Introduction
Breast cancer (BC) is the most commonly diagnosed
cancer among women worldwide and a leading cause of cancer-related
mortality in developed countries (1). According to recent research, BC has
risen to have the second highest mortality rate among cancers
(2). As a disease with a complex,
multifarious genetic and biochemical background, the exact
mechanisms of breast carcinogenesis remain unclear. Hence,
screening for more useful prognostic and predictive markers that
contribute to BC progression is urgently needed to identify more
effective therapies.
Karyopherin (Kpn) proteins, all of which have an
N-terminal RanGTP-binding domain, a C-terminal cargo-binding
domain, and the capacity to bind components of the nuclear pore
complex (NPC), are nuclear transport receptors that function in
transporting cargo proteins and certain RNAs into and out of the
cell nucleus via the NPC (3).
Nuclear import via Kpn β-1 (Kpnβ1) can occur either by Kpnβ1 acting
as an autonomous nuclear transport receptor, or through its
association with an adaptor protein, such as Kpnα (also known as
importin alpha), in which case the import process is known as
classical nuclear import (4). Kpnβ1
is involved in importing proteins, such as receptor tyrosine kinase
2 (ErbB-2) (5), epidermal growth
factor receptor (EGFR) (6), and
fibroblast growth factor 1 (FGF1) (7). Furthermore, several studies have
extended the role of Kpn proteins in the regulation of the cell
cycle, mitosis, and replication (8). Notably, recent studies revealed that
Kpn proteins also have a key role in various cancers. For example,
Kpnα2 expression was found to be associated with gastric cancer
(9), prostate cancer (10), epithelial ovarian carcinoma
(11), BC (12), endometrial cancer (13), hepatocellular carcinoma (14) and esophageal squamous cell carcinoma
(15). Furthermore, Kpn expression
was found to be associated with several malignant tumors such as
cervical cancer (16), malignant
peripheral nerve sheath tumors (17), and head, neck and lung cancer
(18). Accordingly, Kpnβ1 exhibits
clear potential as an anticancer therapeutic target (19). Although Kpn has been reported to be
involved in chromosome stability in BC patients (20), there is no report demonstrating the
function and mechanism of Kpn in the progression and prognosis of
BC, to the best of our knowledge.
The tyrosine kinase receptor Her2 is amplified in
20–30% of human cancers and its overexpression has been associated
with poor patient prognosis (21).
Recently, evidence has highlighted that nuclear Her2 may play a
more aggressive role during tumor progression (22). Nuclear Her2 has been determined to
act as a transcription factor for genes such as cyclin D1, FGF2 and
cyclooxygenase-2 (COX-2) (5).
Despite recent research on the translocation of Her2 to the
nucleus, the mechanism by which Her2 travels from the cell surface
to the nucleus is unclear.
In this study we focused on Kpnβ1 expression in
primary and BC cell lines, its association with clinicopathological
features, and its prognostic value for BC patient survival. This
study provided evidence for a role of Kpnβ1 in contributing to BC
phenotype. Furthermore, we investigated the possible role of Kpnβ1
in the proliferation and apoptosis of BC cell lines. Based on our
findings, we suggest that Kpnβ1 could be a novel therapeutic target
for BC.
Materials and methods
Patients and tissue samples
BC sections were obtained from 140 patients who had
undergone breast surgical resection at the Department of General
Surgery of the Affiliated Hospital of Nantong University between
April 2002 and May 2010. The tissues had been formalin-fixed and
paraffin-embedded for pathological diagnosis and
immunohistochemical study and were authenticated histologically.
The TNM tumor staging and histological grade were performed
according to the World Health Organization guidelines. Patient
information corresponding with these tissues was subsequently
obtained from return patient visits to the hospital and telephone
contact. The clinical features of all the patients, including age,
tumor size, histologic grade, axillary lymph node status, and
histology are shown in Table I.
Fresh BC and normal tissue samples were immediately frozen in
liquid nitrogen after surgical resection and maintained at −80°C.
All human tissues were collected using protocols approved by the
Ethics Committee of the Affiliated Cancer Hospital of Nantong
University (Nantong, Jiangsu, China). The median patient age was 54
years (range, 34–88 years). The median 7-year follow-up time for
the 140 patients was 30 months (range, 6–90 months). Histological
grade was classified as follows: well-differentiated (grade I,
n=17), moderately differentiated (grade II, n=60), and poorly
differentiated (grade III, n=63). The majority of cases included
infiltrating ductal carcinoma (n=109, 78%) and the remaining 22% of
the cases consisted of other types. The main patient
clinicopathological variables are shown in Table I. Informed consent was obtained from
all patients.
 | Table I.Expression of Kpnβ1, Ki-67 and
clinicopathological parameters in 140 breast cancer specimens. |
Table I.
