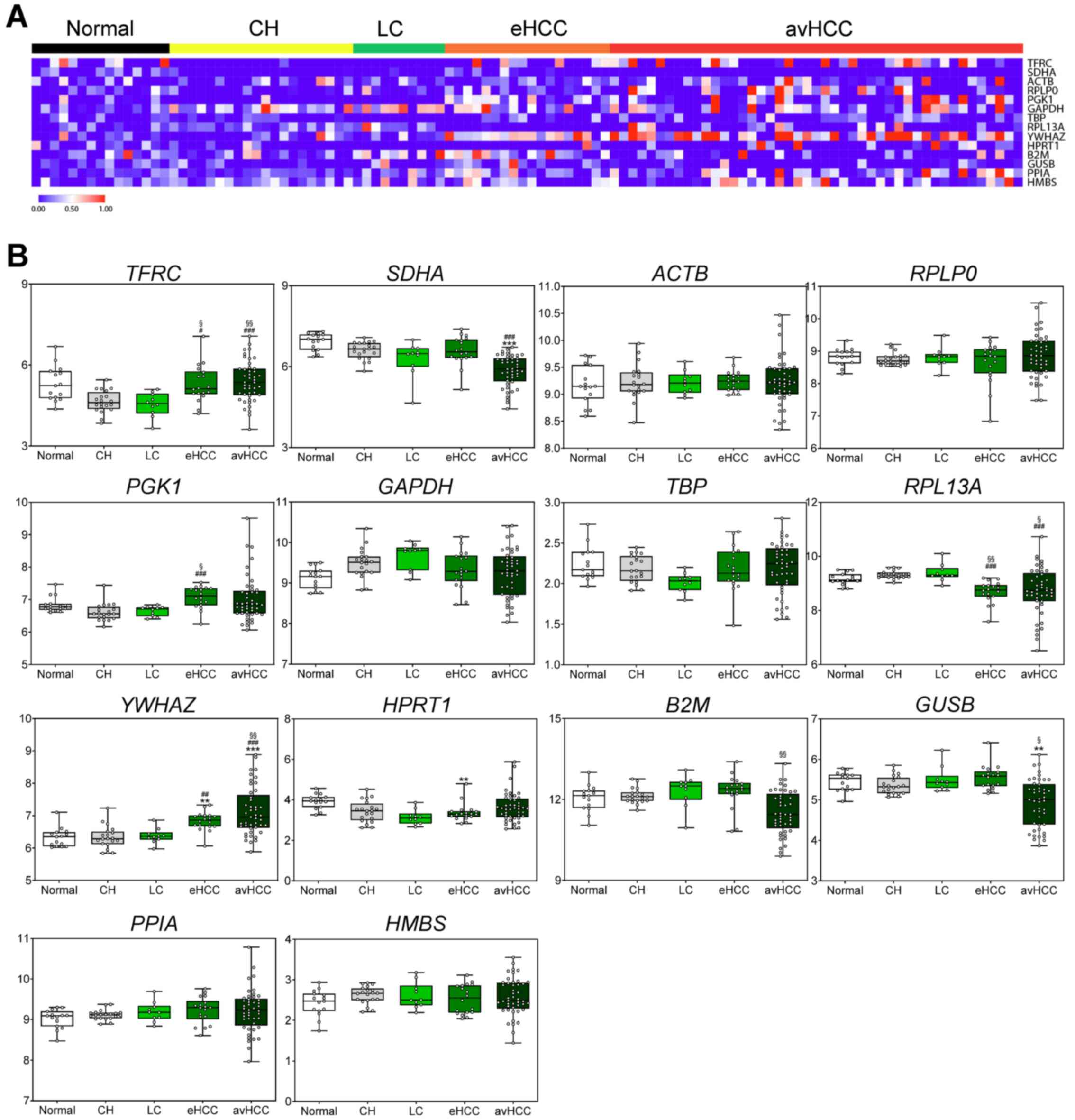
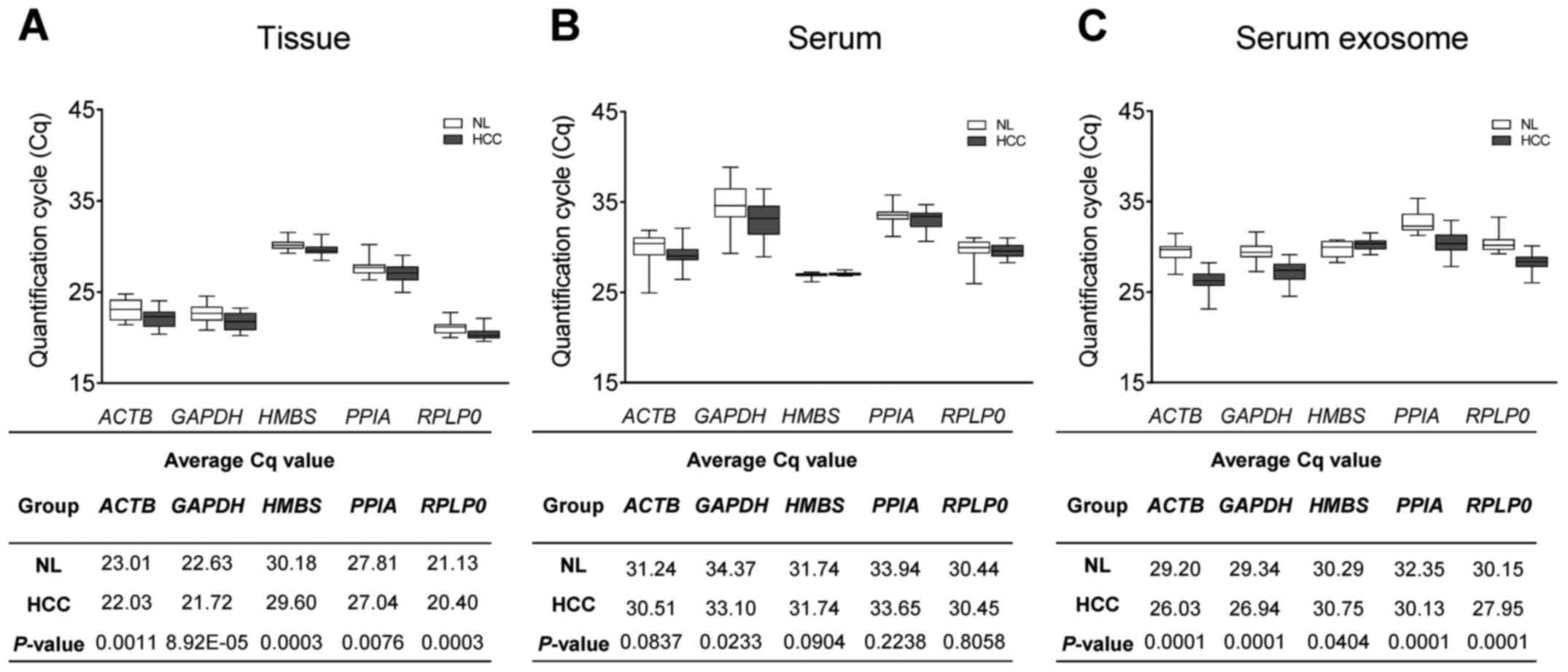
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kim SS, Baek GO, Ahn HR, Sung S, Seo CW,
Cho HJ, Nam SW, Cheong JY and Eun JW: Serum small extracellular
vesicle-derived LINC00853 as a novel diagnostic marker for early
hepatocellular carcinoma. Mol Oncol. 14:2646–2659. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Song P, Feng X, Zhang K, Song T, Ma K,
Kokudo N, Dong J and Tang W: Perspectives on using
des-γ-carboxyprothrombin (DCP) as a serum biomarker: Facilitating
early detection of hepatocellular carcinoma in China. Hepatobiliary
Surg Nutr. 2:227–231. 2013.PubMed/NCBI
|
|
4
|
Schulze K, Imbeaud S, Letouzé E,
Alexandrov LB, Calderaro J, Rebouissou S, Couchy G, Meiller C,
Shinde J, Soysouvanh F, et al: Exome sequencing of hepatocellular
carcinomas identifies new mutational signatures and potential
therapeutic targets. Nat Genet. 47:505–511. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang C, Peng L, Zhang Y, Liu Z, Li W,
Chen S and Li G: The identification of key genes and pathways in
hepatocellular carcinoma by bioinformatics analysis of
high-throughput data. Med Oncol. 34:1012017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gao X, Wang X and Zhang S: Bioinformatics
identification of crucial genes and pathways associated with
hepatocellular carcinoma. Biosci Rep. 38:382018. View Article : Google Scholar
|
|
7
|
Xu L, Wu LF and Deng FY: Exosome: An
Emerging Source of Biomarkers for Human Diseases. Curr Mol Med.
