Introduction
Glioma is the most common tumor type in the central
nervous system with high morbidity and mortality (1). Despite aggressive treatment including
glioma surgery, radiotherapy, chemotherapy, gene therapy,
immunotherapy and other novel biological therapies, the median
survival duration of patients with advanced glioma has not
significantly improved due to its high recurrence and metastasis
rate (2,3). Therefore, an improved understanding of
the biological basis of glioma progression might provide useful
information for the clinical management of this disease.
MicroRNAs (miRNAs) are small, approximately 22
nucleotides in length, non-coding RNAs that negatively regulate
gene expression at a post-translational level by binding to
complementary sequences in the 3′UTRs of targeted mRNAs (4,5).
miRNAs have been reported to be involved in a range of cellular
functions such as differentiation, proliferation, and apoptosis
(6,7). Accumulating evidence supports critical
roles of miRNAs in the progression of different cancers, where they
play a crucial role in tumor development through regulation of
cellular proliferation, invasion, metastasis and apoptosis
(8–10). Many miRNAs have been demonstrated to
function as oncogenes or tumor suppressors in glioma (11,12),
which highlight the implications of miRNAs in diagnosis, treatment,
and prognosis of glioma.
miR-365, a newly discovered miRNA, has been reported
to be involved in tumor progression and development in several
types of human cancers, such as lung cancer (13,14),
melanoma (15), gastric carcinoma
(16), osteosarcoma (17), and colon cancer (18). However, the detail biological
function and underlying molecular mechanism of miR-365 in glioma
remains unclear. Therefore, the goals of the present study were to
investigate the biological function and underlying molecular
mechanism of miR-365 on the carcinogenesis of glioma. Our results
showed that miR-365 expression was significantly downregulated in
glioma cell lines and tissues. We also found that miR-365
overexpression inhibited cell proliferation, migration and invasion
of glioma by targeting PIK3R3. Our results might contribute to the
understanding of the molecular mechanism underlying glioma
pathogenesis.
Materials and methods
Ethics statement
This study was approved by the Medical Ethics
Committee of Jilin University (Changchun, China). All participating
patients provided written informed consent for the use of surgical
samples before surgery. All animals were treated in accordance with
standard guidelines for the care and use of laboratory animals of
Jilin University.
Clinical samples and cell lines
Thirty-six pairs of glioma tissues and their
adjacent normal brain tissues were collected from patients
undergoing surgery at the first of Hospital of Jilin University
from July 2012 to December 2014. All the tissues were snap-frozen
in liquid nitrogen immediately after resection and stored at −80°C
until use.
Primary normal human astrocytes (NHA) and four human
glioma cell lines (U251, U87, U118 and LN18) were from the Type
Culture Collection of the Chinese Academy of Sciences (Shanghai,
China). The cells were routinely cultured in Dulbecco's modified
Eagle's medium (DMEM, Gibco, Grand Island, NY, USA) supplemented
with 10% fetal bovine serum (FBS, Gibco), 100 U/ml penicillin and
100 mg/ml streptomycin (Sigma, St. Louis, MO, USA) at 37°C with 5%
CO2 in a humidified atmosphere.
Isolation of RNA and quantitative
real-time PCR (qRT-PCR)
Total RNA was extracted from the glioma cell lines
(2×106 cells) and glioma tissues (100 mg) with TRIzol
reagent (Invitrogen, Carlsbad, CA, USA) according to the
manufacturer's protocol. The optical density of the RNA samples at
260 nM was quantified by a NanoDrop ND-2000 Spectrophotometer
(NanoDrop Technologies, Houston, TX, USA). For identification of
miR-365 expression, complementary DNA (cDNA) were synthesized using
TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems,
Foster City, CA, USA). qRT-PCR was performed with the TaqMan
MicroRNA Assay for miR-365 and U6 (Ambion, Austin, TX, USA) and
TaqMan Universal Master Mix II without UNG (Ambion) under ABI PRISM
7900 Sequence Detection System (Applied Biosystems). For
determination of PIK3R3 mRNA expression, reverse
transcription (RT) were performed with a M-MLV First Strand kit
(Invitrogen) using Oligo(dT) 20 primers (Invitrogen). qRT-PCR was
performed using SYBR Select Master Mix (Invitrogen) under ABI PRISM
7900 Sequence Detection System. The primes of PIK3R3 and GAPDH used
in this study were according to a previous study (19). The level of mature miR-365 was
normalized relative to U6 endogenous control and PIK3R3 mRNA
expression was normalized relative to GAPDH (endogenous
control) using the 2−∆∆Ct method.
