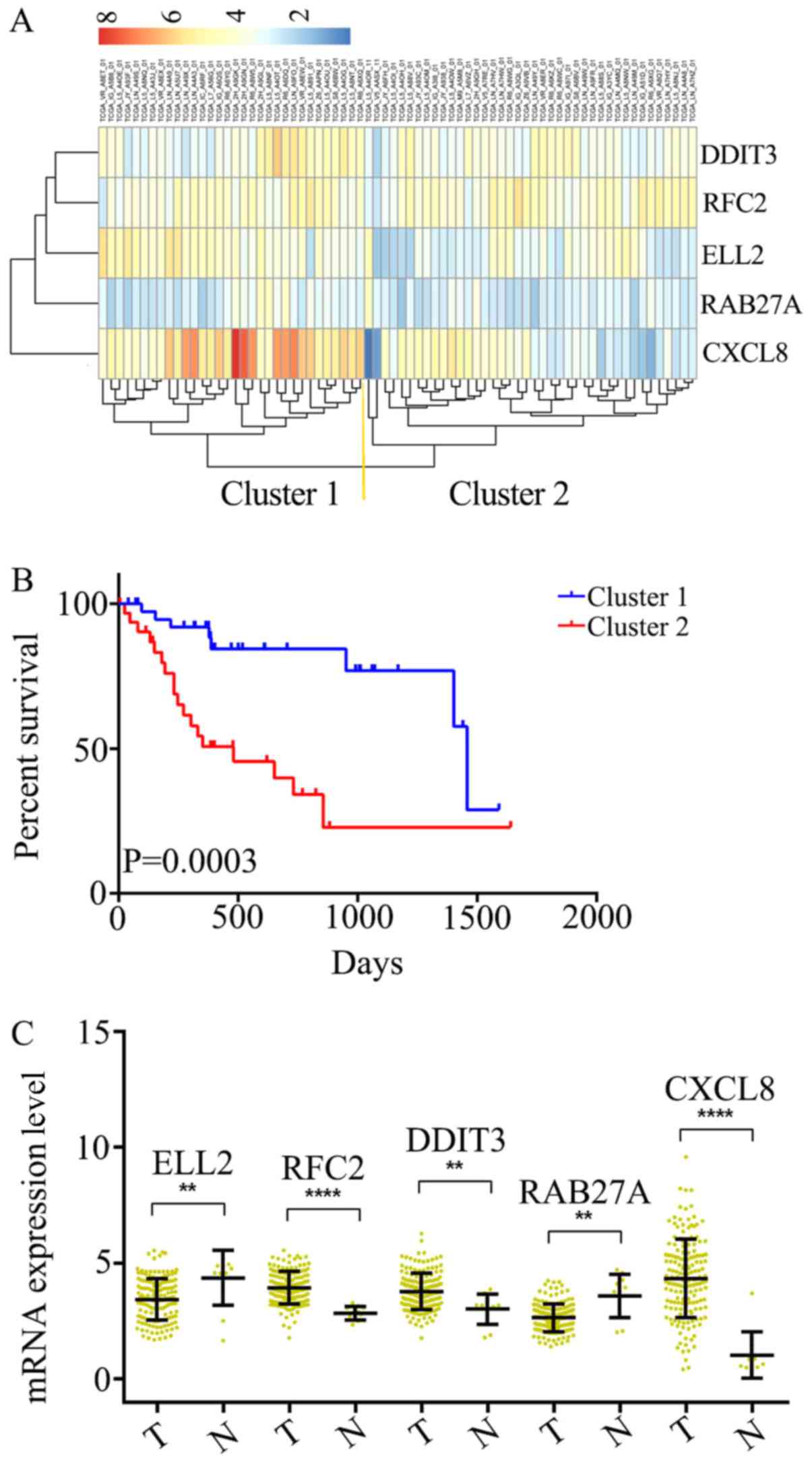
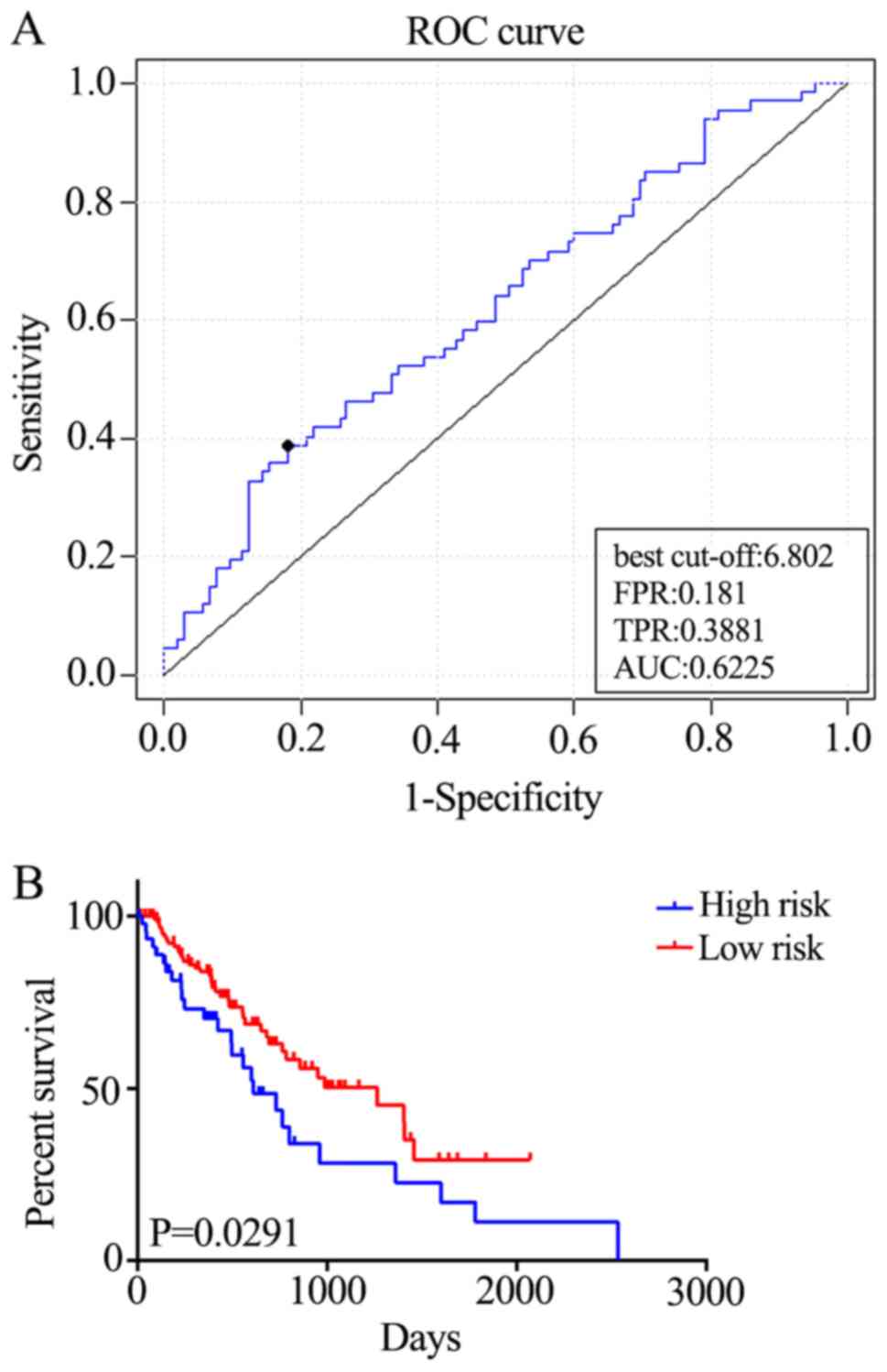