Expression of Kpnβ1, Ki-67 and
clinicopathological parameters in 140 breast cancer specimens.
|
|
| Kpnβ1
expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Parameters | Total | Low | High |
P-valuea | χ2 |
|---|
| Age (years) |
|
|
|
|
|
|
≤50 | 57 | 28 | 29 | 0.975 | 0.001 |
|
>50 | 83 | 41 | 42 |
|
|
| Grade |
|
|
|
|
|
| I | 17 | 11 | 6 | 0.048a | 6.081 |
| II | 60 | 34 | 26 |
|
|
|
III | 63 | 24 | 39 |
|
|
| ER |
|
|
|
|
|
|
Negative | 69 | 36 | 33 | 0.500 | 0.454 |
|
Positive | 71 | 33 | 38 |
|
|
| PR |
|
|
|
|
|
|
Negative | 70 | 34 | 36 | 0.866 | 0.029 |
|
Positive | 70 | 35 | 35 |
|
|
| Her2 |
|
|
|
|
|
|
Negative | 68 | 38 | 30 | 0.129 | 2.203 |
|
Positive | 72 | 31 | 41 |
|
|
| Tumor size |
|
|
|
|
|
|
≤2×2×2 | 77 | 41 | 26 | 0.007a | 7.290 |
|
>2×2×2 | 63 | 28 | 45 |
|
|
| Axillary lymph node
status |
|
|
|
|
|
| N0 | 52 | 31 | 21 | 0.060 | 3.532 |
| Nx | 88 | 38 | 50 |
|
|
| Nerve invasion and
metastasis |
|
|
|
|
|
|
Negative | 85 | 45 | 40 | 0.282 | 1.157 |
|
Positive | 55 | 24 | 31 |
|
|
| Vascular
metastasis |
|
|
|
|
|
|
Negative | 75 | 42 | 33 | 0.088 | 2.914 |
|
Positive | 65 | 27 | 38 |
|
|
| Histology |
|
|
|
|
|
|
Ductal | 109 | 59 | 50 | 0.032a | 4.619 |
|
Others | 31 | 10 | 21 |
|
|
| Ki-67 |
|
|
|
|
|
|
Low | 49 | 18 | 31 | 0.015a | 5.894 |
|
High | 91 | 38 | 53 |
|
|
Antibodies
All antibodies were sourced from Santa Cruz
Biotechnology (Santa Cruz, CA, USA) unless otherwise specified.
Antibodies for immunohistochemistry included anti-Kpnβ1 (sc-11367,
1:200) and anti-Ki-67 (AB9260, 1:100; Millipore, Bedford, MA, USA);
for western blot analysis they included anti-Kpnβ1 (1:500),
anti-proliferating cell nuclear antigen (PCNA; 1:1,000),
anti-β-actin antibody (1:1,000), anti-cyclin D1 antibody (1:1,000)
and anti-GAPDH (sc-7196, 1:1,000); and for immunofluorescent
staining they included anti-Kpnβ1 (1:500), anti-Her2 (1:400), and
anti-actin monoclonal antibody (1:1,000).
Immunohistochemical staining
In brief, tissue slices were dewaxed in xylene,
rehydrated in graded ethanol and endogenous peroxidase activity was
blocked by steeping in 3% methanolic peroxide for 20 min. The
sections were then heated to 121°C in an autoclave for 10 min in
0.1 M citrate buffer (pH 6.0) to retrieve the antigen. After being
rinsed in phosphate-buffered saline (PBS, pH 7.2) for 5 min (three
times), tissue sections were incubated with the rabbit anti-human
Kpnβ1 antibody (diluted 1:200) and the mouse anti-human Ki-67
antibody (diluted 1:500) for 3 h at room temperature. After being
washed with PBS, the peroxidase reaction was visualized by
incubating the sections with DAB [0.1% phosphate-buffered solution
(PBS), 0.02% diaminobenzidine tetrahydrochloride, and 3%
H2O2]. Finally, the sections were
counterstained with hematoxylin, dehydrated through graded alcohol,
and mounted under a cover slip after being rinsed in water.