Cell transfection
miR-365 mimic and corresponding negative control
miRNA (miR-NC) were purchased from Shanghai GenePharma (Shanghai,
China). PIK3R3 overexpression plasmid (pCDNA3.1) was a gift of Dr
Peng Zhang (Jilin University). Transfection was performed using
Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's
instructions.
Proliferation assay
Cell proliferation assay was performed using
3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide
(MTT) (Sigma). Briefly, approx. 2000 transfected cells were seeded
into each well of 96-well plates and cultured for 1–4 days. At the
indicated time (24, 48, 72 and 96 h), 100 µl fresh medium
containing MTT 0.5 mg/ml was added into each well and cultured for
4 h at 37°C, then the medium was replaced with 100 µl of dimethyl
sulfoxide (DMSO, Sigma) and shaken at room temperature for 10 min.
The absorption was measured at 490 nm with a microplate reader
(Thermo Labsystems, Helsinki, Finland).
Invasion and migration and assays
The invasive ability of glioma cells was determined
using 24-well Transwell chambers coated with Matrigel (BD
Biosciences, San Jose, CA, USA). In brief, 1×105
transfected cells in serum-free medium were seeded at in the top
chamber coated with Matrigel and incubated at 37°C in a humidified
incubator containing 5% CO2. The bottom chamber was
filled with medium containing 10% FBS as a chemoattractant. After
24 h of incubation, the non-invaded cells on the upper surface of
the membrane were removed with a cotton swab, cells that invaded to
the underside of the membrane were fixed with 70% ethanol for 30
min and stained with 0.2% crystal violet for 10 min. Photographs
were imaged, and the number was counted in five randomly selected
fields under a light microscope (Olympus, Tokyo, Japan). For
Transwell migration assays, glioma cells were determined using
Transwell chambers without the Matrigel coating.
Dual luciferase reporter assay
The wild-type 3′-UTR segment of PIK3R3, which
contained a putative binding site for miR-365, was amplified from
normal human genomic DNA by PCR and inserted downstream of the
luciferase gene in pGL3-control vector (Promega, Madison, WI, USA),
named as Wt-PIK3R3. A mutant 3-UTR of PIK3R3 contained a mutation
in the complementary seed region of miR-365 was amplified by PCR
and inserted downstream of the luciferase gene in pGL3-control
vector, referred to as Mut-PIK3R3. U87 cells (1×105)
were seeded in 24-well plates and grown to 60–70% confluence. Cells
were then cotransfected with 200 ng Wt-PIK3R3 or Mut-PIK3R3
reporter plasmid, 50 nmol miR-365 mimic or miR-NC, and 20 ng pRL-TK
Renilla plasmid (Promega) using Lipofectamine 2000
(Invitrogen). At 48 h after transfection, both firefly and
Renilla luciferase activities were measured 48 h after
transfection by using the Dual-Luciferase Reporter Assay System
(Promega).
Animal studies
Five-week-old female BALB/c nude mice were purchased
from the Animal Center of Jilin University (Changchun, China). All
animal experiments were performed in accordance with the NIH Guide
for the Care and Use of Laboratory Animals. U87 cells stable
expressing miR-365 or miR-NC were injected subcutaneously into each
side of the posterior flank of the nude mouse (six per group,
2×106 cells for each mouse). Tumor growth was examined
every five day after injection, and tumor volumes were calculated
using the equation V=AxB2/2 (mm3), where A is
the largest diameter and B is the perpendicular diameter. Five
weeks after the implantation, the mice were sacrificed, and the
xenograft tumors were excised, and weighted. Part of tumor tissues
were stored for further analysis.
