments. EB planned and supervised
experiments, and assisted in the preparation of the manuscript for
publication.
Ethics approval and consent to
participate
The Ethics Committee of the Medical University of
Lodz approved the present study (RNN/88/16/KE). Written informed
consent was obtained from the patients for participation in the
study.
Consent for publication
Written informed consent was obtained from the
patients for the publication of their data.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
AML
|
acute myeloid leukemia
|
|
RUNX1
|
runt-related transcription factor
1
|
|
RUNX3
|
runt-related transcription factor
3
|
|
qPCR
|
quantitative polymerase chain
reaction
|
|
dNTP
|
deoxynucleotides
|
|
CN-AML
|
cytogenetically normal acute myeloid
leukemia
|
|
OS
|
overall survival
|
|
EFS
|
event-free survival
|
References
|
1
|
Saultz JN and Garzon R: Acute myeloid
leukemia: A concise review. J Clin Med. 5:pii: E33. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fey MF and Buske C: ESMO Guidelines
Working Group: Acute myeloblastic leukaemias in adult patients:
ESMO Clinical Practice Guidelines for diagnosis, treatment and
follow-up. Ann Oncol. 24 Suppl 6:vi138–vi143. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Thein MS, Ershler WB, Jemal A, Yates JW
and Baer MR: Outcome of older patients with acute myeloid leukemia:
An analysis of SEER data over three decades. Cancer. 119:2720–2727.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Döhner H, Weisdorf DJ and Bloomfield CD:
Acute myeloid leukemia. N Engl J Med. 375:1136–1152. 2015.
View Article : Google Scholar
|
|
5
|
Döhner H, Estey E, Grimwade D, Amadori S,
Appelbaum FR, Büchner T, Dombret H, Ebert BL, Fenaux P, Larson RA,
et al: Diagnosis and management of AML in adults: 2017 ELN
recommendations from an international expert panel. Blood.
129:424–447. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ito Y: Oncogenic potential of the RUNX
gene family: ‘Overview’. Oncogene. 23:4198–4208. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cheng CK, Li L, Cheng SH, Lau KM, Chan NP,
Wong RS, Shing MM, Li CK and Ng MH: Transcriptional repression of
the RUNX3/AML2 gene by the t(8;21) and inv(16) fusion proteins in
acute myeloid leukemia. Blood. 112:3391–3402. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ito Y, Bae SC and Chuang LS: The RUNX
family: Developtal regulators in cancer. Nature Reviews Cancer.
15:81–95. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Medinger M, Lengerke C and Passweg J:
Novel prognostic and therapeutic mutations in acute myeloid
leukemia. Cancer Genomics Proteomics. 13:317–329. 2016.PubMed/NCBI
|
|
10
|
Tracey WD and Speck NA: Potential roles
for RUNX1 and its orthologs in determining hematopoietic cell fate.
Semin Cell Dev Biol. 11:337–342. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
GeneCards® Human Gene Database:
RUNX3 gene. http://www.genecards.org/cgi-bin/carddisp.pl?gene=RUNX3April
24–2017
|
|
12
|
Asou N: The role of a Runt domain
transcription factor AML1/RUNX1 in leukemogenesis and its clinical
implications. Crit Rev Oncol Hematol. 45:129–150. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bangsow C, Rubins N, Glusman G, Bernstein
Y, Negreanu V, Goldenberg D, Lotem J, Ben-Asher E, Lancet D,
Levanon D and Groner Y: The RUNX3 gene-sequence, structure and
regulated expression. Gene. 279:221–232. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Otto F, Stock M, Fliegauf M, Fenaux P,
Preudhomme C and Lübbert M: Absence of somatic mutations within the
Runt domain of AML2/RUNX3 in acute myeloid leukaemia. Leukemia.
17:1677–1678. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lau QC, Raja E, Salto-Tellez M, Liu Q, Ito
K, Inoue M, Putti TC, Loh M, Ko TK, Huang C, et al: RUNX3 is
frequently inactivated by dual mechanisms of protein
mislocalization and promoter hypermethylation in breast cancer.
Cancer Res. 66:6512–6520. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bae SC and Choi JK: Tumor suppressor
activity of RUNX3. Oncogene. 23:4336–4340. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Guo C, Ding J, Yao L, Sun L, Lin T, Song
Y, Sun L and Fan D: Tumor suppressor gene Runx3 sensitizes gastric
cancer cells to chemotherapeutic drugs by downregulating Bcl-2,
MDR-1 and MRP-1. Int J Cancer. 116:155–160. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kabel A, Zamzami F, Al-Talhi M, Al-Dwila K
and Hamza R: Acute myeloid leukemia: A focus on risk factors,
clinical presentation, diagnosis and possible lines of management.
