Introduction
Colorectal cancer (CRC) is the second most common
cancer in women and the third most common in men in developed
countries (1). Current monoclonal
antibody-based therapies have improved the prognosis of CRC
patients. These agents include bevacizumab, an inhibitor of the
vascular endothelial growth factor (VEGF), and cetuximab, an
epidermal growth factor receptor (EGFR) inhibitor. However,
long-term survival remains low for metastatic CRC. A better
understanding of the molecular mechanisms underlying CRC is
essential for the development of novel therapeutic strategies.
microRNAs (miRNAs) are endogenous single-stranded
non-coding RNAs of 19–25 nucleosides in length that are generated
by Dicer, an RNase III enzyme. miRNAs regulate gene expression at
the post-transcriptional level by the degradation of mRNA and
translational repression. miRNAs play essential roles in diverse
processes, including development, proliferation, differentiation
and apoptosis (2–4). miRNAs function as negative regulators
of gene expression, and the overexpression of oncogenic miRNAs can
contribute to tumorigenesis by promoting cellular proliferation and
evasion of apoptosis.
In CRC, the dysregulation of miRNAs has been
reported to influence carcinogenesis, invasion and metastasis. The
first study of miRNA expression in CRC in 2003 identified miR-143
and miR-145 as novel dysregulated miRNAs (5). Since then, numerous miRNAs have been
reported that are related to CRC prognosis.
Alternative expression of miR-181a has been reported
in a number of cancers. miR-181a is a prognostic marker in acute
myeloid leukemia (6) and non-small
cell lung cancer (7). The screening
of miRNAs implicated in other cancers has shown that miR-181a is
upregulated in CRC; however, there has been no assessment of its
correlation with clinicopathological and prognostic factors. Since
miR-181a has been reported to be overexpressed in TP53 null cells
(8), miR-181a may also be involved
in the carcinogenesis of CRC.
In the present study, we investigated miR-181a as an
oncogenic miRNA in CRC. We demonstrate that miR-181a has clinical
significance as a prognostic indicator in CRC. Our data also
suggest that the expression of the tumor suppressor protein,
phosphatase and tensin homolog (PTEN), may be suppressed by
miR-181a, resulting in the suppression of the PTEN/Akt pathway.
Materials and methods
Clinical tissue samples
This study included 162 patients who underwent
resection of primary CRC at Kyushu University Hospital (Beppu,
Japan) or at its affiliated hospitals between 1992 and 2000.
Resected tissue samples were immediately cut and embedded in
Tissue-Tek OCT compound (Sakura Finetechnical Co., Ltd., Tokyo,
Japan), frozen in liquid nitrogen and stored at −80°C until RNA
extraction. The clinicopathological factors and clinical stage were
classified using the criteria of the International Union Against
Cancer (9). All sample data,
including age, gender, tumor size and depth, lymph node metastasis,
vascular invasion, distant metastasis, clinical stage and
histological grade, were obtained from the clinical and
pathological records. The mean follow-up period was 35.6 months.
None of the patients had pre-operative chemotherapy or irradiation.
After surgery, patients with stage III/IV tumors were treated with
standard 5-fluorouracil-based chemotherapy. The Human Ethics Review
Committee of Osaka University (Osaka, Japan) and Kyusyu University
(Ohita, Japan) approved the use of the resected samples. The
reporting recommendations for tumor marker prognostic studies
(REMARK) guidelines for tumor marker studies were used for the
preparation of this study (10).
Evaluation of miR-181a expression in
clinical samples
For the quantitative real-time PCR (qRT-PCR)
analysis of miR-181a, cDNA was synthesized from 10 ng
of total RNA using TaqMan™ MicroRNA hsa-miR-181a-specific primers
(Applied Biosystems, Tokyo, Japan) and the TaqMan™ MicroRNA Reverse
Transcription kit (Applied Biosystems). Reverse transcription
conditions were as described previously (11). miRNA expression was determined
relatively to the expression of RNU6B (control) and was analyzed
using the ΔΔCt method (12).
Cell cultures
The Colo 201 colon cancer cell line was maintained
in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% FBS
(Sigma-Aldrich, St. Louis, MO), 100 U/ml penicillin and 100 μg/ml
streptomycin in a humidified atmosphere with 5% CO2 at
37°C. Cell count was assessed using a hemocytometer.
