Introduction
Renal cell carcinoma (RCC) is the seventh most
common cancer globally, and alongside an increasing incidence and
mortality rate, ~144,000 individuals succumb to the disease each
year (1,2). Renal clear cell carcinoma (KIRC) is the
most common histological subtype accounting for ~80% of all RCC
cases (3), and patients with
advanced RCC have a 5-year survival rate of <30% in the United
States between 2008 and 2014 (4).
Currently, the treatment of RCC remains a major challenge due to
the poor response to conventional radiotherapy and chemotherapy
(5,6). Therefore, novel therapeutic approaches
and diagnostic biomarkers are required to improve the prognosis of
patients with RCC.
Long non-coding RNAs (lncRNAs) are a class of
mRNA-like transcripts >200 nucleotides in length that lack
protein coding capability (7).
lncRNAs serve important roles in the regulation of gene expression
at multiple levels, including chromatin remodeling, transcriptional
regulation, post-transcriptional processing, mRNA translation and
protein stability (8). lncRNA
dysregulation has been associated with the occurrence and
progression of malignant tumors, including lung cancer, cervical
cancer and colorectal cancer, by regulating various cellular
processes in tumor cells, such as proliferation, migration,
differentiation, apoptosis and the cell cycle (9).
Metastasis-associated lung adenocarcinoma transcript
1 (MALAT1), also known as nuclear-enriched abundant transcript 2,
was one of the first lncRNA molecules discovered with a designated
role in cancer (10,11). The MALAT1 transcript is ~8,000
nucleotides in length and is highly conserved among mammals
(12,13). Accumulating evidence has shown that
MALAT1 is upregulated in various cancer types, such as lung cancer,
esophageal squamous cell carcinoma, gastric cancer, glioma and
KIRC, and is reported to promote tumor cell hyperproliferation and
metastasis (11,14–17). The
nomenclature of MALAT1 is based on its function in regulating lung
cancer metastasis; however, the molecular mechanisms underlying the
MALAT1-mediated regulation of cell migration are still largely
unclear.
A major mechanism by which MALAT1 exerts its
biological functions is regulating RNA processing (18). Tripathi et al (18) reported that MALAT1 may regulate RNA
splicing by interacting with several splicing factors, such as
serine/arginine-rich splicing factor 1 and serine/arginine-rich
splicing factor 3. A previous study demonstrated that MALAT1 binds
multiple subunits of the RNA spliceosome, such as
serine/arginine-rich splicing factor 7, ATP-dependent RNA helicase
A and splicing factor U2AF2 (19).
RNA sequencing conducted by Engreitz et al (20) showed that MALAT1 could indirectly
interact with a number of pre-mRNAs including pre-cofilin-1
(pre-CFL1) mediated by proteins. CFL1 is an actin binding protein
that regulates F-actin severing and depolymerization, a critical
step for cytoskeleton dynamics during cellular migration (21,22). A
previous study have shown that CFL1 expression is highly associated
with cell locomotion and invasion (23). However, to the best of our knowledge,
the relationship between MALAT1 and CFL1 has not been investigated,
and it is unknown whether CFL1 is an important downstream factor
for the regulation of MALAT1-induced cellular migration.
In the present study, we studied the role of MALAT1
in the in the regulation of cell migration in RCC cells and
investigated the underlying molecular mechanism.
Materials and methods
Cell culture and transfection
Human renal cancer cell lines ACHN and 786-O were
obtained from the American Type Culture Collection. The cells were
cultured in Minimum Essential Medium or Dulbecco's modified Eagle's
medium (both Corning, Inc.) supplemented with 10% fetal bovine
serum (FBS) (Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin/streptomycin (100 µg/ml) at 37°C (5% CO2) in
a humidified culture incubator. For small interfering (si)RNA
knockdown experiments, MALAT1 siRNA (si-MALAT1-1 and si-MALAT1-2)
and scrambled negative control siRNA (Scr) were purchased from
Guangzhou RiboBio, Co., Ltd, and the siRNA sequences are displayed
in Table SI. Cells cultured in
6-well plates at 40% confluence (4×105 cells. per plate)
were transfected using X-tremeGENE siRNA Transfection Reagent
(Roche Diagnostics, Inc.) and were harvested after 48 h of
incubation at 37°C. For rescue experiments, the entire cloned CFL1
sequence (accession number: NM_005507.3) was inserted into the
GV358 vector (GeneChem, Inc.). MALAT1 knockdown cells were
incubated at 37°C with 2 µg CFL1 expression plasmid GV358-CFL1 and
X-tremeGENE HP DNA Transfection Reagent (Roche Diagnostics, Inc.)
and harvested after 36 h.
Reverse transcription-quantitative
(RT-q)PCR
Total RNA was isolated from transfected ACHN and
786-O cells using TRIzol® reagent (Invitrogen, Thermo
Fisher Scientific, Inc.). For the detection of mRNA, a Fast Quant
RT kit (TianGen Biochemical Technology, Co., Ltd.) was used to
reverse transcribe 5 µg total RNA into cDNA according to the
manufacturer's protocols. The reaction conditions for reverse
transcription were: 42°C for 6 min, 42°C for 1 h and 95°C for 10
min. qPCR reactions was performed with the SuperReal SYBR Green
PreMix (TianGen Biotech, Co., Ltd.) using a 7500 Fast Real-Time PCR
system (Applied Biosystems; Thermo Fisher Scientific, Inc.) and the
following reaction conditions: 30 sec at 94°C, followed by 40
cycles of 5 sec at 94°C and 1 min at 60°C. Relative mRNA levels of
the target genes were normalized using the reference gene GAPDH and
assessed using the 2−∆∆Cq method (24). The primer sequences used for RT-qPCR
are displayed in Table SII.