Cancer Res Treat. 5:62–67. 2017.
|
|
19
|
Silver N, Best S, Jiang J and Thein SL:
Selection of housekeeping genes for gene expression studies in
human reticulocytes using real-time PCR. BMC Mol Biol. 7:332006.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fu L, Fu H, Tian L, Xu K, Hu K, Wang J,
Wang J, Jing H, Shi J and Ke X: High expression of RUNX1 is
associated with poorer outcomes in cytogenetically normal acute
myeloid leukemia. Oncotarget. 7:15828–15839. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lam K and Zhang DE: RUNX1 and RUNX1-ETO:
Roles in hematopoiesis and leukemogenesis. Front Biosci (Landmark
Ed). 17:1120–1139. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ge T, Yin M, Yang M, Liu T and Lou G:
MicroRNA-302b suppresses human epithelial ovarian cancer cell
growth by targeting RUNX1. Cell Physiol Biochem. 34:2209–2220.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ferrari N, Mohammed ZM, Nixon C, Mason SM,
Mallon E, McMillan DC, Morris JS, Cameron ER, Edwards J and Blyth
K: Expression of RUNX1 correlates with poor patient prognosis in
triple negative breast cancer. PLoS One. 9:e1007592014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Silva FP, Morolli B, Storlazzi CT, Anelli
L, Wessels H, Bezrookove V, Kluin-Nelemans HC and Giphart-Gassler
M: Identification of RUNX1/AML1 as a classical tumor suppressor
gene. Oncogene. 22:538–547. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Osato M: Point mutations in the RUNX1/AML1
gene: Another actor in RUNX leukemia. Oncogene. 23:4284–4296. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ben-Ami O, Friedman D, Leshkowitz D,
Goldenberg D, Orlovsky K, Pencovich N, Lotem J, Tanay A and Groner
Y: Addiction of t(8;21) and inv(16) acute myeloid leukemia to
native RUNX1. Cell Rep. 4:1131–1143. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Goyama S, Schibler J, Cunningham L, Zhang
Y, Rao Y, Nishimoto N, Nakagawa M, Olsson A, Wunderlich M, Link KA,
et al: Transcription factor RUNX1 promotes survival of acute
myeloid leukemia cells. J Clin Invest. 123:3876–3888. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Goyama S, Huang G, Kurokawa M and Mulloy
JC: Posttranslational modifications of RUNX1 as potential
anticancer targets. Oncogene. 34:3483–3492. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wilkinson AC, Ballabio E, Geng H, North P,
Tapia M, Kerry J, Biswas D, Roeder RG, Allis CD, Melnick A, et al:
RUNX1 is a key target in t(4;11) leukemias that contributes to gene
activation through an AF4-MLL complex interaction. Cell Rep.
3:116–127. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Inoue K, Ozaki S, Shiga T, Ito K, Masuda
T, Okado N, Iseda T, Kawaguchi S, Ogawa M, Bae SC, et al: RUNX3
controls the axonal projection of proprioceptive dorsal root
ganglion neurons. Nat Neurosci. 5:946–954. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Taniuchi I, Osato M, Egawa T, Sunshine MJ,
Bae SC, Komori T, Ito Y and Littman DR: Differential requirements
for Runx proteins in CD4 repression and epigenetic silencing during
T lymphocyte development. Cell. 111:621–633. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li QL, Ito K, Sakakura C, Fukamachi H,
Inoue Ki, Chi XZ, Lee KY, Nomura S, Lee CW, Han SB, et al: Causal
relationship between the loss of RUNX3 expression and gastric
cancer. Cell. 109:113–124. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jiang Y, Tong D, Lou G, Zhang Y and Geng
J: Expression of RUNX3 gene, methylation status and
clinicopathological significance in breast cancer and breast cancer
cell lines. Pathobiology. 75:244–251. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lacayo NJ, Meshinchi S, Kinnunen P, Yu R,
Wang Y, Stuber CM, Douglas L, Wahab R, Becton DL, Weinstein H, et
al: Gene expression profiles at diagnosis in de novo child-hood AML
patients identify FLT3 mutations with good clinical outcomes.
Blood. 104:2646–2654. 2004. View Article : Google Scholar : PubMed/NCBI
|