Transfection of miR-181a precursor
(pre-miR-181a)
Cells were grown to 60–80% confluence and
transfected with 5 nmol/l of pre-miR-181a or negative control
oligonucleotides (Applied Biosystems) using the siPORT™ NeoFX
Transfection Agent (Ambion, Inc., Austin, TX). miR-181a expression
was analyzed in the cells 24 and 48 h after transfection, and PTEN
expression was analyzed 24 h after transfection.
Evaluation of PTEN mRNA expression in the
Colo 201 cell line
Total RNA was extracted from Colo 201 cells using
the modified acid-guanidine-phenol-chloroform method described
previously (13). Total RNA (8 μg)
was reverse-transcribed to cDNA with M-MLV RT (Invitrogen,
Carlsbad, CA). The PCR primer sequences for PTEN mRNA were as
follows: PTEN forward, 5′-GAGGGATAAAACACCATG-3′ and reverse,
5′-AGGGGTAGGATGTGAACCAGTA-3′. Glyceraldehyde-3-phosphate
dehydrogenase (GAPDH) was used as the internal control, and the
GAPDH primers were as follows: GAPDH forward,
5′-TTGGTATCGTGGAAGGACTCTA-3′ and reverse, 5′-TGTCATATTTGGCAGGTT-3′.
Real-time monitoring of PCR was performed with the
LightCycler® system (Roche Applied Science,
Indianapolis, IN) and SYBR-Green I dye (Roche Diagnostics, Tokyo,
Japan). Monitoring was performed according to the instructions of
the manufacturer as described previously (14). In brief, a master mixture was
prepared on ice that contained 1 μM of cDNA, 2 μl of DNA Master
SYBR-Green I mix, 50 ng of primers and 24 μl of 25 mmol/l
MgCl2. The final volume was adjusted to 20 μl with
water. qRT-PCR was performed as described previously (15). The concentrations of unknown samples
were calculated by plotting their crossing points against the
standard curve and dividing by the value for GAPDH.
Statistical analysis
The qRT-PCR data were analyzed with JMP®
8 (SAS Institute, Cary, NC). The overall survival rates were
calculated by the Kaplan-Meier method and were measured starting on
the day of surgery. The differences between groups were estimated
using the χ2 test, the Student’s t-test or the log-rank
test. For multivariate analysis, the Cox proportional hazards
regression model was used. A probability level (P-value) of 0.05
was used to determine statistically significant diferences.
Results
Clinicopathological significance of
miR-181a expression in CRC
In this study, the CRC cases were classified into
two groups, a low expression group (n=82) with miR-181a levels less
than the median expression level and a high expression group (n=80)
with miR-181a expression above the median level. We compared
overall (Fig. 1A) and disease-free
survival (Fig. 1B) between the
groups. Patients in the high expression group had significantly
poorer prognosis than those in the low expression group (overall
survival, P=0.011; disease-free survival, P=0.010). We also
compared miR-181a expression in normal colon mucosa and CRC tissues
and found no significant difference (Fig. 2). We then analyzed
clinicopathological factors and miR-181a expression. There were no
significant differences related to miR-181a expression as regards
age, gender, histological grade, tumor size, tumor depth, lymph
node metastasis, lymphatic invasion, venous invasion, liver
metastasis, peritoneal metastasis or stage (Table I).
 | Table ImiR-181a expression and
clinicopathological characteristics in colorectal cancer patients
(n=162). |
Table I
miR-181a expression and
clinicopathological characteristics in colorectal cancer patients
(n=162).