19:387–394. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li W, Li C, Zhou T, Liu X, Liu X, Li X and
Chen D: Role of exosomal proteins in cancer diagnosis. Mol Cancer.
16:1452017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Corrado C, Raimondo S, Chiesi A, Ciccia F,
De Leo G and Alessandro R: Exosomes as intercellular signaling
organelles involved in health and disease: Basic science and
clinical applications. Int J Mol Sci. 14:5338–5366. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wong CH and Chen YC: Clinical significance
of exosomes as potential biomarkers in cancer. World J Clin Cases.
7:171–190. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Regev-Rudzki N, Wilson DW, Carvalho TG,
Sisquella X, Coleman BM, Rug M, Bursac D, Angrisano F, Gee M, Hill
AF, et al: Cell-cell communication between malaria-infected red
blood cells via exosome-like vesicles. Cell. 153:1120–1133. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gachon C, Mingam A and Charrier B:
Real-time PCR: What relevance to plant studies? J Exp Bot.
55:1445–1454. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wong ML and Medrano JF: Real-time PCR for
mRNA quantitation. Biotechniques. 39:75–85. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huggett J, Dheda K, Bustin S and Zumla A:
Real-time RT-PCR normalisation; strategies and considerations.
Genes Immun. 6:279–284. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kozera B and Rapacz M: Reference genes in
real-time PCR. J Appl Genet. 54:391–406. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang X, Ding L and Sandford AJ: Selection
of reference genes for gene expression studies in human neutrophils
by real-time PCR. BMC Mol Biol. 6:42005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Razavi SA, Afsharpad M, Modarressi MH,
Zarkesh M, Yaghmaei P, Nasiri S, Tavangar SM, Gholami H,
Daneshafrooz A and Hedayati M: Validation of reference genes for
normalization of relative qRT-PCR studies in papillary thyroid
carcinoma. Sci Rep. 9:152412019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zampieri M, Ciccarone F, Guastafierro T,
Bacalini MG, Calabrese R, Moreno-Villanueva M, Reale A, Chevanne M,
Bürkle A and Caiafa P: Validation of suitable internal control
genes for expression studies in aging. Mech Ageing Dev. 131:89–95.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dheda K, Huggett JF, Bustin SA, Johnson
MA, Rook G and Zumla A: Validation of housekeeping genes for
normalizing RNA expression in real-time PCR. BioTechniques.
37:112–119. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu Y, Qin Z, Cai L, Zou L, Zhao J and
Zhong F: Selection of internal references for qRT-PCR assays of
human hepatocellular carcinoma cell lines. Biosci Rep. 37:372017.
View Article : Google Scholar
|
|
21
|
Fu LY, Jia HL, Dong QZ, Wu JC, Zhao Y,
Zhou HJ, Ren N, Ye QH and Qin LX: Suitable reference genes for
real-time PCR in human HBV-related hepatocellular carcinoma with
different clinical prognoses. BMC Cancer. 9:492009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Waxman S and Wurmbach E: De-regulation of
common housekeeping genes in hepatocellular carcinoma. BMC
Genomics. 8:2432007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cicinnati VR, Shen Q, Sotiropoulos GC,
Radtke A, Gerken G and Beckebaum S: Validation of putative
reference genes for gene expression studies in human hepatocellular
carcinoma using real-time quantitative RT-PCR. BMC Cancer.
8:3502008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu S, Zhu P, Zhang L, Ding S, Zheng S,
Wang Y and Lu F: Selection of reference genes for RT-qPCR analysis
in tumor tissues from male hepatocellular carcinoma patients with
hepatitis B infection and cirrhosis. Cancer Biomark. 13:345–349.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gao Q, Wang XY, Fan J, Qiu SJ, Zhou J, Shi
YH, Xiao YS, Xu Y, Huang XW and Sun J: Selection of reference genes
for real-time PCR in human hepatocellular carcinoma tissues. J
Cancer Res Clin Oncol. 134:979–986. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Benz F, Roderburg C, Vargas Cardenas D,
Vucur M, Gautheron J, Koch A, Zimmermann H, Janssen J,
Nieuwenhuijsen L, Luedde M, et al: U6 is unsuitable for
normalization of serum miRNA levels in patients with sepsis or
liver fibrosis. Exp Mol Med. 45:e422013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang Y, Li T, Qiu Y, Zhang T, Guo P, Ma
X, Wei Q and Han L: Serum microRNA panel for early diagnosis of the
onset of hepatocellular carcinoma. Medicine (Baltimore).