Western blotting
Transiently transfected cells were lysed using 2%
sodium dodecyl sulfate (Beijing Solarbio Science and Technology,
Co., Ltd.) supplemented with cOmplete™ protease inhibitor cocktail
(Roche Diagnostics, Inc.) for 10 min on ice. Total protein was
quantified using a BCA quantification kit (Invitrogen; Thermo
Fisher Scientific, Inc.). A total of 30 µg protein per lane was
loaded onto a 10% gel, resolved using SDS-PAGE and then transferred
to activated polyvinylidene difluoride membranes (PVDF) (0.2 µm
pore size, EMD Millipore). After blocking with 5% skim milk for 1 h
at room temperature (RT), the PVDF membranes were incubated with
rabbit anti-human primary antibodies against GAPDH (1:1,000
dilution, cat. no. ab181602, Abcam), β-actin (1:1,000 dilution,
cat. no. ab8227, Abcam) or CFL1 (1:1,000 dilution, cat. no.
ab42824, Abcam) at 4°C overnight. After washing with TBST, the
blots were incubated with goat anti-rabbit IgG heavy chain (H) +
light chain (L) HRP secondary antibody (1:1,000 dilution; cat. no.
S0001; Affinity Biosciences) for 1 h at RT. Finally, the blots were
developed using enhanced chemiluminescence luminol reagents
(Pierce; Thermo Fisher Scientific, Inc.) and the protein bands were
analyzed using ImageJ software version 1.8.0 (National Institutes
of Health).
Clinical specimens
To investigate the expression of MALAT1 and CFL1
using RT-qPCR in human RCC tissues, 20 malignant renal cancer
tissue specimens were collected from 15 male and 5 female patients
(median age, 65 years; age range, 45–75 years). The tissues were
surgically removed at the Second Hospital of Tianjin Medical
University with written consent from the patients, and with
approval from the Tianjin Medical University Second Hospital
Medical Ethics Committee (Tianjin, China). All patients received
radical nephrectomy with no preoperative or postoperative adjuvant
therapy. No additional inclusion or exclusion criteria were used.
All specimens were evaluated by two experienced pathologists
independently to ensure that they were correctly identified as
KIRC. Diagnostic criteria for KIRC include grossly circumscribed
mass, sheets and nests of cells surrounded by extensive capillary
network, predominantly composed of clear cells, nuclei ranging from
round and regular at low grade to pleomorphic at high grade,
dysplasia of adjacent non-carcinomatous tubules in some cases, and
multiple and/or familial clear cell carcinomas may be seen in von
Hippel Lindau syndrome (25). The
collected tumor tissues were frozen in liquid nitrogen immediately
and stored at −80°C. Clinicopathological information of the
patients is presented in Table
SIII.
Confocal imaging
Renal cancer cells were plated at 3×104
per well into 12-well plates containing sterile glass coverslips.
After incubation for 24 h at 37°C, the cells were fixed with 4%
paraformaldehyde at RT for 10 min and permeabilized with 0.2%
Triton X-100 (Beijing Solarbio Science and Technology, Co., Ltd.)
for 10 min. After being washed three times with phosphate buffered
saline (PBS), the cells were blocked with 3% bovine serum albumin
(Beijing Solarbio Science and Technology, Co., Ltd.) for 1 h at RT.
Next, the cells were incubated with rhodamine conjugated phalloidin
(Invitrogen; Thermo Fisher Scientific, Inc.) for 30 min at RT.
After being washed three times with PBS, cell nuclei were stained
with 4′,6-diamidino-2-phenylindole for 10 min at RT. The stained
cells were visualized using confocal laser scanning microscopy at
×400 magnification (Olympus Corporation).
Actin polymerization assay
Renal cancer cells were seeded at 3×105
cells per well in 6-well plates. After incubation for 24 h at 37°C,
the cells were fixed with 4% paraformaldehyde at RT for 10 min,
permeabilized with 0.1% Triton X-100 for 20 min at RT, and then
washed three times with F-actin buffer (10 mM HEPES, 20 mM
KH2PO4, 5 mM EGTA and 2 mM MgCl2).
Next, the cells were incubated with rhodamine conjugated phalloidin
(dilution, 1:40; cat. no. R415; Thermo Fisher Scientific, Inc.) at
RT for 60 min. Phalloidin was extracted using 100% methanol, and
the fluorescence intensity was quantified using a microplate reader
(BioTek Instruments, Inc.).
Wound healing assay
Following 48 h transfection, ACHN and 786-O cells
were cultured in 6-well plates until confluent. The cells were
scratched in the middle of the plate using a 10 µl sterile pipette
tip and washed three times with PBS to remove detached cells and
debris. The cells were cultured in medium containing 1% FBS for an
additional 24 h. Images were captured at 0 and 24 h, and the
migration distance was measured at 200× magnification using a light
microscope (Olympus Corporation).
Invasion assay
An invasion assay was performed using 24-well
Transwell chambers (Corning, Inc.) containing polycarbonate
membranes (pore size, 8 µm) precoated with Matrigel at 37°C for 30
min (BD Biosciences). After starvation overnight, 1×105
transfected cells in 200 µl serum-free medium were loaded into the
upper chambers, and 600 µl medium containing 10% FBS was added to
the lower chambers as a chemoattractant. After incubation for 24 h
at 37°C, non-invasive cells on the upper membrane surface were
removed with wet cotton swabs, and the cells that had invaded to
the lower membrane surface were fixed with 4% paraformaldehyde at
RT (Beijing Solarbio Science and Technology, Co., Ltd.) and stained
with 0.01% crystal violet solution (Sigma-Aldrich; Merck KGaA) at
RT for 10 min. The migratory cells were counted manually in six
randomly selected regions at 200× magnification using a light
microscope (Olympus Corporation).