Western blotting
Western blot analysis was performed as previously
described (20). Briefly, cultured
cells or tissues were harvested and lysed in ice-cold RIPA buffer
(Beyotime, Jiangsu, China) according to the manufacturer's
instructions. The total concentrations of protein were determined
using the BCA Protein assay kit (Beyotime). Equal amounts of
proteins (30 µg) were separated by 10% sodium
dodecylsulfate-polyacrylamide gels (SDS-PAGE, Pierce, Rockford, IL,
USA) and transferred onto nitrocellulose membranes (Millipore,
Madison, WI, USA). After blocking with 5% non-fat dry milk in TBST
(20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% Tween-20), the membrane
was incubated with mouse anti-human PIK3R3 monoclonal antibody
(1:1000; Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA), mouse
anti-human AKT monoclonal antibody (1:1000; Santa Cruz
Biotechnology Inc.), mouse anti-human p-AKT monoclonal antibody
(1:1000; Santa Cruz Biotechnology Inc.), mouse anti-human mTOR
monoclonal antibody (1:1000; Santa Cruz Biotechnology Inc.), mouse
anti-human p-mTOR monoclonal antibody (1:1000; Santa Cruz
Biotechnology Inc.) and mouse anti-human GAPDH monoclonal antibody
(1:5000; Santa Cruz Biotechnology Inc.) at 4°C overnight. The
membrane was incubated with the goat anti-mouse IgG conjugated to
horseradish peroxidase antibody (1:5000; Santa Cruz Biotechnology
Inc.) at room temperature for 2 h. The protein bland was observed
using a chemiluminescent detection system (ECL, Thermo Scientific,
Rockford, IL, USA) and exposed to X-ray film (Thermo Fisher
Scientific).
Statistical analysis
All data are presented as the mean ± SD (standard
deviation) from at least three independent experiments. The SPSS
software package (version 18.0, SPSS Inc.; Chicago, IL, USA) was
used to perform the statistical analysis. A two tailed Student's
t-test was used to evaluate the significance of differences between
two groups. ANOVA was employed to analyze the significance of
differences in more than two groups. The relationship between
miR-365 and PIK3R3 expressions was tested using Spearman's
correlation analysis. The significance level was set as
P<0.05.
Results
miR-365 is downregulated in human
glioma cell lines and tissues
To examine levels of miR-365 in glioma, we first
measured the expression of miR-365 in four human glioma cell lines
(U251, U87, U118 and LN18) and normal human astrocytes (NHA) by
qRT-PCR. As shown in Fig. 1A, the
expression of miR-365 was markedly downregulated in all four human
HCC cell lines U251, U87, U118 and LN18 compared with the NHA cell
line. We also evaluated miR-365 expression in 36 glioma tissues and
adjacent normal tissues. Consistent with the results from cell
lines, miR-365 levels were significantly decreased in glioma
tissues compared with normal tissues (Fig. 1B). These results indicated that
miR-365 was downregulated in glioma.
miR-365 inhibits glioma cell
proliferation, migration and invasion
To explore the possible biological functions of
miR-365 in glioma, we transfected miR-365 mimic or miR-NC into U87
cells, which has lower expression of miR-365 (Fig. 1A), then transfection efficiency were
determined by qRT-PCR. The results showed that miR-365 expression
was higher in U87 cells transfected with miR-365 mimic compared to
cells transfected with miR-NC (Fig.