96:e56422017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Keerthikumar S, Chisanga D, Ariyaratne D,
Al Saffar H, Anand S, Zhao K, Samuel M, Pathan M, Jois M,
Chilamkurti N, et al: ExoCarta: A web-based compendium of exosomal
cargo. J Mol Biol. 428:688–692. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Marrero JA, Kulik LM, Sirlin CB, Zhu AX,
Finn RS, Abecassis MM, Roberts LR and Heimbach JK: Diagnosis,
staging, and management of hepatocellular carcinoma: 2018 practice
guidance by the American Association for the Study of Liver
Diseases. Hepatology. 68:723–750. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Drazic A, Aksnes H, Marie M, Boczkowska M,
Varland S, Timmerman E, Foyn H, Glomnes N, Rebowski G, Impens F, et
al: NAA80 is actin's N-terminal acetyltransferase and regulates
cytoskeleton assembly and cell motility. Proc Natl Acad Sci USA.
115:4399–4404. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ercolani L, Florence B, Denaro M and
Alexander M: Isolation and complete sequence of a functional human
glyceraldehyde-3-phosphate dehydrogenase gene. J Biol Chem.
263:15335–15341. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tisdale EJ: Glyceraldehyde-3-phosphate
dehydrogenase is phosphorylated by protein kinase Ciota/lambda and
plays a role in microtubule dynamics in the early secretory
pathway. J Biol Chem. 277:3334–3341. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bung N, Roy A, Chen B, Das D, Pradhan M,
Yasuda M, New MI, Desnick RJ and Bulusu G: Human
hydroxymethylbilane synthase: Molecular dynamics of the pyrrole
chain elongation identifies step-specific residues that cause AIP.
Proc Natl Acad Sci USA. 115:E4071–E4080. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wei Y, Jinchuan Y, Yi L, Jun W, Zhongqun W
and Cuiping W: Antiapoptotic and proapoptotic signaling of
cyclophilin A in endothelial cells. Inflammation. 36:567–572. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Davis TL, Walker JR, Campagna-Slater V,
Finerty PJ, Paramanathan R, Bernstein G, MacKenzie F, Tempel W,
Ouyang H, Lee WH, et al: Structural and biochemical
characterization of the human cyclophilin family of peptidyl-prolyl
isomerases. PLoS Biol. 8:e10004392010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Remacha M, Jimenez-Diaz A, Santos C,
Briones E, Zambrano R, Rodriguez Gabriel MA, Guarinos E and
Ballesta JP: Proteins P1, P2, and P0, components of the eukaryotic
ribosome stalk. New structural and functional aspects. Biochem Cell
Biol. 73:959–968. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Pfaffl MW, Tichopad A, Prgomet C and
Neuvians TP: Determination of stable housekeeping genes,
differentially regulated target genes and sample integrity:
BestKeeper - Excel-based tool using pair-wise correlations.
Biotechnol Lett. 26:509–515. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sen MK, Hamouzová K, Košnarová P, Roy A
and Soukup J: Identification of the most suitable reference gene
for gene expression studies with development and abiotic stress
response in Bromus sterilis. Sci Rep. 11:133932021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chia TS, Wong KF and Luk JM: Molecular
diagnosis of hepatocellular carcinoma: trends in biomarkers
combination to enhance early cancer detection. Hepatoma Res.
5:92019.
|
|
41
|
Matos LL, Trufelli DC, de Matos MG and da
Silva Pinhal MA: Immunohistochemistry as an important tool in
biomarkers detection and clinical practice. Biomark Insights.
5:9–20. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Manole E, Bastian AE, Popescu D,
Constantin C, Mihai S, Gaina G, Codrici E and Neagu M: Immunoassay
techniques highlighting biomarkers in immunogenetic diseases.
Immunogenetics. Nov 5–2018.(Epub ahead of print).
|
|
43
|
Li H, Bai R, Zhao Z, Tao L, Ma M, Ji Z,
Jian M, Ding Z, Dai X, Bao F, et al: Application of droplet digital
PCR to detect the pathogens of infectious diseases. Biosci Rep.