Immunohistochemical evaluation
All immunostained sections were randomly evaluated
by three independent observers in a blinded manner using a Leica
fluorescence microscope (Leica Microsystems GmbH, Wetzlar,
Germany). Five fields of view were chosen per slide and at least
500 cells were counted per view at high power. To evaluate the
immunoreaction of Kpnβ1, the staining intensity was estimated in
comparison to the control and scored as follows: I, the reaction
was not easily perceived from the background or <5% of tumor
cells were stained; II, 5–30% of tumor cells were positively
stained; and III, >30% of tumor cells were positively-stained
(Xue et al, 2013).
Western blot analysis
Before immunoblotting, tissues were immediately
homogenized in lysis buffer [1% NP-40, 50 mmol/l Tris (pH 7.5), 5
mmol/l EDTA, 1% SDS, 1% sodium deoxycholate, 1% Triton X-100, 1
mmol/l PMSF, 10 mg/ml aprotinin and 1 mg/ml leupeptin] and the
cells were washed three times with ice-cold PBS and resuspended in
2X lysis buffer (50 mM Tris-HCl, 120 mM NaCl, 0.5% NP-40, 100 mM
NaF, 200 mM Na3VO4, and protease inhibitor
mixture). The cells were then denatured at 100°C for 15 min. Total
protein concentration was then determined with a Bio-Rad protein
assay (Bio-Rad Laboratories, Hercules, CA, USA). All protein
samples were stored at −20°C. For sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), samples were
denatured at 100°C for 3 min. The proteins were then separated with
SDS-PAGE and subsequently transferred to polyvinylidene difluoride
filter (PVDF) membranes (Millipore). The membranes were then
blocked in 5% skimmed-milk in Tris-buffered saline-Tween (TBST: 20
mM Tris, 150 mM NaCl, 0.05% Tween-20) for 2 h at room temperature,
and then incubated with primary antibodies overnight at 4°C for 6–8
h at room temperature. The membranes were then washed with TBST
three times (5 min/wash) and incubated with horseradish
peroxidase-linked IgG secondary antibodies for 2 h at room
temperature. The protein bands were detected using the enhanced
chemiluminescence (ECL) detection system (Pierce, Rockford, IL,
USA) and the band intensities were assessed using ImageJ analysis
software (Wayne Rasband; National Institutes of Health, Bethesda,
MD, USA).
Cell culture and cell cycle
analysis
Two human BC cell lines, SKBR-3 and MDA-MB-231, were
obtained from the Cell Bank of the Chinese Academy of Sciences
(Shanghai, China) and used in this study. The MDA-MB-231 cell line
was maintained in Dulbeccos modified Eagles medium (DMEM;
Gibco-BRL, Grand Island, NY, USA) supplemented with 10%
heat-inactivated fetal bovine serum (HI-FBS), 2 mM L-glutamine, and
100 U/ml penicillin-G and incubated at 37°C and 5% CO2.
The SKBR-3 cell line was maintained in Mccoys 5A medium (Gibco-BRL)
supplemented with 10% HI-FBS, 2 mM L-glutamine, and 100 U/ml
penicillin-G and incubated at 37°C and 0% CO2.
Starvation and re-feeding was used to
imitate the cell cycle
First, DMEM or Mccoys 5A medium without FBS was used
to incubate the MDA-MB-231 or SKBR-3 cells, respectively, for 48 h
to synchronize cells, and was then replaced by complete medium.
Subsequently, the cells were rapidly harvested at specified
time-points and fixed in 70% ethanol for at least 24 h at −20°C,
and then incubated with 1 mg/ml RNase A for 20 min at 37°C. The
cells were then stained with 0.5% Tween-20/propidium iodide (PI, 50
mg/ml) in PBS and analyzed using a Becton-Dickinson flow cytometer
(BD FACScan; Becton-Dickinson, San Jose, CA, USA) and CellQuest
acquisition and analysis programs.
RNA interference of Kpnβ1
Small interference RNAs (siRNA) were designed and
chemically synthesized by GeneChem (Shanghai, China). The
Kpnβ1-specific siRNA target sequences were as follows:
Kpnβ1-siRNA#0, 5-GAGATCGAAGACTA ACAAA-3; Kpnβ1-siRNA#1,
5-CAGTGTAGTTGTTCGA GAT-3; Kpnβ1-siRNA#2, 5-ACGAGAAGTCAAGAAC TAT-3;
and non-specific scrambled siRNA sequence, 5-GCTGTTAGTGAGCTAAGTA-3.