2A). To demonstrate the effect of miR-365 on glioma growth, MTT
assay was performed in glioma cells. As shown in Fig. 2B, overexpression of miR-365 in U87
attenuated cell proliferation from 48 to 96 h after transfection
(Fig. 2B). We also investigated the
effect of miR-365 overexpression on the migration and invasion
abilities of glioma cells by Transwell assay. We found that miR-365
overexpression significantly decreased the migration and invasion
capacity of U87 cells compared to miR-NC group (Fig. 2C and D). These results demonstrated
that miR-365 suppressed glioma cell proliferation, migration and
invasion.
PIK3R3 is a target of miR-365 in
glioma cells
To understand how miR-365 suppresses glioma growth,
migration and invasion, bioinformatics (miRTarBase and TargetScan)
were used to identify the target of miR-365. PIK3R3 was chosen as a
target of miR-365, since it has a binding sequence for miR-365 at
position (67–73) of 3′UTR (Fig.
3A). To verify PIK3R3 as a direct target of miR-363, luciferase
activity assay was performed. The results showed that miR-365
significantly inhibited the luciferase activity of wild-type 3′-UTR
of PIK3R3 in U87 cells, but luciferase activity in mutant-type
3′-UTR of PIK3R3 in U87 cells was unchanged (Fig. 3B). Moreover, we found that
overexpression of miR-365 significantly suppressed PIK3R3
expression on mRNA and protein levels (Fig. 3C and D). These results indicated
that PIK3R3 is a direct target of miR-365 in glioma cells.
PIK3R3 expression was upregulated and
inversely correlated with miR-365 expression in glioma tissues
Further experiments were performed to investigate
the expression of PIK3R3 in glioma tissues and adjacent normal
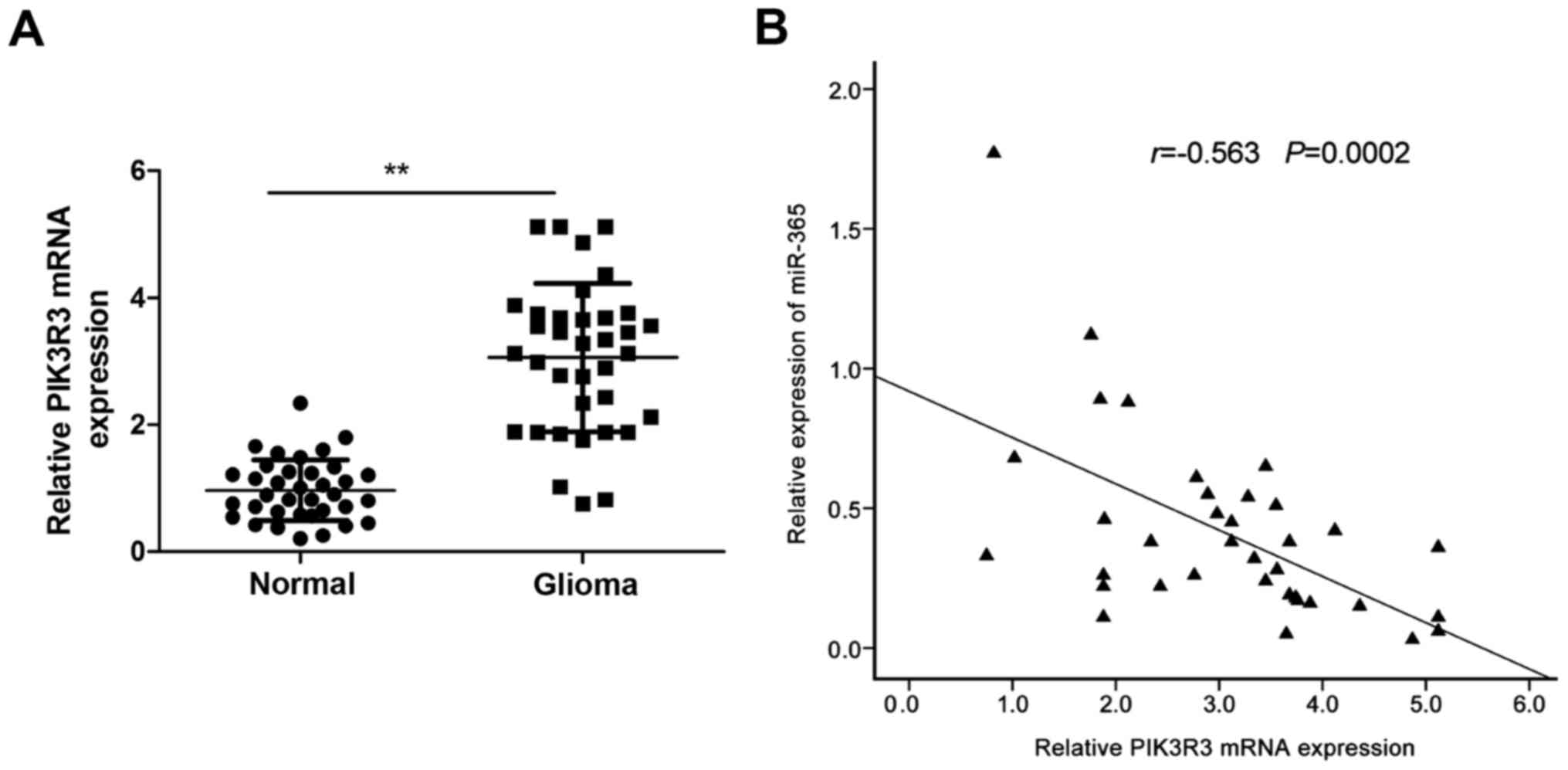
tissues by qRT-PCR. The result showed that the PIK3R3 mRNA
expression was upregulated in glioma tissues compared with adjacent
normal tissues (Fig. 4A). In
addition, Spearman's correlation analysis showed a reversed
correlation between miR-365 expression levels and PIK3R3 mRNA