38:382018. View Article : Google Scholar
|
|
44
|
Macgregor PF and Squire JA: Application of
microarrays to the analysis of gene expression in cancer. Clin
Chem. 48:1170–1177. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sanders R, Mason DJ, Foy CA and Huggett
JF: Considerations for accurate gene expression measurement by
reverse transcription quantitative PCR when analysing clinical
samples. Anal Bioanal Chem. 406:6471–6483. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Schulze K, Nault JC and Villanueva A:
Genetic profiling of hepatocellular carcinoma using next-generation
sequencing. J Hepatol. 65:1031–1042. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Soung YH, Ford S, Zhang V and Chung J:
Exosomes in cancer diagnostics. Cancers (Basel). 9:92017.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Gorji-Bahri G, Moradtabrizi N, Vakhshiteh
F and Hashemi A: Validation of common reference genes stability in
exosomal mRNA-isolated from liver and breast cancer cell lines.
Cell Biol Int. 45:1098–1110. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Dai Y, Cao Y, Köhler J, Lu A, Xu S and
Wang H: Unbiased RNA-Seq-driven identification and validation of
reference genes for quantitative RT-PCR analyses of pooled cancer
exosomes. BMC Genomics. 22:272021. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Guo C, Liu S, Wang J, Sun MZ and Greenaway
FT: ACTB in cancer. Clin Chim Acta. 417:39–44. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Barber RD, Harmer DW, Coleman RA and Clark
BJ: GAPDH as a housekeeping gene: Analysis of GAPDH mRNA expression
in a panel of 72 human tissues. Physiol Genomics. 21:389–395. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhan C, Zhang Y, Ma J, Wang L, Jiang W,
Shi Y and Wang Q: Identification of reference genes for qRT-PCR in
human lung squamous-cell carcinoma by RNA-Seq. Acta Biochim Biophys
Sin (Shanghai). 46:330–337. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ceelen L, De Spiegelaere W, David M, De
Craene J, Vinken M, Vanhaecke T and Rogiers V: Critical selection
of reliable reference genes for gene expression study in the HepaRG
cell line. Biochem Pharmacol. 81:1255–1261. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Medrano G, Guan P, Barlow-Anacker AJ and
Gosain A: Comprehensive selection of reference genes for
quantitative RT-PCR analysis of murine extramedullary hematopoiesis
during development. PLoS One. 12:e01818812017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Krzystek-Korpacka M, Diakowska D, Bania J
and Gamian A: Expression stability of common housekeeping genes is
differently affected by bowel inflammation and cancer: Implications
for finding suitable normalizers for inflammatory bowel disease
studies. Inflamm Bowel Dis. 20:1147–1156. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Majidzadeh-A K, Esmaeili R and Abdoli N:
TFRC and ACTB as the best reference genes to quantify Urokinase
Plasminogen Activator in breast cancer. BMC Res Notes. 4:2152011.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Lv Y, Zhao SG, Lu G, Leung CK, Xiong ZQ,
Su XW, Ma JL, Chan WY and Liu HB: Identification of reference genes
for qRT-PCR in granulosa cells of healthy women and polycystic
ovarian syndrome patients. Sci Rep. 7:69612017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Shah KN and Faridi JS: Estrogen,
tamoxifen, and Akt modulate expression of putative housekeeping
genes in breast cancer cells. J Steroid Biochem Mol Biol.
125:219–225. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Ohl F, Jung M, Radonić A, Sachs M, Loening
SA and Jung K: Identification and validation of suitable endogenous
reference genes for gene expression studies of human bladder
cancer. J Urol. 175:1915–1920. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Li T, Diao H, Zhao L, Xing Y, Zhang J, Liu
N, Yan Y, Tian X, Sun W and Liu B: Identification of suitable
reference genes for real-time quantitative PCR analysis of hydrogen
peroxide-treated human umbilical vein endothelial cells. BMC Mol
Biol. 18:102017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Bruce KD, Sihota KK, Byrne CD and
Cagampang FR: The housekeeping gene YWHAZ remains stable in a model
of developmentally primed non-alcoholic fatty liver disease. Liver
Int. 32:1315–1321. 2012. View Article : Google Scholar : PubMed/NCBI
|