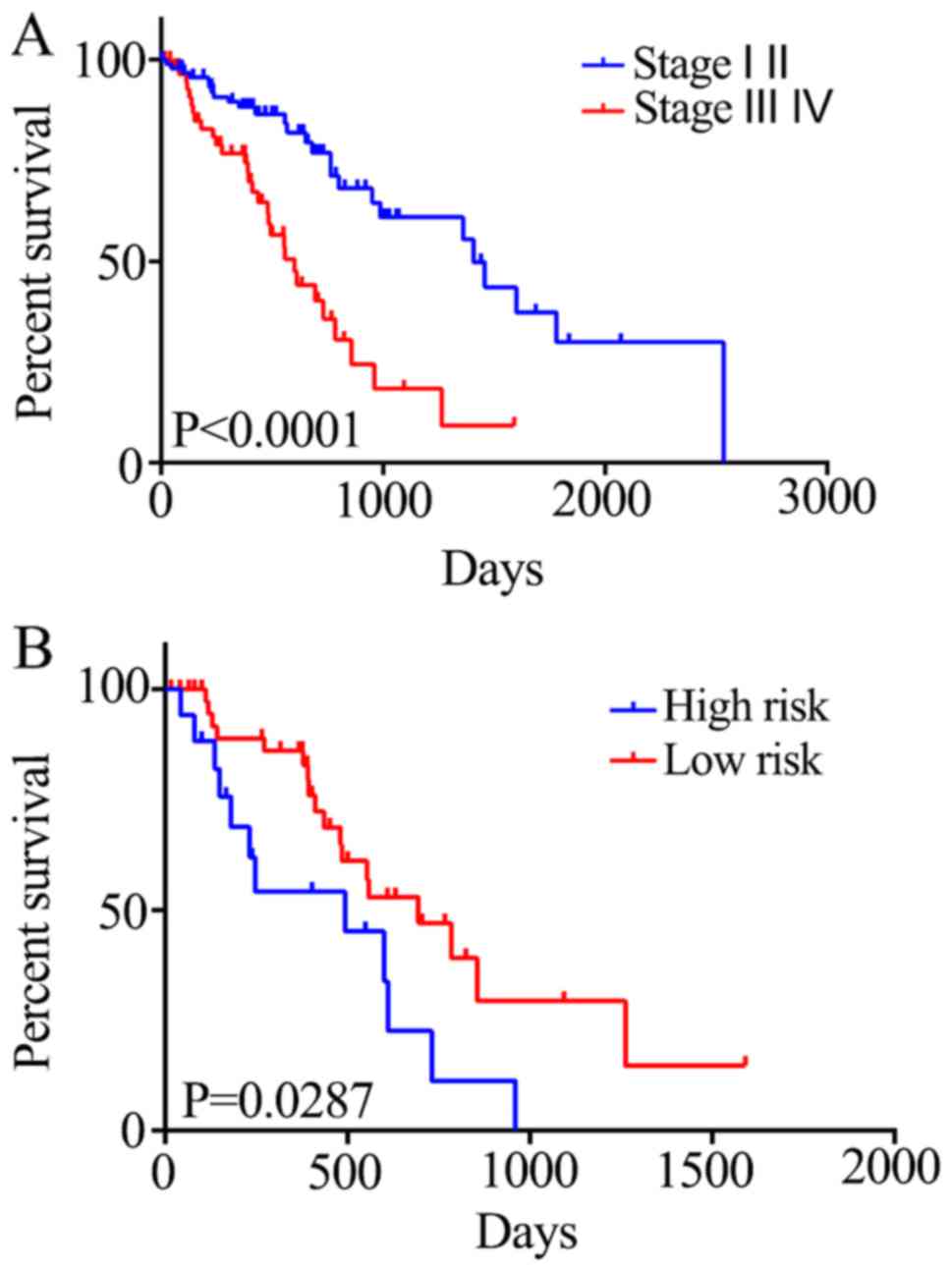
|
1
|
McGuire S: World Cancer Report 2014.
Geneva, Switzerland: World Health Organization, International
Agency for Research on Cancer, WHO Press, 2015. Adv Nutr.