levels in glioma tissues (r= −0.563, P=0.0002; Fig. 4B).
Overexpression of PIK3R3 rescues the
inhibition effect of miR-365 in glioma
To further illustrate whether miR-365 affects human
glioma cell proliferation, migration and invasion through PIK3R3,
U87 cells were co-transfected with miR-365 or miR-NC and the
overexpression PIK3R3 plasmid. Western blot analysis showed that
miR-365 overexpression significantly decreased PIK3R3 protein
expression, while overexpression PIK3R3 plasmid restored PIK3R3
expression (Fig. 5A). In addition,
we found that PIK3R3 overexpression was able to counteract the
inhibitory effect on cell proliferation (Fig. 5B), migration (Fig. 5C), and cell invasion (Fig. 5D) in glioma cells induced by miR-365
overexpression. These data indicated that miR-365 exerts
suppressive role in glioma by repressing PIK3R3.
Upregulation of miR-365 suppresses
tumor growth in mice
To investigate the role of miR-365 in tumor growth
in vivo, U87 cells (2×106), stable overexpression
of miR-365 mimic or miR-NC were injected subcutaneously into nude
mice and the mice were monitored closely for tumor growth. As shown
in Fig. 6A, tumor growth was slower
in miR-365 overexpression group than that of miR-NC overexpression
(Fig. 6A). At 35 days after
injection, mice was sacrified and tumor tissues were excised and
weighed. We found that tumor weight was decreased in miR-365
overexpression group compared to miR-NC overexpression group
(Fig. 6B). In addition, miR-365 and
PIK3R3 expression were determined in tumor xenograft tumors by
qRT-PCR and western blotting, respectively. Compared to miR-NC
overexpression group, miR-365 expression was increased (Fig. 6C), and PIK3R3 protein expression was
decreased in miR-365 overexpression group (Fig. 6D). These results suggested that
miR-365 suppressed glioma growth in vivo by repressing
PIK3R3.
miR-365 regulates the AKT/mTOR
signaling pathway
PIK3R3 has been showed to be involved in tumor
progression by regulating AKT/mTOR signal pathway (21–23).