Bioinformatics analysis
Gene expression and clinical data were obtained from
The Cancer Genome Atlas (TCGA) database (https://www.cancer.gov/tcga) via the TCGAbioLinks
version 2.12.2 (26) Bioconductor R
package in R (27) by RStudio
(version 3.6.0) (28). The following
tumor types were selected: Bladder urothelial carcinoma (BLCA),
breast invasive carcinoma (BRCA), colon adenocarcinoma (COAD),
esophageal carcinoma, glioblastoma multiforme, head and neck
squamous cell carcinoma (HNSC), KIRC, renal papillary cell
carcinoma (KIRP), liver hepatocellular carcinoma (LIHC), lung
adenocarcinoma, lung squamous cell carcinoma (LUSC), pancreatic
adenocarcinoma, pheochromocytoma and paraganglioma, prostate
adenocarcinoma (PRAD), rectum adenocarcinoma (READ), sarcoma, skin
cutaneous melanoma, stomach adenocarcinoma (STAD), thyroid
carcinoma, thymoma (THYM) and uterine corpus endometrial carcinoma.
The raw read counts per gene of the gene expression data were
normalized to Transcripts Per Million (29). Differential expression analyses of
MALAT1 and CFL1 between tumor and normal samples were conducted
using the ggplot2 version 3.2.0 (http://ggplot2.tidyverse.org) package integrated with
ggpubr version 0.2.1 (https://rpkgs.datanovia.com/ggpubr/) package in R.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes
(KEGG) pathway enrichment analysis were conducted by
clusterProfiler R/bioconductor package (v3.12.0) (30) in R.
Statistical analysis
Prism version 7.0 (GraphPad Software, Inc.) was used
to plot mean and standard deviation data. Statistical analysis was
performed by two-tailed unpaired Student's t-test between two
groups or one-way analysis of variance followed by Tukey's post hoc
test between multiple groups. Wilcoxon rank-sum test was used for
statistical analysis of TCGA data, when patients were stratified
into high expression group and low expression group, based on the
33 and 66 percentiles of the two RNAs (low, 1–33 percentile; high,
66–100 percentile). Overall survival (OS) analysis was performed
using the survival R package version 2.43.3 (https://github.com/therneau/survival). Survival
analysis was performed using the Kaplan-Meier method and compared
using the log-rank test. The correlation between MALAT1 and
CFL1 mRNA expression was evaluated using Pearson's
coefficient. P<0.05 was considered to indicate a statistically
significant difference.
Results
Pan-cancer analysis of MALAT1
expression
To acquire the expression pattern of MALAT1 across
different cancer types, RNA sequencing datasets from 21 of the most
common types of cancer were obtained from TCGA database. The
results indicated that MALAT1 is significantly upregulated in eight
different types of cancer compared with normal tissues, including
COAD (3-fold change, P<0.001), HNSC (1.6-fold change,
P<0.001), KIRC (2.1-fold change, P<0.001), KIRP (1.2-fold
change, P=0.041); LIHC (2.9-fold change, P<0.001), PRAD
(1.5-fold change, P<0.001), READ (2.4-fold change, P=0.01) and
STAD (fold-change: 1.7; P=0.003). KIRC was one of the cancer types
with the most significant upregulation of MALAT1. Surprisingly,
MALAT1 was also decreased in three types of cancer: BLCA (0.7-fold
change, P=0.032), BRCA (0.9-fold change; P=0.004) and LUSC
(0.8-fold change, P=0.036) (all Fig.
1). The expression of MALAT1 was not significantly changed in
the other ten types of cancer. These results suggest that the
expression of MALAT1 is cancer type-dependent and may play distinct
biological roles in different types of cancer.
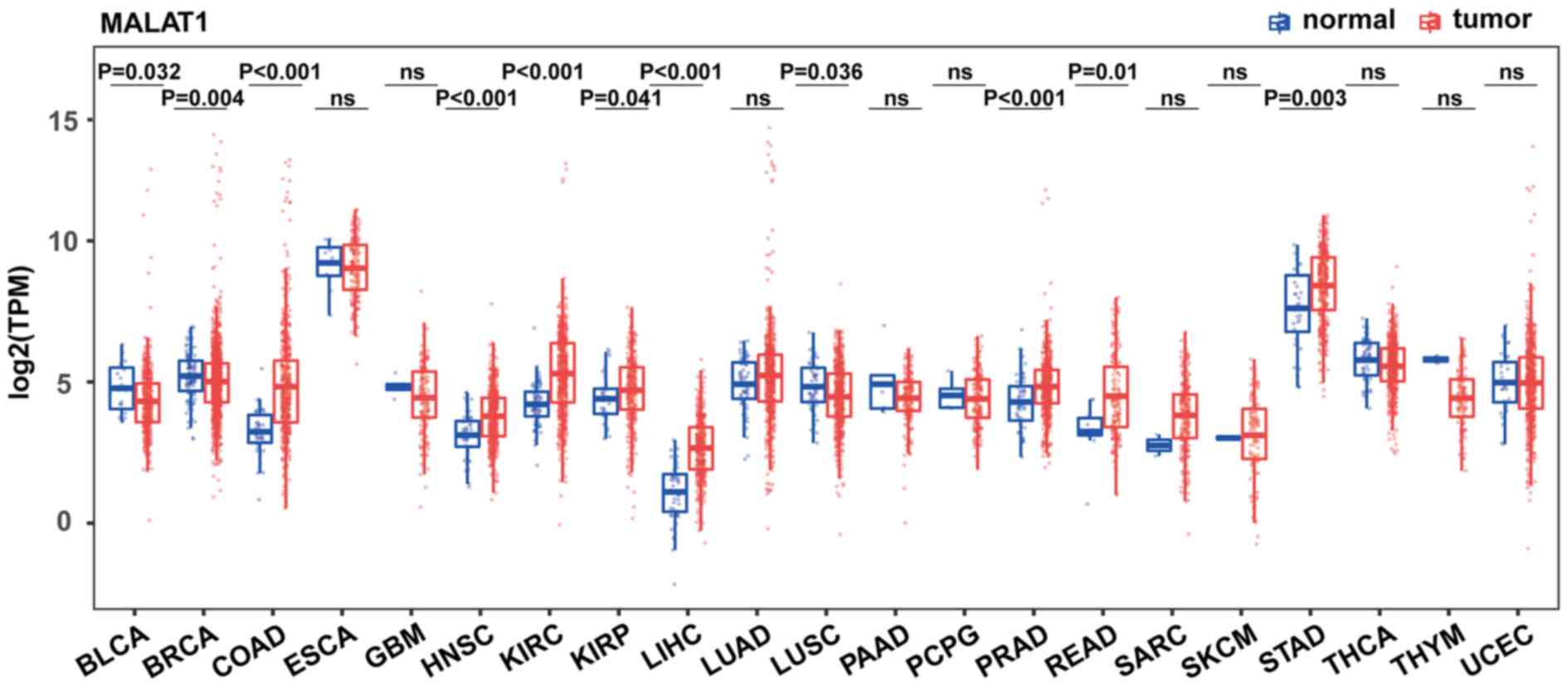 | Figure 1.Comparison of MALAT1 expression in
tumors and corresponding normal tissues from TCGA. Expression of
MALAT1 was measured using log2(TPM). Wilcoxon rank-sum
test was used for statistical analysis and the data were not
paired. MALAT1, metastasis-associated lung adenocarcinoma
transcript 1; TPM, Transcripts Per Million; TCGA, The Cancer Genome
Atlas; BLCA, bladder urothelial carcinoma; BRCA, breast invasive