| Characteristic | Low expression group
(n=82)
n (%) | High expression group
(n=80)
n (%) | P-value |
|---|
| Age (mean ± SD) | 67.0±1.20 | 66.7±1.21 | 0.83 |
| Gender |
| Male | 48 (58.6) | 50 (62.5) | 0.61 |
| Female | 34 (41.4) | 30 (37.5) | |
| Histological
grade |
| Well and moderately
differentiated | 77 (93.9) | 72 (90.0) | 0.36 |
| Poorly
differentiated and other grades | 5 (6.1) | 8 (10.0) | |
| Size (mm) |
| <33 (small) | 13 (17.1) | 19 (23.7) | 0.3 |
| >31 (large) | 63 (82.9) | 61 (76.3) | |
| Tumor deptha |
| m/sm | 9 (11.0) | 12 (15.0) | 0.45 |
| mp/ss/se/si | 73 (89.0) | 68 (85.0) | |
| Lymph node
metastasis |
| Absent | 45 (54.9) | 45 (56.3) | 0.86 |
| Present | 37 (45.1) | 35 (43.7) | |
| Lymphatic
invasion |
| Absent | 48 (58.5) | 43 (53.8) | 0.54 |
| Present | 34 (41.5) | 37 (46.2) | |
| Venous invasion |
| Absent | 65 (79.3) | 66 (82.5) | 0.6 |
| Present | 17 (20.7) | 14 (17.5) | |
| Liver metastasis |
| Absent | 72 (87.8) | 68 (85.0) | 0.6 |
| Present | 10 (12.2) | 12 (15.0) | |
| Peritoneal
dissemination |
| Absent | 79 (96.3) | 77 (96.3) | 0.98 |
| Present | 3 (3.7) | 3 (3.7) | |
| Srage |
| 0–I | 18 (22.0) | 23 (28.8) | 0.32 |
| III–IV | 64 (78.0) | 57 (71.2) | |
Upregulated expression of miR-181a is
associated with overall survival
Univariate analysis for overall survival showed that
several clinicopathological factors, including histological grade,
tumor size, lymph node metastasis, lymphatic invasion, venous
invasion and liver metastasis, and miR-181a expression, were
significant predictors of poor prognosis (Table II). Multivariate analysis showed
that miR-181a expression was an independent and significant
prognostic factor for overall survival (relative risk, 1.83; 95%
CI, 1.26–2.76; P=0.0013); histological grade, lymph node
metastasis, lymphatic invasion, venous invasion and liver
metastasis were independent and significant prognostic indicators
as well (Table II).
 | Table IIUnivariate and multivariate analyses
of overall survival in colorectal cancer patients (n=162). |
