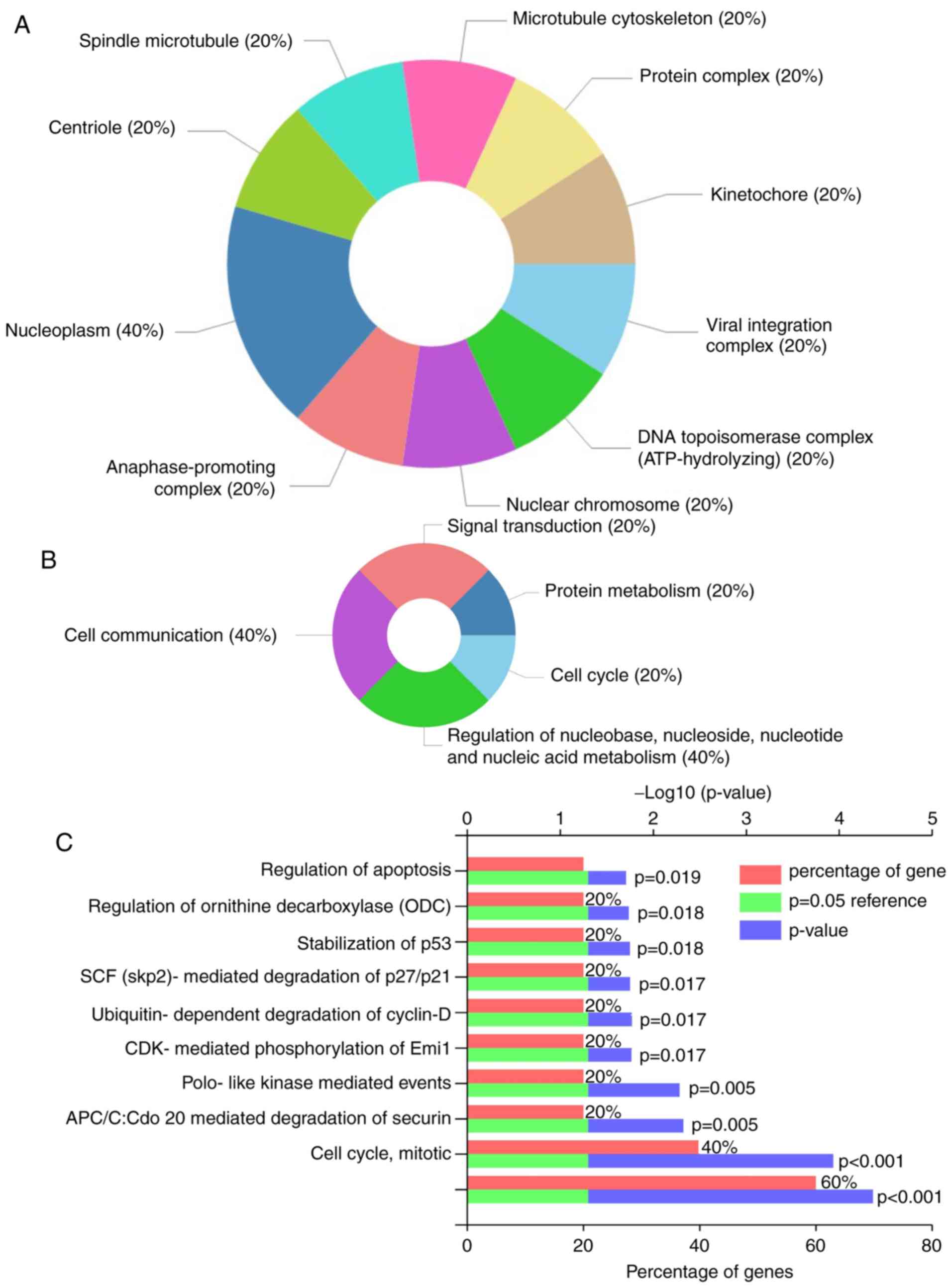
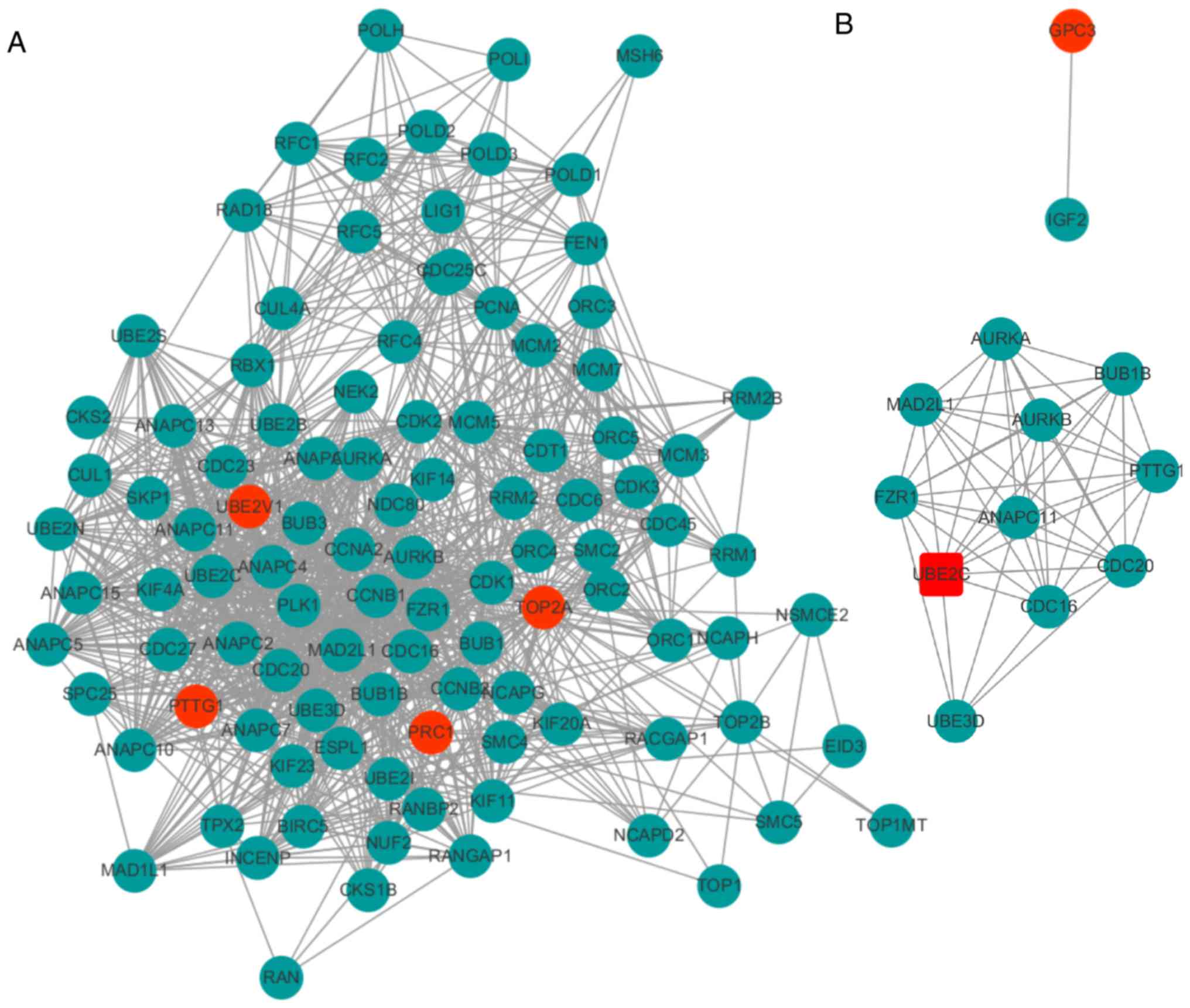
|
1
|
Neumann H, Longo D, Fauci A, Kasper D,
Hauser S, Jameson J and Loscalzo J: Harrison's Principles of
Internal Medicine. 2011.