carcinoma; COAD, colon adenocarcinoma; ESCA, esophageal carcinoma;
GBM, glioblastoma multiforme; HNSC, head and neck squamous cell
carcinoma; KIRC, clear cell kidney carcinoma; KIRP, kidney renal
papillary cell carcinoma; LIHC, liver hepatocellular carcinoma;
LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma;
PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and
paraganglioma; PRAD, prostate adenocarcinoma; READ, rectum
adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD,
stomach adenocarcinoma; THCA, thyroid carcinoma; THYM, thymoma;
UCEC, uterine corpus endometrial carcinoma; ns, not
significant. |
Functional analysis of MALAT1 binding
pre-mRNAs
MALAT1 is a major component of nucleus speckles
(18) and indirectly binds with a
number of pre-mRNAs through protein intermediates, thereby
regulating pre-mRNA expression through RNA splicing (20). To better understand the biological
function of MALAT1, GO and KEGG pathway enrichment analysis was
performed on the pre-mRNAs that were identified to bind with
MALAT1. Functional enrichment analysis revealed that the top-ranked
biological processes included ‘mRNA processing’ and ‘RNA splicing’
(Fig. S1A). KEGG pathway enrichment
analysis showed that pre-mRNAs that bound to MALAT1 were
significantly enriched in ‘spliceosome’ pathways with the highest
protein count as 20 (Fig. S1B).
This result further supports the role of MALAT1 in RNA splicing.
Pre-CFL1 mRNA also had a marked enrichment among the 488
observed pre-mRNAs (fold-19.79 enrichment) (20). CFL1 is a critical modulator of
cytoskeleton rearrangement during cell migration (21,22), and
the present findings suggest that MALAT1 may be directly involved
in cell migration by regulating pre-CFL1 splicing.
MALAT1 regulates the expression of
CFL1
Next, the association between MALAT1 and CFL1 was
examined. MALAT1 siRNA knockdown was investigated in ACHN and 786-O
cells, and MALAT1 downregulation of was subsequently confirmed
using RT-qPCR (Fig. 2A and B).
MALAT1 knockdown inhibited CFL1 expression at both the mRNA and
protein levels compared with respective control cells in both ACHN
(Fig. 2A) and 786-O cells. (Fig. 2B). si-MALAT1-2 had a higher knockdown
efficiency compared with si-MALAT1-1 and was used for the following
experiments. In addition, levels of pre-CFL1 mRNA were
examined using primers that targeted the junction of the first exon
and intron (Fig. 2C). Knockdown of
MALAT1 decreased the levels of CFL1 mRNA in both ACHN
(P<0.001, Fig. 2D) and 786-O
cells (P=0.008; Fig. 2E) but not
pre-CFL1. Taken together, these results suggest that
MALAT-knockdown inhibits CFL1 expression at both the RNA and
protein level by post-transcriptional regulation.
MALAT1-knockdown elevates F-actin
accumulation
To establish how MALAT1 modulates the cytoskeleton
through CFL1, the overexpression efficiency of CFL1 plasmid
in ACHN was first evaluated (at mRNA level, P<0.001; at protein
level, P=0.003; Fig. S2A) and 786-O
cells (at mRNA level, P=0.004; at protein level, P<0.001;
Fig. S2B). Then, rescue experiments
were performed by first knocking down MALAT1 with siRNA and then
overexpressing CFL1. The efficiency of transfection was evaluated
using RT-qPCR and western blotting in both ACHN (Fig. 3A) and 786-O cells (Fig. 3B). Western blotting showed that
MALAT1 knockdown did not affect the total amount of β-actin
(Fig. 3C). A fluorescence staining
assay was performed to measure the levels of F-actin in the cells
and observed marked accumulation of F-actin near the cell membrane
in MALAT1 knockdown cells as compared with the corresponding
control cells (Fig. 3D). To verify
that the accumulation of F-actin was mediated by CFL1, a rescue
experiment was performed in MALAT1-knockdown cells with the CFL1
vector. Restoring the expression of CFL1 in MALAT1-knockdown cells
decreased F-actin accumulation to a similar level as the control
cells. In addition, the levels of F-actin in the cells were
quantified and showed a consistent result with the fluorescence
staining assay (Fig. 3A-D).