Table II
Univariate and multivariate analyses
of overall survival in colorectal cancer patients (n=162).
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|---|
| Characteristic | RR | 95% CI | P-value | RR | 95% CI | P-value |
|---|
| Age (>65/<64
years) | 0.88 | 0.63–1.26 | 0.485 | - | - | - |
| Gender
(female/male) | 0.91 | 0.63–1.27 | 0.579 | - | - | - |
| miR-181a expression
(high/low) | 1.59 | 1.11–2.35 | 0.0102 | 1.83 | 1.26–2.76 | 0.0013 |
| Histological grade
(poorly differentiated and other grades/well and moderately
differentiated) | 2.38 | 1.44–3.61 | 0.0016 | 2.06 | 1.18–3.41 | 0.0133 |
| Tumor size
(>31/<30 mm) | 2.37 | 1.30–5.89 | 0.0022 | 1.3 | 0.67–3.31 | 0.4804 |
| Lymph node
metastasis (positive/negative) | 2.81 | 1.87–4.60 | <0.0001 | 1.62 | 1.02–2.79 | 0.0427 |
| Lymphatic invasion
(positive/negative) | 2.18 | 1.52–3.23 | <0.0001 | 1.91 | 1.24–3.04 | 0.0033 |
| Venous invasion
(positive/negative) | 2.1 | 1.47–2.95 | 0.0001 | 1.85 | 1.25–2.73 | 0.0025 |
| Liver metastasis
(positive/negative) | 2.65 | 1.86–3.73 | <0.0001 | 2.48 | 1.65–3.74 | <0.0001 |
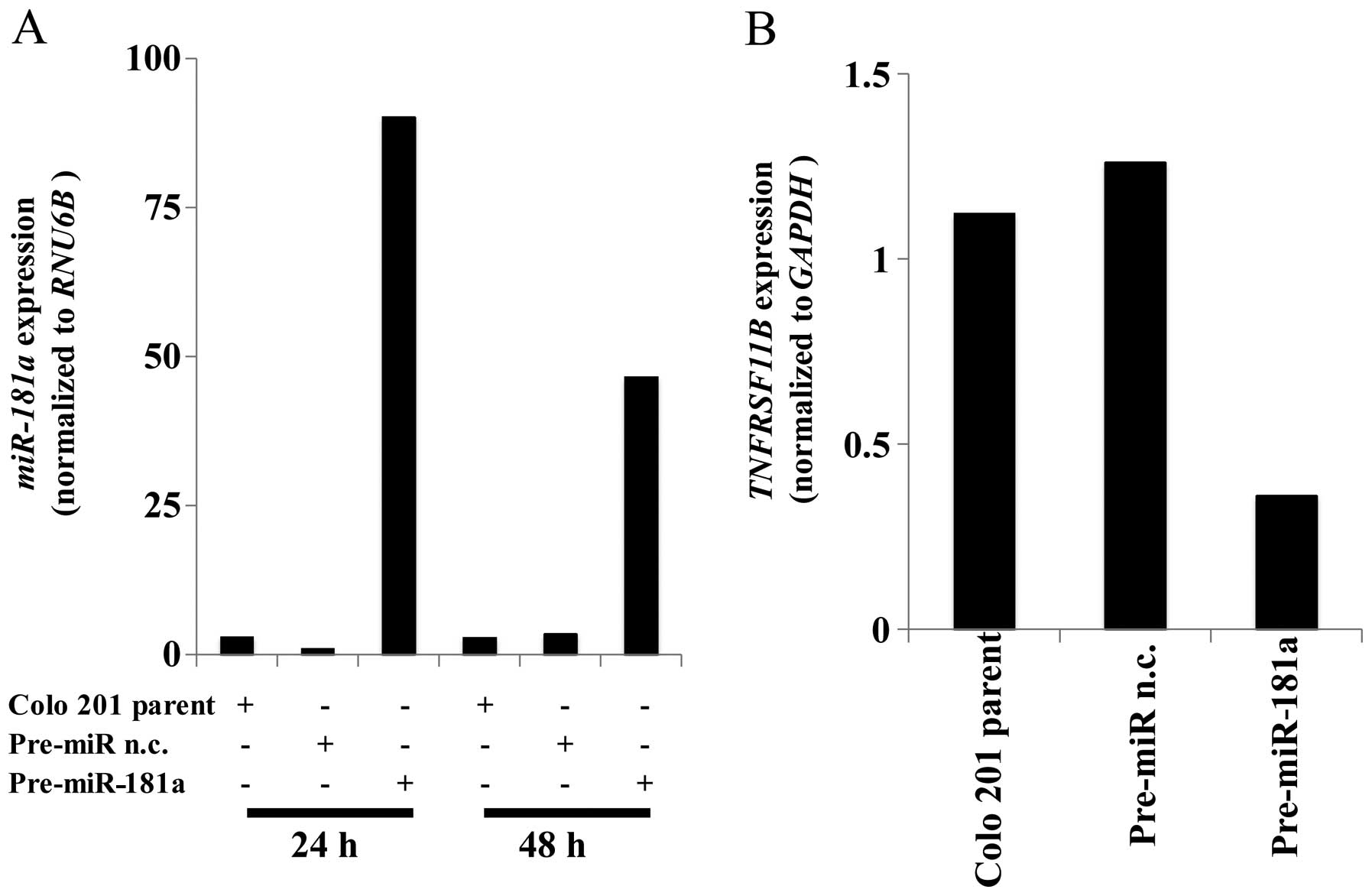
miR-181a regulates PTEN expression
We then examined which genes were controlled by
miR-181a expression in vitro. Using the in silico
miRNA target prediction tools, miRanda and TargetScan, we
discovered the sequences of the miR-181a binding sites in
transcripts encoding PTEN. To investigate this further, we analyzed
miR-181a expression in several colon cancer cell lines. qRT-PCR
analysis showed that miR-181a expression in Colo 201 cell lines was
lower than other cell lines (data not shown). We then transfected
pre-miR-181a into Colo 201 cells and analyzed miR-181a expression
using qRT-PCR. miR-181a levels were significantly higher 24 and 48
h in the transfected cells than in the cells treated with a pre-miR
negative control construct (Fig.
3A). The expression levels of PTEN were significantly
suppressed in the cells that overexpressed miR-181a (Fig. 3B).