Since PIK3R3 was confirmed as a target of miR-365 in the above
results, we investigated whether miR-365 could regulate the
AKT/mTOR pathway. We detected AKT, p-AKT, mTOR and p-mTOR protein
expression in glioma cells and xenograft tumor tissues from nude
mice. We found that miR-365 overexpression significantly inhibited
phosphorylation of AKT and mTOR in glioma cells and xenograft tumor
tissues. Total AKT and mTOR protein levels did not change. These
data might indicate that miR-365 inhibit cell proliferation and
invasion of glioma through indirectly regulating the AKT/mTOR
signaling pathway (Fig. 7).
Discussion
Recently number of miRNAs have been identified to
function as a tumor suppressor or an oncogene in glioma by
regulating their target molecule (11,12).
For example, Zhu et al reported that miR-217 inhibited
proliferation, colony formation, migration and invasion of glioma
cells by repressing Runx2 (20).
Chen et al found that overexpression of miR-19a by a miR-19a
mimic promoted glioma cell proliferation and invasion by targeting
the Ras homolog family member B (RhoB) (24). Peng et al also found that
miR-506 was downregulated in glioma tissues and cell lines, and
functions as a novel tumor suppressor to inhibit the proliferation,
colony formation, migration and invasion of glioma cells in
vitro, and suppress glioma tumor growth in vivo by
targeting STAT3 (25). In the
present study, our results revealed that miR-365 expression was
significantly downregulated in glioma tissues and cell lines
compared to adjacent normal tissues and the normal cell line. Our
results also demonstrated that miR-365 overexpression significantly
decreased cell proliferation, migration and invasion of glioma
in vitro, and suppressed tumor growth in vivo. These
data suggest that miR-365 may play crucial role in glioma
development.
Aberrant expression of miR-365 has been found in
various human cancers. In gastric cancer (16), cutaneous squamous cell carcinoma
(26) and pancreatic cancer
(27), miR-365 expression is
frequent upregulated and acts as an oncogene. On the contrary, in
lung cancer (13,14), melanoma (15), osteosarcoma (17), and colon cancer (18), miR-365 expression was downregulated
and functions as a tumor suppressor. However, the function and
relevant mechanisms of miR-365 in glioma remains unclear. Herein,
we found that miR-365 expression was significantly downregulated in
glioma tissues and cell lines, and that miR-365 significantly
inhibited cell proliferation, invasion and migration of glioma
cells, and suppressed tumor growth in nude mice. These results
suggested that miR-365 might function as a tumor suppressor in
glioma.
PIK3R3 (phosphoinositide-3-kinase regulatory subunit
3), a member of the phosphatidylinositol 3-kinase (PI3K) family,
has been suggested to play crucial roles in diverse biological
processes, such as cell proliferation, differentiation,
carcinogenesis and tumor angiogenesis (28,29).
Accumulating evidence suggested that PIK3R3 was involved in tumor
development and progression, and functions as an oncogene in
multiple cancers, including ovarian cancer (22), gastric cancer (30), hepatocellular carcinoma (21), lung cancer (22), colorectal cancer (31), and breast cancer (28). It has been shown that PIK3R3 could
regulate the AKT/mTOR pathway, which contribute to promotion of
cancer progression (21–23). In glioma, it has been found that
overexpression of PIK3R3 can promote growth of glioma cells in
vitro (32), which imply PIK3R3
as an oncogene in glioma. In this study, PIK3R3 was proved to be a
direct target of miR-365 in glioma cells, and its mRNA expression
was inversely correlated with miR-365 expression in clinical glioma
tissues. Overexpression of PIK3R3 in U87 cells reversed the
inhibition effected on proliferation, migration and invasion
induced by miR-365 overexpression. Of note, miR-365 was able to
regulate PIK3R3 and its downstream protein p-AKT, p-mTOR expression
in vitro and in vivo, which are key participants in
the AKT/mTOR pathway. These findings might imply that miR-365
exerts it tumor-suppressing functions in glioma by regulating the
PIK3R3/AKT/ mTOR signal pathway.