Knockdown of MALAT1 significantly increased the level of F-actin,
which was reduced to control levels in rescued cells (Fig. 3E). Taken together, these results
demonstrate that MALAT1-knockdown inhibited F-actin
depolymerization and induced F-actin accumulation near the cell
membrane thorough suppressing CFL1 expression.
 | Figure 3.Reverse transcription-quantitative
PCR and western blotting analyses of CFL1 in MALAT1-downregulated
(A) ACHN and (B) 786-O cells transfected with CFL1 plasmids and the
corresponding control cells. The data are presented as the mean ±
SD (n=3) and one-way ANOVA followed by Tukey's post hoc test was
used for statistical analysis. (C) Western blots of CFL1 and
β-actin in MALAT1-downregulated ACHN (upper panel) and 786-O (lower
panel) cells and their corresponding control cells. Data are shown
as mean ± SD (n=3) and unpaired Student's t-test was used for
statistical analysis. (D) Confocal imaging of cellular F-actin in
cells transfected with MALAT1 siRNA or MALAT1 siRNA and CFL1
vector. F-actin was stained with rhodamine conjugated phalloidin.
Upper panel, ACHN; lower panel, 786-O. Scale bar, 20 µm. (E)
Quantification of F-actin levels using an actin polymerization
assay in cells transfected with MALAT1 siRNA or MALAT1 siRNA and
CFL1 vector. F-actin was stained with rhodamine conjugated
phalloidin. Data are expressed as mean ± SD (n=3) and one-way ANOVA
followed by Tukey's post hoc test was used for statistical
analysis. MALAT1, metastasis-associated lung adenocarcinoma
transcript 1; ns, not significant; CFL1, cofilin-1; SD, standard
deviation; Scr, scramble; si, small interfering; ANOVA, analysis of
variance. |
MALAT1 regulates the migration and
invasiveness of RCC cells through CFL1
To determine whether MALAT1 is required for the
migration and invasion of RCC cells, the effect of MALAT1-knockdown
was examined in ACHN and 786-O cells using cellular migration
assays. In the wound assay, 1% FBS was used to sustain cell
vitality. MALAT1 siRNA knockdown inhibited the migration and
invasiveness of RCC cells compared with their corresponding control
cells. To confirm the role of CFL1 in MALAT1-mediated cell
mobility, the expression of CFL1 in MALAT-knockdown cells was
restored, and showed that the migration and invasion capacities of
these cells were elevated to similar levels as in the corresponding
control cells (Fig. 4). Taken
together, these results suggest that MALAT1 downregulation inhibits
RCC cell migration and invasiveness through CFL1.
 | Figure 4.MALAT1 regulates the migration and
invasion of renal cell carcinoma cells through CFL1. (A) Cell
migration assay of siMALAT1 cells and cells transfected with both
siMALAT1 and CFL1 vector. Left panel, ACHN; right panel, 786-O.
Representative images of the cells at 0 and 24 h are shown.
Magnification, ×100. (B) Quantification of cell migration assay.
Migration distances are shown as the mean ± SD of three independent
analyses, and one-way ANOVA followed by Tukey's post hoc test was
used for statistical analysis. Left panel, ACHN; right panel,
786-O. (C) Invasion assay of the kidney renal clear cell carcinoma
cells with indicated treatments. Left panel, ACHN; right panel,
786-O. Representative images are shown. Magnification, ×200. Scale
bar, 100 µm. (D) Quantification of the invasion assay. Results are
shown as the mean ± SD of three independent analyses and one-way
ANOVA followed by Tukeys post hoc test was used for statistical
analysis Upper panel, ACHN; lower panel, 786-O. ns, not
significant; MALAT1, metastasis-associated lung adenocarcinoma
transcript 1; si, small interfering; CFL1, cofilin-1; SD, standard
deviation; ANOVA, analysis of variance. |
Clinical analyses of MALAT1 and
CFL1
Finally, the clinical implications of MALAT1 and
CFL1 were investigated. Levels of MALAT1 and CFL1
mRNA were analyzed using RT-qPCR in 20 KIRC tumor tissues. The
results showed a positive correlation (r=0.8131, P<0.001)
between MALAT1 and CFL1 (Fig. 5A), which further confirmed the
present hypothesis. By analyzing the mRNA sequencing data from TCGA
database, it was found that the levels of CFL1 mRNA were
significantly increased in 15 common types of cancer, including
KIRC (1.2-fold change; P<0.001) and significantly decreased only
in COAD (P<0.001) as compared with the normal tissues (Fig. 5B). In addition, Kaplan-Meier survival
analyses revealed that both MALAT1 (P<0.001) and CFL1
(P<0.001) were associated with poor overall survival time in
patients with KIRC (Fig. 5C and D).