Cells were transfected with 100 nmol/l of siRNA duplexes using
Lipofectamine Plus reagent (Invitrogen, Carlsbad, CA, USA)
according to the manufacturer's protocol.
Plate colony formation assay
SKBR-3 and MDA-MB-231 cells pretreated with
Kpnβ1-siRNA and non-specific scrambled siRNA (2,000 cells/plate)
were cultured in 5 ml of DMEM supplemented with 10% FBS in a 6-cm
plate. After 14 days, the colonies were washed with PBS, fixed with
methanol for 30 min, and stained with crystal violet for 30 min.
Clearly visible colonies (>50 mm in diameter) were counted as
positive for growth.
Cell proliferation assay
Cell Counting Kit-8 (CCK-8; Dojindo Laboratories,
Kumamoto, Japan) was employed to assess cell proliferation. In
brief, the cells were seeded into a 96-well cell culture cluster
plate (Corning Incorporated, Corning, NY, USA) at a concentration
of 2×104 cells/well in a volume of 100 µl and grown
overnight. The cells were then incubated with CCK-8 reagents for 2
h at 37°C and the absorbance was quantified on an automated plate
reader. Each condition was performed in triplicate and each
experiment was repeated three times.
Flow cytometric analysis of cell
apoptosis
SKBR-3 cells transfected with Kpnβ1-siRNA and
control-siRNA were cultured for 48 h and harvested. Muse™ Annexin V
and Dead Cell reagent (60 µl, part no. 4700-1485, 100 tests/bottle)
was then added to each tube with 60 µl of cell suspension. After
incubation for 20 min at room temperature in the dark, the
apoptosis assay was performed using the Muse™ Cell Analyzer
(Millipore) according to the manufacturers instructions.
Coimmunoprecipitation
Cellular extracts were lysed in lysis buffer (150 mM
NaCl, 1 mM EDTA, 20 mM Tris (pH 7.5), 0.5% NP-40, 1 mM NaF, 1 mM
MNa3VO4, 1 mM PMSF, and 2 g/ml aprotinin) and
incubated with primary antibodies at 4°C followed by incubation
with protein G-sepharose. The immunocomplexes were washed three
times with lysis buffer. For single immunoprecipitation, the bound
proteins were eluted by boiling the samples in SDS sample buffer
containing 2-mercaptoethanol. For sequential double
immunoprecipitation, the bound proteins were eluted from the
sepharose beads by boiling for 3 min in 25 l of SDS lysis buffer
(20 mM Tris (pH 7.5), 50 mM NaCl, 1% SDS, and 1 mM dithiothreitol).
Samples were cooled and the supernatants were diluted with 225 l of
lysis buffer containing the appropriate antibodies and incubated
overnight at 4°C. The immunocomplexes were then precipitated with
protein A-Sepharose beads, washed with lysis buffer, and
resuspended in SDS sample buffer. The eluted proteins were then
boiled for 5 min and subjected to SDS-PAGE.
Immunofluorescent staining assay
Following prior treatment, the cells were fixed with
paraformaldehyde for 40 min at room temperature, permeabilized for
15 min with 1% Triton-X, blocked for 2 h with 1% BSA in PBS at 4°C,
and incubated overnight at 4°C with anti-Kpnβ1 monoclonal and
anti-Her2 antibodies. The cells were then washed three times with
PBS followed by incubation for 2 h with anti-rabbit IgG secondary
antibody, anti-mouse IgG secondary antibody, or anti-actin
monoclonal antibody. Nuclear staining was achieved with DAPI
(4,6-diamidino-2-phenylindole; fluorescence) before mounting. After
being washed with PBS, the slides were air-dried, mounted with
anti-fading mounting reagent, and examined under a fluorescent
microscope.
Statistical analysis
Data were analyzed by SPSS 17.0 software (SPSS,
Inc., Chicago, IL, USA). Statistical significance of the
correlations between Kpnβ1 and Ki-67 expression and the
clinicopathological features were analyzed using the Chi-squared
test. The Kaplan-Meier method was used to analyze survival curves
and the log-rank test. Multivariate analysis was performed using
Coxs proportional hazards model. The results are expressed as means
± standard deviation (SD). P<0.05 was considered to indicate a
statistically significant result.