Our study provides evidence that miR-365 expression
was downregulated in glioma tissues and cell lines, and that
miR-365 suppressed glioma cell proliferation, migration, and
invasion in vitro, as well as glioma growth in vivo
by directly targeting PIK3R3 and indirectly regulating the AKT/mTOR
pathway. These results suggest that miR-365 could serve as a
potential novel target for future glioma therapy.
References
|
1
|
Reardon DA, Rich JN, Friedman HS and
Bigner DD: Recent advances in the treatment of malignant
astrocytoma. J Clin Oncol. 24:1253–1265. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Clarke J, Butowski N and Chang S: Recent
advances in therapy for glioblastoma. Arch Neurol. 67:279–283.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Grauer OM, Wesseling P and Adema GJ:
Immunotherapy of diffuse gliomas: Biological background, current
status and future developments. Brain Pathol. 19:674–693. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fabian MR, Sonenberg N and Filipowicz W:
Regulation of mRNA translation and stability by microRNAs. Annu Rev
Biochem. 79:351–379. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Guo H, Ingolia NT, Weissman JS and Bartel
DP: Mammalian microRNAs predominantly act to decrease target mRNA
levels. Nature. 466:835–840. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Almeida MI, Reis RM and Calin GA: MicroRNA
history: Discovery, recent applications, and next frontiers. Mutat
Res. 717:1–8. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Farazi TA, Spitzer JI, Morozov P and
Tuschl T: miRNAs in human cancer. J Pathol. 223:102–115. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
McManus MT: MicroRNAs and cancer. Semin
Cancer Biol. 13:253–258. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
England B, Huang T and Karsy M: Current
understanding of the role and targeting of tumor suppressor p53 in
glioblastoma multiforme. Tumour Biol. 34:2063–2074. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tivnan A and McDonald KL: Current progress
for the use of miRNAs in glioblastoma treatment. Mol Neurobiol.
48:757–768. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Qi J, Rice SJ, Salzberg AC, Runkle EA,
Liao J, Zander DS and Mu D: MiR-365 regulates lung cancer and
developmental gene thyroid transcription factor 1. Cell Cycle.
11:177–186. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sun R, Liu Z, Ma G, Lv W, Zhao X, Lei G
and Xu C: Associations of deregulation of mir-365 and its target
mRNA TTF-1 and survival in patients with NSCLC. Int J Clin Exp
Pathol. 8:2392–2399. 2015.PubMed/NCBI
|
|
15
|
Bai J, Zhang Z, Li X and Liu H:
MicroRNA-365 inhibits growth, invasion and metastasis of malignant
melanoma by targeting NRP1 expression. Int J Clin Exp Pathol.
8:4913–4922. 2015.PubMed/NCBI
|
|
16
|
Guo SL, Ye H, Teng Y, Wang YL, Yang G, Li
XB, Zhang C and Yang X, Yang ZZ and Yang X: Akt-p53-miR-365-cyclin
D1/cdc25A axis contributes to gastric tumorigenesis induced by PTEN
deficiency. Nat Commun. 4:25442013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gao J, Zhao P, Chen X, Wang W, Li Y, Xi W,
Zhang W, Hu P, Wang T and Shan L: miR-365 inhibits proliferation
and promotes apoptosis of SOSP9607 osteosarcoma cells. Xi Bao Yu
Fen Zi Mian Yi Xue Za Zhi. 32:44–48. 2016.(In Chinese). PubMed/NCBI
|
|
18
|
Nie J, Liu L, Zheng W, Chen L, Wu X, Xu Y,
Du X and Han W: microRNA-365, down-regulated in colon cancer,
inhibits cell cycle progression and promotes apoptosis of colon
cancer cells by probably targeting Cyclin D1 and Bcl-2.
Carcinogenesis. 33:220–225. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu K, Li X, Cao Y, Ge Y, Wang J and Shi
B: MiR-132 inhibits cell proliferation, invasion and migration of
hepatocellular carcinoma by targeting PIK3R3. Int J Oncol.