Taken together, these data suggest that MALAT1 and
CFL1 expression are positively correlated in KIRC tumor
tissues, and that both MALAT1 and CFL1 were
upregulated and associated with poor prognosis.
 | Figure 5.Correlation and survival analyses of
MALAT1 and CFL1 expression. (A) Correlation between MALAT1
and CFL1 mRNA expression in RCC tissues. The data were
expressed as mean ± SD (n=20). (B) Comparison of CFL1 expression in
tumors and corresponding normal tissues. Expression of CFL1 was
measured using log2(TPM). Data were obtained from TCGA database,
and the data were not paired. Wilcoxon rank-sum test was used for
statistical analysis. Kaplan-Meier survival analysis of (C)
MALAT1 and (D) CFL1 mRNA in patients with KIRC from
TCGA database using log-rank test. Patients were stratified into
high expression group and low expression group based on the 33 and
66 percentiles of the two RNAs (low, 1–33 percentile; high, 66–100
percentile). ns, not significant; MALAT1, metastasis-associated
lung adenocarcinoma transcript 1; CFL1, cofilin-1; SD, standard
deviation; TPM, Transcripts Per Million; TCGA, The Cancer Genome
Atlas; BCLA, bladder urothelial carcinoma; BRCA, breast invasive
carcinoma; COAD, colon adenocarcinoma, HNSC, esophageal carcinoma,
glioblastoma multiforme, head and neck squamous cell carcinoma;
KIRC, clear cell kidney carcinoma; KIRP, renal papillary cell
carcinoma; LIHC, liver hepatocellular carcinoma; LUAD, lung
adenocarcinoma; LUSC, lung squamous cell carcinoma; PAAD,
pancreatic adenocarcinoma; PCPG, pheochromocytoma and
paraganglioma; PRAD, prostate adenocarcinoma; READ rectum
adenocarcinoma; SARC, sarcoma; STAD, skin cutaneous melanoma,
stomach adenocarcinoma; THCA, thyroid carcinoma; THYM, thymoma;
UCEC, uterine corpus endometrial carcinoma. |
Discussion
In the past decade, lncRNA MALAT1 has been
implicated in the progression of several cancer types, including
lung cancer, bladder cancer, hepatocellular carcinoma and cervical
cancer (11,12,31–35).
Most studies have highlighted the pro-oncogenic role of MALAT1;
however, it is shown to suppress tumor progression in some cancer
types. The pan-cancer analysis performed in the present study
showed that the expression levels of MALAT1 increase in the
majority of cancer types, but is downregulated in several types,
such as BRCA and LUSC. A recent study showed that MALAT1 inhibits
breast cancer metastasis by binding and inactivating the
pro-metastatic transcription factor TEAD (36). Additionally, an earlier study
revealed that MALAT1 expression in metastatic adenocarcinoma is
several folds higher compared with that in non-metastatic
adenocarcinoma, but that the relative expression is decreased in
squamous cell carcinomas (12).
These studies indicate that the roles of MALAT1 may be cancer type-
and histological subtype-dependent.
A number of studies have demonstrated that MALAT1
functions in the regulation of different hallmarks of cancer,
especially cell proliferation and apoptosis. For example, MALAT1
promoted cell proliferation and inhibited apoptosis by suppressing
tumor necrosis factor receptor-associated factor 6 expression via
sponging microRNA (miR)-146b-5p in hepatocellular carcinoma
(37). It was also reported that
MALAT1 regulated cellular proliferation and apoptosis by binding to
unmethylated polycomb 2 protein and promoting E2F1 SUMOylation
(38). A previous study also
demonstrated that MALAT1 promotes cell proliferation and inhibits
apoptosis by suppressing p53 activation (19). Overall, these studies suggest that
MALAT1 may be involved in various tumor-associated cellular
processes via distinct mechanisms.
The underlying mechanism by which MALAT1 regulates
the metastatic phenotype of different types of cancer is still
largely unclear (11,14–17).
Several studies have demonstrated the role of MALAT1 in
epithelial-mesenchymal transition (EMT). For example, MALAT1 is
reported to promote EMT by sponging miR-126-5p and thereby
increasing the expression of metastasis-associated molecules, such
as vascular endothelial growth factor A, snail family
transcriptional repressor 2 and twist family bHLH transcription
factor 1 in colorectal cancer (39).
Hirata et al (32) also
reported that MALAT1 promoted cell invasion through interacting
with enhancer of zeste 2 polycomb repressive complex 2 subunit and
inhibited E-cadherin expression (32). In our previous study, MALAT1 mediated
tumor growth by regulating the activity of p53 (19). However, it is unknown whether MALAT1
can directly act on the cytoskeleton. In the present study, MALAT1
knockdown decreased CFL1 expression at both the mRNA and protein
level without affecting the abundance of its pre-mRNA. A possible
explanation for this is that MALAT1 regulates the alternative
splicing of pre-CFL1, resulting in higher levels of unstable
CFL1 transcripts that are immediately degraded. In an ongoing
project in the School of Basic Medical Sciences at Tianjin Medical
University, it was observed that MALAT knockdown markedly affected
the landscape of alternative splicing (data not shown). However,
the pool of pre-mRNA is affected by three factors; transcription,
degradation and splicing (40).
Further evidence is required to determine the contribution of each
of these factors in the results of the present study, by
experiments such as RNA sequencing.
CFL1 is a critical molecule that regulates
cytoskeletal dynamics by depolymerizing actin filaments, and is
required for cytokinesis and cell movement (21,23,41),
while the loss of CFL1 increases F-actin accumulation (42). Although a previous study implicated
CFL1 in cell proliferation (43),
substantial research has showed that CFL1 is more directly
associated with the cytoskeleton and cell migration (21,23,41).
Therefore, the role of CFL1 in cell migration was the focus of the
present study. Furthermore, Wang et al (22) demonstrated that CFL1 was associated
with cell invasion and cancer metastasis. The present data
demonstrated that MALAT1 promotes cell migration and invasion by
modulating the level of F-actin through regulation of the
expression of CFL1 in RCC cells (Fig.