7:418–419. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Enzinger PC and Mayer RJ: Esophageal
cancer. N Engl J Med. 349:2241–2252. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Luo J, Wang W, Tang Y, Zhou D, Gao Y,
Zhang Q, Zhou X, Zhu H, Xing L and Yu J: mRNA and methylation
profiling of radioresistant esophageal cancer cells: The
involvement of Sall2 in acquired aggressive phenotypes. J Cancer.
8:646–656. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang W, Pang Q, Yan C, Wang Q, Yang J, Yu
S, Liu X, Yuan Z, Wang P and Xiao Z: Induction of PD-L1 expression
by epidermal growth factor receptor-mediated signaling in
esophageal squamous cell carcinoma. Onco Targets Ther. 10:763–771.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang W, Yan S, Liu M, Zhang G, Yang S, He
S, Bai J, Quan L, Zhu H, Dong Y and Xu N: Beta-catenin/TCF pathway
plays a vital role in selenium induced-growth inhibition and
apoptosis in esophageal squamous cell carcinoma (ESCC) cells.
Cancer Lett. 296:113–122. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xiang J, Zang W, Che J, Chen K and Hang J:
Regulation network analysis in the esophageal squamous cell
carcinoma. Eur Rev Med Pharmacol Sci. 16:2051–2056. 2012.PubMed/NCBI
|
|
7
|
Ashktorab H, Kupfer SS, Brim H and
Carethers JM: Racial disparity in gastrointestinal cancer risk.
Gastroenterology. 153:910–923. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Quackenbush J: Microarray analysis and
tumor classification. N Engl J Med. 354:2463–2472. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang L, Wang C, Wang J, Huang X and Cheng
Y: A novel systemic immune-inflammation index predicts survival and
quality of life of patients after curative resection for esophageal
squamous cell carcinoma. J Cancer Res Clin Oncol. 143:2077–2086.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Salmaninejad A, Zamani MR, Pourvahedi M,
Golchehre Z, Hosseini Bereshneh A and Rezaei N: Cancer/testis
antigens: Expression, regulation, tumor invasion, and use in
immunotherapy of cancers. Immunol Invest. 45:619–640. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lin ZW, Gu J, Liu RH, Liu XM, Xu FK, Zhao
GY, Lu CL and Ge D: Genome-wide screening and co-expression network
analysis identify recurrence-specific biomarkers of esophageal
squamous cell carcinoma. Tumour Biol. 35:10959–10968. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Barrett T, Troup DB, Wilhite SE, Ledoux P,
Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M and
Edgar R: NCBI GEO: Mining tens of millions of expression
profiles-database and tools update. Nucleic Acids Res 35 (Database
Issue). D760–D765. 2007. View Article : Google Scholar
|
|
13
|
James G, Witten D, Hastie T and Tibshirani
R: An introduction to statistical learning with applications in R.
Springer-Verlag New York; 2013
|
|
14
|
Cho HJ, Ami Y, Kim S, Kang J, Seung-Mo H
and Kang: Robust likelihood-based survival modeling with microarray
data. J Statist Software. 29:1–16. 2008.
|
|
15
|
Eisen MB, Spellman PT, Brown PO and
Botstein D: Cluster analysis and display of genome-wide expression
patterns. Proc Natl Acad Sci USA. 95:14863–14868. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Therneau T: A package for survival
analysis in S. R package version 2.37–7. 2014.
|
|
17
|
Simon R, Lam A, Li MC, Ngan M, Menenzes S
and Zhao Y: Analysis of gene expression data using BRB-ArrayTools.
Cancer Inform. 3:11–17. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ogata H, Goto S, Sato K, Fujibuchi W, Bono
H and Kanehisa M: KEGG: Kyoto encyclopedia of genes and genomes.
Nucleic Acids Res. 27:29–34. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang J, Vasaikar S, Shi Z, Greer M and
Zhang B: WebGestalt 2017: A more comprehensive, powerful, flexible
and interactive gene set enrichment analysis toolkit. Nucleic Acids
Res 45 (W1). W130–W137. 2017. View Article : Google Scholar
|
|
20
|
Wu C, Irizarry R, MacDonald J and Gentry
J: gcrma: Background adjustment using sequence information.