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liang HW, Yang X, Wen DY, Gao L, Zhang XY,
Ye ZH, Luo J, Li ZY, He Y, Pang YY and Chen G: Utility of
miR-133a-3p as a diagnostic indicator for hepatocellular carcinoma:
An investigation combined with GEO, TCGA, meta-analysis and
bioinformatics. Mol Med Rep. 17:14692018.PubMed/NCBI
|
|
4
|
Yang N, Ekanem NR, Sakyi CA and Ray SD:
Hepatocellular carcinoma and microRNA: New perspectives on
therapeutics and diagnostics. Adv Drug Deliv Rev. 81:62–74. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kulasingam V and Diamandis EP: Strategies
for discovering novel cancer biomarkers through utilization of
emerging technologies. Nat Clin Pract Oncol. 5:588–599. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Vogelstein B, Papadopoulos N, Velculescu
VE, Zhou S, Diaz LA Jr and Kinzler KW: Cancer genome landscapes.
Science. 339:1546–1558. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Guo Y, Bao Y, Ma M and Yang W:
Identification of key candidate genes and pathways in colorectal
cancer by integrated bioinformatical analysis. Int J Mol Sci.
18:E7222017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liang L, Gao L, Zou XP, Huang ML, Chen G,
Li JJ and Cai XY: Diagnostic significance and potential function of
miR-338-5p in hepatocellular carcinoma: A bioinformatics study with
microarray and RNA sequencing data. Mol Med Rep. 17:2297–2312.
2018.PubMed/NCBI
|
|
9
|
Zeng T, Wang D, Chen J, Chen K, Yu G, Chen
Q, Liu Y, Yan S, Zhu L, Zhou H, et al: AF119895 regulates NXF3
expression to promote migration and invasion of hepatocellular
carcinoma through an interaction with miR-6508-3p. Exp Cell Res.
363:129–139. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gan TQ, Xie ZC, Tang RX, Zhang TT, Li DY,
Li ZY and Chen G: Clinical value of miR-145-5p in NSCLC and
potential molecular mechanism exploration: A retrospective study
based on GEO, qRT-PCR, and TCGA data. Tumour Biol.
39:10104283176916832017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang L, Huang L, Liang H, Zhang R, Chen
G, Pang Y and Feng Z: Clinical value and potential targets of
miR-224-5p in hepatocellular carcinoma validated by a TCGA- and
GEO- based study. Int J Clin Exp Pathol. 10:9970–9989. 2017.
|
|
12
|
Grinchuk OV, Yenamandra SP, Iyer R, Singh
M, Lee HK, Lim KH, Chow PK and Kuznetsov VA: Tumor-adjacent tissue
co-expression profile analysis reveals pro-oncogenic ribosomal gene
signature for prognosis of resectable hepatocellular carcinoma. Mol
Oncol. 12:89–113. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang H, Huo X, Yang XR, He J, Cheng L,
Wang N, Deng X, Jin H, Wang N, Wang C, et al: STAT3-mediated
upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer
metastasis by regulating SOX4. Mol Cancer. 16:1362017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen X, Cheung ST, So S, Fan ST, Barry C,
Higgins J, Lai KM, Ji J, Dudoit S, Ng IO, et al: Gene expression
patterns in human liver cancers. Mol Biol Cell. 13:1929–1939. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Roessler S, Long EL, Budhu A, Chen Y, Zhao
X, Ji J, Walker R, Jia HL, Ye QH, Qin LX, et al: Integrative
genomic identification of genes on 8p associated with
hepatocellular carcinoma progression and patient survival.
Gastroenterology. 142:957–966. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Roessler S, Jia HL, Budhu A, Forgues M, Ye
QH, Lee JS, Thorgeirsson SS, Sun Z, Tang ZY, Qin LX and Wang XW: A
unique metastasis gene signature enables prediction of tumor
relapse in early-stage hepatocellular carcinoma patients. Cancer
Res. 70:10202–10212. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dhanasekaran R, Limaye A and Cabrera R:
Hepatocellular carcinoma: Current trends in worldwide epidemiology,
risk factors, diagnosis, and therapeutics. Hepat Med. 4:19–37.