6).
In conclusion. the present study identified a novel
mechanism by which MALAT1 regulates RCC cell migration and
invasion. TCGA database and RT-qPCR analyses further demonstrated a
positive correlation between MALAT1 and CFL1. Moreover, increased
expression of these two molecules in RCC tumor tissues were
associated with poorer overall patient survival. Functional and
mechanistic analyses suggested that MALAT1 knockdown inhibited
renal cancer cell migration by inhibiting CFL1 expression. These
findings may provide a potentially novel therapeutic target for RCC
treatment.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from The
National Natural Science Foundation of China (grant nos. 21974094,
21575103, 81872078 and 81772945); The Natural Science Foundation of
Tianjin (grant nos. 18JCYBJC25200 and 18JCYBJC26700), the Young
Elite Scientists Sponsorship Program (grant no. TJSQNTJ-2017-10)
and the Scientific Research Foundation for the Returned Overseas
Chinese Scholars (grant no. 2016015).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request or available in The Cancer Genome Atlas repository,
https://www.cancer.gov/tcga.
Authors' contributions
YLZ wrote the manuscript and conducted experiments;
XG assisted in performing the experiments. RBC designed the study
and revised the manuscript. HW and XG analyzed and interpreted the
data. YW and DY collected the tumor tissues and processed the
samples for RT-qPCR analysis, and also assisted in writing and
revising the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Tianjin Medical University Second Hospital Medical
Ethics Committee (Tianjin, China) approved the present study
(approval no. KY2020K019) and all participants provided written
informed consent for opt-in.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
RCC
|
renal cell carcinoma
|
|
lncRNA
|
long non-coding RNA
|
|
MALAT1
|
metastasis-associated lung
adenocarcinoma transcript 1
|
|
CFL1
|
cofilin-1
|
|
TCGA
|
The Cancer Genome Atlas
|
|
KIRC
|
renal clear cell carcinoma
|
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Source, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lopez-Beltran A, Carrasco JC, Cheng L,
Scarpelli M, Kirkali Z and Montironi R: 2009 update on the
classification of renal epithelial tumours in adults. Int J Urol.
16:432–443. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bedke J, Gauler T, Grünwald V, Hegele A,
Herrmann E, Hinz S, Janssen J, Schmitz S, Schostak M, Tesch H, et
al: Systemic therapy in metastatic renal cell carcinoma. World J
Urol. 35:179–188. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Brown C: Targeted therapy: An elusive
cancer target. Nature. 537:S106–S108. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ulitsky I and Bartel DP: LincRNAs:
Genomics, evolution, and mechanisms. Cell. 154:26–46. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang L, Lin C, Jin C, Yang JC, Tanasa B,
Li W, Merkurjev D, Ohgi KA, Meng D, Zhang J, et al:
LncRNA-dependent mechanisms of androgen receptor-regulated gene
activation programs. Nature. 500:598–602. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wilusz JE: Long noncoding RNAs: Re-writing
dogmas of RNA processing and stability. Biochim Biophys Acta.
1859:128–138. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gutschner T, Hämmerle M and Diederichs S:
MALAT1-a paradigm for long noncoding RNA function in cancer. J Mol
Med (Berl). 91:791–801. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gutschner T, Hämmerle M, Eissmann M, Hsu
J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, et al:
The noncoding RNA MALAT1 is a critical regulator of the metastasis
phenotype of lung cancer cells. Cancer Res. 73:1180–1189. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ji P, Diederichs S, Wang W, Böing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel noncoding RNA, and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Guttman M, Amit I, Garber M, French C, Lin
MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al:
Chromatin signature reveals over a thousand highly conserved large
non-coding RNAs in mammals. Nature. 458:223–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hu L, Wu Y, Tan D, Meng H, Wang K, Bai Y
and Yang K: Up-regulation of long noncoding RNA MALAT1 contributes
to proliferation and metastasis in esophageal squamous cell
carcinoma. J Exp Clin Cancer Res. 34:72015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Okugawa Y, Toiyama Y, Hur K, Toden S,
Saigusa S, Tanaka K, Inoue Y, Mohri Y, Kusunoki M, Boland CR and
Goel A: Metastasis-associated long non-coding RNA drives gastric
cancer development and promotes peritoneal metastasis.
Carcinogenesis. 35:2731–2739. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Park JY, Lee JE, Park JB, Yoo H, Lee SH
and Kim JH: Roles of long non-coding RNAs on tumourigenesis and
glioma development. Brain Tumour Res Treat. 2:1–6. 2014. View Article : Google Scholar
|
|
17
|
Xiao H, Tang K, Liu P, Chen K, Hu J, Zeng
J, Xiao W, Yu G, Yao W, Zhou H, et al: LncRNA MALAT1 functions as a
competing endogenous RNA to regulate ZEB2 expression by sponging
miR-200s in clear cell kidney carcinoma. Oncotarget. 6:38005–38015.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tripathi V, Ellis JD, Shen Z, Song DY, Pan
Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al: The
nuclear-retained noncoding RNA MALAT1 regulates alternative
splicing by modulating SR splicing factor phosphorylation. Mol
Cell. 39:925–938. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen R, Liu Y, Zhuang H, Yang B, Hei K,
Xiao M, Hou C, Gao H, Zhang X, Jia C, et al: Quantitative
proteomics reveals that long non-coding RNA MALAT1 interacts with
DBC1 to regulate p53 acetylation. Nucleic Acids Res. 45:9947–9959.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Engreitz JM, Sirokman K, McDonel P,
Shishkin AA, Surka C, Russell P, Grossman SR, Chow AY, Guttman M
and Lander ES: RNA-RNA interactions enable specific targeting of
noncoding RNAs to nascent pre-mRNAs and chromatin sites. Cell.