Discussion
The resutls from the present study showed that
miR-181a expression was an independent significant prognostic
factor in CRC. miR-181a expression did not correlate with
clinicopathological parameters, some of which were prognostic
factors. These data suggest that miR-181a can affect prognosis via
unknown mechanisms. Additional experiments showed that PTEN was a
candidate target gene of miR-181a.
miR-181a has been shown to be upregulated in thyroid
cancer (16,17) and non-small cell lung cancer
(7) and downregulated in glioma
cells (18). Kanaan et al
(19) reported that miR-181a is
upregulated during the progression from non-neoplasia to dysplasia
in inflammatory bowel disease-associated CRC, whereas miR-181a
expression decreases when dysplasia develops into cancer. In the
present study, we found no differences in miR-181a expression in
normal and cancer tissue. Xi et al (8) used miRNA expression array analysis to
compare HCT-116 and null-p53 HCT-116 cells and found that miR-181a
was controlled in part by the p53 gene. These findings show
that miR-181a may be related to the carcinogenesis of CRC. However,
the upstream regulators of miR-181a expression remain unknown. In
this study, we analyzed miR-181a expression and prognosis in CRC
and found that miR-181a upregulation correlated with worse
prognosis. miR-181a expression was not associated with
clinicopathological factors. Although miR-181a expression could not
be used as a detection marker for CRC, miR-181a might be a new
independent prognostic factor in CRC patients.
The reported target genes of miR-181a in acute
myeloid leukemia are HOXA7, HOXA9, HOXA11 and PBX3 (20). Pekarsky et al (21) suggested that T cell
leukemia/lymphoma 1 (TCL1) expression in chronic lymphocytic
leukemia is regulated, at least in part, by miR-181a and that
miR-181a may be used as a therapeutic target. In the present study,
we detected a different candidate target gene for miR-181a in CRC,
namely, PTEN. Since miR-181a expression wadids not correlate with
clinicopathological parameters, miR-181a may influence the
malignant potential of CRC by controlling PTEN expression.
PTEN is a key tumor suppressor gene, and the loss of
PTEN gene expression has been implicated in the carcinogenesis of
numerous types of cancer. Specifically, the loss of PTEN expression
has been linked to a poorer prognosis in gliomas (22), endometrial carcinomas (23), prostate carcinomas (24), gastric carcinomas (25), as well as CRC (26). Furthermore, PTEN plays a critical
role in apoptosis, cell cycle arrest, cell migration and cell
spreading (27). The subcellular
localization of PTEN has been related to tumor progression, and it
is thought that nuclear PTEN regulates cell cycle arrest while
cytoplasmic PTEN regulates apoptosis (28). Therefore, miR-181a, which may
control PTEN expression, may be a therapeutic target in CRC.
In conclusion, our results show that miR-181a
expression does not correlate with clinicopathological
characteristics. However, the data suggest that miR-181a expression
may be a useful prognostic marker in CRC patients. Further studies
are required to investigate the potential of miR-181a as a
therapeutic target in CRC.
Acknowledgements
This study was supported by a Grant-in-Aid for
Cancer Research from the Ministry of Education, Culture, Science,
Sports and Technology of Japan.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
2
|
Brennecke J, Hipfner DR, Stark A, Russell
RB and Cohen SM: bantam encodes a developmentally regulated
microRNA that controls cell proliferation and regulates the
proapoptotic gene hid in Drosophila. Cell. 113:25–36. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lee RC, Feinbaum RL and Ambros V: The
C. elegans heterochronic gene lin-4 encodes small RNAs with
antisense complementarity to lin-14. Cell. 75:843–854. 1993.
|
|
4
|
Reinhart BJ, Slack FJ, Basson M, et al:
The 21-nucleotide let-7 RNA regulates developmental timing
in Caenorhabditis elegans. Nature. 403:901–906. 2000.
View Article : Google Scholar
|
|
5
|
Michael MZ, SM OC, van Holst Pellekaan NG,
Young GP and James RJ: Reduced accumulation of specific microRNAs
in colorectal neoplasia. Mol Cancer Res. 1:882–891. 2003.PubMed/NCBI
|
|
6
|
Schwind S, Maharry K, Radmacher MD, et al:
Prognostic significance of expression of a single microRNA,
miR-181a, in cytogenetically normal acute myeloid leukemia: a
Cancer and Leukemia Group B study. J Clin Oncol. 28:5257–5264.
2010. View Article : Google Scholar
|
|
7
|
Gao W, Yu Y, Cao H, et al: Deregulated
expression of miR-21, miR-143 and miR-181a in non small cell lung
cancer is related to clinicopathologic characteristics or patient
prognosis. Biomed Pharmacother. 64:399–408. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xi Y, Shalgi R, Fodstad O, Pilpel Y and Ju
J: Differentially regulated micro-RNAs and actively translated
messenger RNA transcripts by tumor suppressor p53 in colon cancer.