47:1585–1593. 2015.PubMed/NCBI
|
|
20
|
Zhu Y, Zhao H, Feng L and Xu S:
MicroRNA-217 inhibits cell proliferation and invasion by targeting
Runx2 in human glioma. Am J Transl Res. 8:1482–1491.
2016.PubMed/NCBI
|
|
21
|
Cao G, Dong W, Meng X, Liu H, Liao H and
Liu S: MiR-511 inhibits growth and metastasis of human
hepatocellular carcinoma cells by targeting PIK3R3. Tumour Biol.
36:4453–4459. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xu L, Wen Z, Zhou Y, Liu Z, Li Q, Fei G,
Luo J and Ren T: MicroRNA-7-regulated TLR9 signaling-enhanced
growth and metastatic potential of human lung cancer cells by
altering the phosphoinositide-3-kinase, regulatory subunit 3/Akt
pathway. Mol Biol Cell. 24:42–55. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang L, Huang J, Yang N, Greshock J,
Liang S, Hasegawa K, Giannakakis A, Poulos N, O'Brien-Jenkins A,
Katsaros D, et al: Integrative genomic analysis of
phosphatidylinositol 3′-kinase family identifies PIK3R3 as a
potential therapeutic target in epithelial ovarian cancer. Clin
Cancer Res. 13:5314–5321. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen Q, Guo W, Zhang Y, Wu Y and Xiang J:
MiR-19a promotes cell proliferation and invasion by targeting RhoB
in human glioma cells. Neurosci Lett. 628:161–166. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Peng T, Zhou L, Zuo L and Luan Y: MiR-506
functions as a tumor suppressor in glioma by targeting STAT3. Oncol
Rep. 35:1057–1064. 2016.PubMed/NCBI
|
|
26
|
Zhou M, Liu W, Ma S, Cao H, Peng X, Guo L,
Zhou X, Zheng L, Guo L, Wan M, et al: A novel onco-miR-365 induces
cutaneous squamous cell carcinoma. Carcinogenesis. 34:1653–1659.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hamada S, Masamune A, Miura S, Satoh K and
Shimosegawa T: MiR-365 induces gemcitabine resistance in pancreatic
cancer cells by targeting the adaptor protein SHC1 and
pro-apoptotic regulator BAX. Cell Signal. 26:179–185. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Klahan S, Wu MS, Hsi E, Huang CC, Hou MF
and Chang WC: Computational analysis of mRNA expression profiles
identifies the ITG family and PIK3R3 as crucial genes for
regulating triple negative breast cancer cell migration. BioMed Res
Int. 2014:5365912014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xia X, Cheng A, Akinmade D and Hamburger
AW: The N-terminal 24 amino acids of the p55 gamma regulatory
subunit of phosphoinositide 3-kinase binds Rb and induces cell
cycle arrest. Mol Cell Biol. 23:1717–1725. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhou J, Chen GB, Tang YC, Sinha RA, Wu Y,
Yap CS, Wang G, Hu J, Xia X, Tan P, et al: Genetic and
bioinformatic analyses of the expression and function of PI3K
regulatory subunit PIK3R3 in an Asian patient gastric cancer
library. BMC Med Genomics. 5:342012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang G, Yang X, Li C, Cao X, Luo X and Hu
J: PIK3R3 induces epithelial-to-mesenchymal transition and promotes
metastasis in colorectal cancer. Mol Cancer Ther. 13:1837–1847.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Soroceanu L, Kharbanda S, Chen R, Soriano
RH, Aldape K, Misra A, Zha J, Forrest WF, Nigro JM, Modrusan Z, et
al: Identification of IGF2 signaling through
phosphoinositide-3-kinase regulatory subunit 3 as a
growth-promoting axis in glioblastoma. Proc Natl Acad Sci USA.
104:3466–3471. 2007. View Article : Google Scholar : PubMed/NCBI
|