159:188–199. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dawe HR, Minamide LS, Bamburg JR and
Cramer LP: ADF/cofilin controls cell polarity during fibroblast
migration. Curr Biol. 13:252–257. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang W, Eddy R and Condeelis J: The
cofilin pathway in breast cancer invasion and metastasis. Nat Rev
Cancer. 7:429–440. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bravo-Cordero JJ, Magalhaes MA, Eddy RJ,
Hodgson L and Condeelis J: Functions of cofilin in cell locomotion
and invasion. Nat Rev Mol Cell Biol. 14:405–415. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Warren AY and Harrison D: WHO/ISUP
classification, grading and pathological staging of renal cell
carcinoma: Standards and controversies. World J Urol. 36:1913–1926.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Colaprico A, Silva TC, Olsen C, Garofano
L, Cava C, Garolini D, Sabedot TS, Malta TM, Pagnotta SM,
Castiglioni I, et al: TCGAbiolinks: An R/Bioconductor package for
integrative analysis of TCGA data. Nucleic Acids Res. 44:e712016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
R Core Team (2012), . R: A language and
environment for statistical computing. R Foundation for Statistical
Computing. (Vienna, Austria). ISBN 3-900051-07-0, URL
http://www.R-project.org/.
|
|
28
|
RStudio Team (2015), . RStudio: Integrated
Development for R. RStudio, Inc. (Boston, MA). URL
http://www.rstudio.com/.
|
|
29
|
Rooney MS, Shukla SA, Wu CJ, Getz G and
Hacohen N: Molecular and genetic properties of tumours associated
with local immune cytolytic activity. Cell. 160:48–61. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yu G, Wang LG, Han Y and He QY:
ClusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang HM, Yang FQ, Chen SJ, Che J and
Zheng JH: Upregulation of long non-coding RNA MALAT1 correlates
with tumour progression and poor prognosis in clear cell renal cell
carcinoma. Tumour Biol. 36:2947–2955. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hirata H, Hinoda Y, Shahryari V, Deng G,
Nakajima K, Tabatabai ZL, Ishii N and Dahiya R: Long noncoding RNA
MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and
interacts with miR-205. Cancer Res. 75:1322–1331. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Han Y, Liu Y, Zhang H, Wang T, Diao R,
Jiang Z, Gui Y and Cai Z: Hsa-miR-125b suppresses bladder cancer
development by down-regulating oncogene SIRT7 and oncogenic long
non-coding RNA MALAT1. FEBS Lett. 587:3875–3882. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Malakar P, Shilo A, Mogilevsky A, Stein I,
Pikarsky E, Nevo Y, Benyamini H, Elgavish S, Zong X, Prasanth KV
and Karni R: Long noncoding RNA MALAT1 promotes hepatocellular
carcinoma development by SRSF1 upregulation and mTOR activation.
Cancer Res. 77:1155–1167. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tripathi V, Shen Z, Chakraborty A, Giri S,
Freier SM, Wu X, Zhang Y, Gorospe M, Prasanth SG, Lal A and
Prasanth KV: Long noncoding RNA MALAT1 controls cell cycle
progression by regulating the expression of oncogenic transcription
factor B-MYB. PLoS Genet. 9:e10033682013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kim J, Piao HL, Kim BJ, Yao F, Han Z, Wang
Y, Xiao Z, Siverly AN, Lawhon SE, Ton BN, et al: Long noncoding RNA
MALAT1 suppresses breast cancer metastasis. Nat Genet.
50:1705–1715. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li C, Miao R, Liu S, Wan Y, Zhang S, Deng
Y, Bi J, Qu K, Zhang J and Liu C: Down-regulation of miR-146b-5p by
long noncoding RNA MALAT1 in hepatocellular carcinoma promotes
cancer growth and metastasis. Oncotarget. 8:28683–28695. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yang L, Lin C, Liu W, Zhang J, Ohgi KA,
Grinstein JD, Dorrestein PC and Rosenfeld MG: ncRNA- and Pc2
methylation-dependent gene relocation between nuclear structures
mediates gene activation programs. Cell. 147:773–788. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sun Z, Ou C, Liu J, Chen C, Zhou Q, Yang
S, Li G, Wang G, Song J, Li Z, et al: YAP1-induced MALAT1 promotes
epithelial-mesenchymal transition and angiogenesis by sponging
miR-126-5p in colorectal cancer. Oncogene. 38:2627–2644. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Audibert A, Weil D and Dautry F: In vivo
kinetics of mRNA splicing and transport in mammalian cells. Mol
Cell Biol. 22:6706–6718. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen Q and Pollard TD: Actin filament
severing by cofilin is more important for assembly than
constriction of the cytokinetic contractile ring. J Cell Biol.
195:485–498. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kanellos G, Zhou J, Patel H, Ridgway RA,
Huels D, Gurniak CB, Sandilands E, Carragher NO, Sansom OJ, Witke
W, et al: ADF and cofilin1 control actin stress fibres, nuclear
integrity, and cell survival. Cell Rep. 13:1949–1964. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Pang D, Yang C, Li C, Zou Y, Feng B, Li L,
Liu W, Luo Q, Chen Z and Huang C: Polyphyllin II inhibits liver
cancer cell proliferation, migration and invasion through
downregulated cofilin activity and the AKT/NF-κB pathway. Biol
Open. 9:bio0468542020. View Article : Google Scholar : PubMed/NCBI
|




