2012.PubMed/NCBI
|
|
18
|
Guo X, Wang Z, Zhang J, Xu Q, Hou G, Yang
Y, Dong C, Liu G, Liang C, Liu L, et al: Upregulated KPNA2 promotes
hepatocellular carcinoma progression and indicates prognostic
significance across human cancer types. Acta Biochim Biophys Sin
(Shanghai). 51:285–292. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tong H, Liu X, Li T, Qiu W, Peng C, Shen B
and Zhu Z: INTS8 accelerates the epithelial-to-mesenchymal
transition in hepatocellular carcinoma by upregulating the TGF-beta
signaling pathway. Cancer Manag Res. 11:1869–1879. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Aziz K, Limzerwala JF, Sturmlechner I,
Hurley E, Zhang C, Jeganathan KB, Nelson G, Bronk S, Velasco RF,
van Deursen EJ, et al: Ccne1 overexpression causes chromosome
instability in liver cells and liver tumor development in Mice.
Gastroenterology. 2019. View Article : Google Scholar
|
|
21
|
Bose MV, Gopisetty G, Selvaluxmy G and
Rajkumar T: Dominant negative Ubiquitin-conjugating enzyme E2C
sensitizes cervical cancer cells to radiation. Int J Radiat Biol.
88:629–634. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu YC: Ubiquitin ligases and the immune
response. Annu Rev Immunol. 22:81–127. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Williamson A, Wickliffe KE, Mellone BG,
Song L, Karpen GH and Rape M: Identification of a Physiological E2
module for the human anaphase-promoting complex. Proc Natl Acad
USA. 106:18213–11821. 2009. View Article : Google Scholar
|
|
24
|
Reddy SK, Rape M, Margansky WA and
Kirschner MW: Ubiquitination by the anaphase-promoting complex
drives spindle checkpoint inactivation. Nature. 446:921–926. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
van Ree JH, Jeganathan KB, Malureanu L and
van Deursen JM: Overexpression of the E2 ubiquitin-conjugating
enzyme UbcH10 causes chromosome missegregation and tumor formation.
J Cell Biol. 188:83. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Morikawa T, Kawai T, Abe H, Kume H, Homma
Y and Fukayama M: UBE2C is a marker of unfavorable prognosis in
bladder cancer after radical cystectomy. Int J Clin Exp Pathol.
6:1367–1374. 2013.PubMed/NCBI
|
|
27
|
Rawat A, Gopal G, Selvaluxmy G and
Rajkumar T: Inhibition of ubiquitin conjugating enzyme UBE2C
reduces proliferation and sensitizes breast cancer cells to
radiation, doxorubicin, tamoxifen and letrozole. Cell Oncol
(Dordr). 36:459–467. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hao Z, Zhang H and Cowell J:
Ubiquitin-conjugating enzyme UBE2C: Molecular biology, role in
tumorigenesis, and potential as a biomarker. Tumor Biol.
33:723–730. 2012. View Article : Google Scholar
|
|
29
|
Zhang Y, Han T, Wei G and Wang Y:
Inhibition of microRNA-17/20a suppresses cell proliferation in
gastric cancer by modulating UBE2C expression. Oncol Rep.
33:2529–2536. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chang L, Zhang Z, Yang J, McLaughlin SH
and Barford D: Atomic structure of the APC/C and its mechanism of
protein ubiquitination. Nature. 522:450–454. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xie C, Powell C, Yao M, Wu J and Dong Q:
Molecules in focus: Ubiquitin-conjugating enzyme E2C: A potential
cancer biomarker. Int J Biochem Cell Biol. 47:113–118. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rowinsky EK and Donehower RC: Paclitaxel
(taxol). N Engl J Med. 332:1004–1014. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gascoigne KE and Taylor SS: Article:
Cancer cells display profound intra- and interline variation
following prolonged exposure to antimitotic drugs. Cancer Cell.
14:111–123. 2008. View Article : Google Scholar : PubMed/NCBI
|