Clin Cancer Res. 12:2014–2024. 2006. View Article : Google Scholar
|
|
9
|
Sobin LH and Fleming ID: TNM
Classification of Malignant Tumors, fifth edition (1997). Union
Internationale Contre le Cancer and the American Joint Committee on
Cancer. Cancer. 80:1803–1804. 1997. View Article : Google Scholar
|
|
10
|
McShane LM, Altman DG, Sauerbrei W, et al:
REporting recommendations for tumour MARKer prognostic studies
(REMARK). Br J Cancer. 93:387–391. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nishida N, Yamashita S, Mimori K, et al:
MicroRNA-10b is a prognostic indicator in colorectal cancer and
confers resistance to the chemotherapeutic agent 5-fluorouracil in
colorectal cancer cells. Ann Surg Oncol. Feb 10–2012.(Epub ahead of
print).
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mori M, Mimori K, Yoshikawa Y, et al:
Analysis of the gene-expression profile regarding the progression
of human gastric carcinoma. Surgery. 131:S39–S47. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ogawa K, Utsunomiya T, Mimori K, et al:
Clinical significance of human kallikrein gene 6 messenger RNA
expression in colorectal cancer. Clin Cancer Res. 11:2889–2893.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yamamoto H, Kondo M, Nakamori S, et al:
JTE-522, a cyclooxygenase-2 inhibitor, is an effective
chemopreventive agent against rat experimental liver fibrosis1.
Gastroenterology. 125:556–571. 2003.PubMed/NCBI
|
|
16
|
He H, Jazdzewski K, Li W, et al: The role
of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad
Sci USA. 102:19075–19080. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pallante P, Visone R, Ferracin M, et al:
MicroRNA deregulation in human thyroid papillary carcinomas. Endocr
Relat Cancer. 13:497–508. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shi L, Cheng Z, Zhang J, et al:
hsa-mir-181a and hsa-mir-181b function as tumor suppressors in
human glioma cells. Brain Res. 1236:185–193. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kanaan Z, Rai SN, Eichenberger MR, et al:
Differential microRNA expression tracks neoplastic progression in
inflammatory bowel disease-associated colorectal cancer. Hum Mutat.
33:551–560. 2012. View Article : Google Scholar
|
|
20
|
Li Z, Huang H, Li Y, et al: Up-regulation
of a HOXA-PBX3 homeobox-gene signature following down-regulation of
miR-181 is associated with adverse prognosis in patients with
cytogenetically-abnormal AML. Blood. 119:2314–2324. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pekarsky Y, Santanam U, Cimmino A, et al:
Tcl1 expression in chronic lymphocytic leukemia is regulated by
miR-29 and miR-181. Cancer Res. 66:11590–11593. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kondo Y, Hollingsworth EF and Kondo S:
Molecular targeting for malignant gliomas (Review). Int J Oncol.
24:1101–1109. 2004.PubMed/NCBI
|
|
23
|
Xu B, Yao Q and Dai SZ: Detection of
mutation and protein expression of PTEN gene in endometrial
carcinoma. Ai Zheng. 23:69–73. 2004.(In Chinese).
|
|
24
|
Koksal IT, Dirice E, Yasar D, et al: The
assessment of PTEN tumor suppressor gene in combination with
Gleason scoring and serum PSA to evaluate progression of prostate
carcinoma. Urol Oncol. 22:307–312. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zheng HC, Sun JM, Li XH, Yang XF, Zhang YC
and Xin Y: Role of PTEN and MMP-7 expression in growth, invasion,
metastasis and angiogenesis of gastric carcinoma. Pathol Int.
53:659–666. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Goel A, Arnold CN, Niedzwiecki D, et al:
Frequent inactivation of PTEN by promoter hypermethylation in
microsatellite instability-high sporadic colorectal cancers. Cancer
Res. 64:3014–3021. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Weng L, Brown J and Eng C: PTEN induces
apoptosis and cell cycle arrest through
phosphoinositol-3-kinase/Akt-dependent and -independent pathways.
Hum Mol Genet. 10:237–242. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chung JH and Eng C: Nuclear-cytoplasmic
partitioning of phosphatase and tensin homologue deleted on
chromosome 10 (PTEN) differentially regulates the cell cycle and
apoptosis. Cancer Res. 65:8096–8100. 2005. View Article : Google Scholar
|