2005.
|
|
21
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu H, Liu C, Xu M, Guo M, Xu S and Xie M:
Prognostic value of the number of negative lymph nodes in
esophageal carcinoma without lymphatic metastasis. Thorac Cancer.
9:1129–1135. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yildirim BA, Torun N, Guler OC and Onal C:
Prognostic value of metabolic tumor volume and total lesion
glycolysis in esophageal carcinoma patients treated with definitive
chemoradiotherapy. Nucl Med Commun. 39:553–563. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li JC, Zhao YH, Wang XY, Yang Y, Pan DL,
Qiu ZD, Su Y and Pan JJ: Clinical significance of the expression of
EGFR signaling pathway-related proteins in esophageal squamous cell
carcinoma. Tumour Biol. 35:651–657. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Qin YR, Qiao JJ, Chan TH, Zhu YH, Li FF,
Liu H, Fei J, Li Y, Guan XY and Chen L: Adenosine-to-inosine RNA
editing mediated by ADARs in esophageal squamous cell carcinoma.
Cancer Res. 74:840–851. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang J, Chen Z, Tang Z, Huang J, Hu X and
He J: RNA editing is induced by type I interferon in esophageal
squamous cell carcinoma. Tumour Biol. 39:10104283177085462017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sakai M, Sohda M, Miyazaki T, Suzuki S,
Sano A, Tanaka N, Inose T, Nakajima M, Kato H and Kuwano H:
Significance of karyopherin-{alpha} 2 (KPNA2) expression in
esophageal squamous cell carcinoma. Anticancer Res. 30:851–856.
2010.PubMed/NCBI
|
|
28
|
Yuan Y, Xue L and Fan F: Screening of
differentially expressed genes related to esophageal squamous cell
carcinoma and functional analysis with DNA microarrays. Int J
Oncol. 44:1163–1170. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Anastas JN and Moon RT: WNT signalling
pathways as therapeutic targets in cancer. Nat Rev Cancer.
13:11–26. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
McBride WH, Iwamoto KS, Syljuasen R,
Pervan M and Pajonk F: The role of the ubiquitin/proteasome system
in cellular responses to radiation. Oncogene. 22:5755–5773. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang BY, Lin PY, Wu SC, Chen HS, Hsu PK,
Shih CS, Liu CY, Liu CC and Chen YL: Comparison of pathologic stage
in patients receiving esophagectomy with and without preoperative
chemoradiation therapy for esophageal SCC. J Natl Compr Canc Netw.
12:1697–1705. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li L, Zhao L, Lin B, Su H, Su M, Xie D,
Jin X and Xie C: Adjuvant Therapeutic modalities following
three-field lymph node dissection for stage II/III esophageal
squamous cell carcinoma. J Cancer. 8:2051–2059. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang L, Li W, Lyu X, Song Y, Mao Y, Wang
S and Huang J: Adjuvant chemotherapy with paclitaxel and cisplatin
in lymph node-positive thoracic esophageal squamous cell carcinoma.
Chin J Cancer Res. 29:149–155. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tong Q, Wang XL, Li SB, Yang GL, Jin S,
Gao ZY and Liu XB: Combined detection of IL-6 and IL-8 is
beneficial to the diagnosis of early stage esophageal squamous cell
cancer: A preliminary study based on the screening of serum markers
using protein chips. Onco Targets Ther. 11:5777–5787. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kunizaki M, Hamasaki K, Wakata K, Tobinaga
S, Sumida Y, Hidaka S, Yasutake T, Miyazaki T, Matsumoto K,
Yamasaki T, et al: Clinical value of serum p53 antibody in the
diagnosis and prognosis of esophageal squamous cell carcinoma.
Anticancer Res. 38:1807–1813. 2018.PubMed/NCBI
|