Results
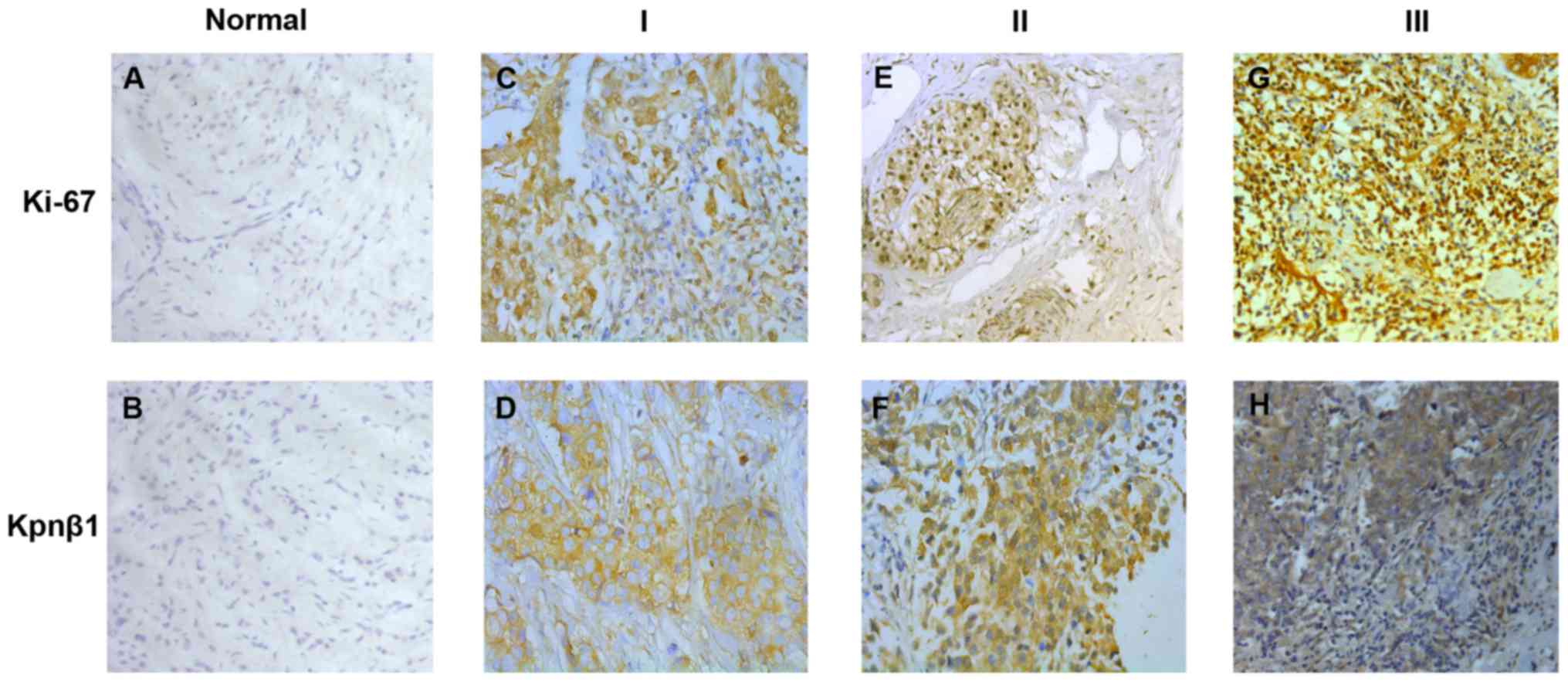
The expression of Kpnβ1 and Ki-67 in
human malignant breast tissues and cell lines
To identify whether Kpnβ1 was associated with BC,
immunohistochemistry was applied to detect the expression and
distribution of Kpnβ1 and Ki-67 in paraffin-embedded mammary tissue
sections screened from 140 patients. For statistical analysis of
Kpnβ1 and Ki-67 expression levels, we defined Kpnβ1 antibody
specificity and scored Kpnβ1 staining as weakly positive (I),
moderately positive (II), and strongly positive (III) based on the
percentage of positively stained cells and staining intensity. When
considering the characteristics of the data, we then classified
group I as low expression and groups II and III as high expression.
The results of these experiments revealed that Kpnβ1 was mainly
located in the cytoplasm with high expression of Kpnβ1
significantly consistent with high expression of nuclear Ki-67
(Fig. 1).
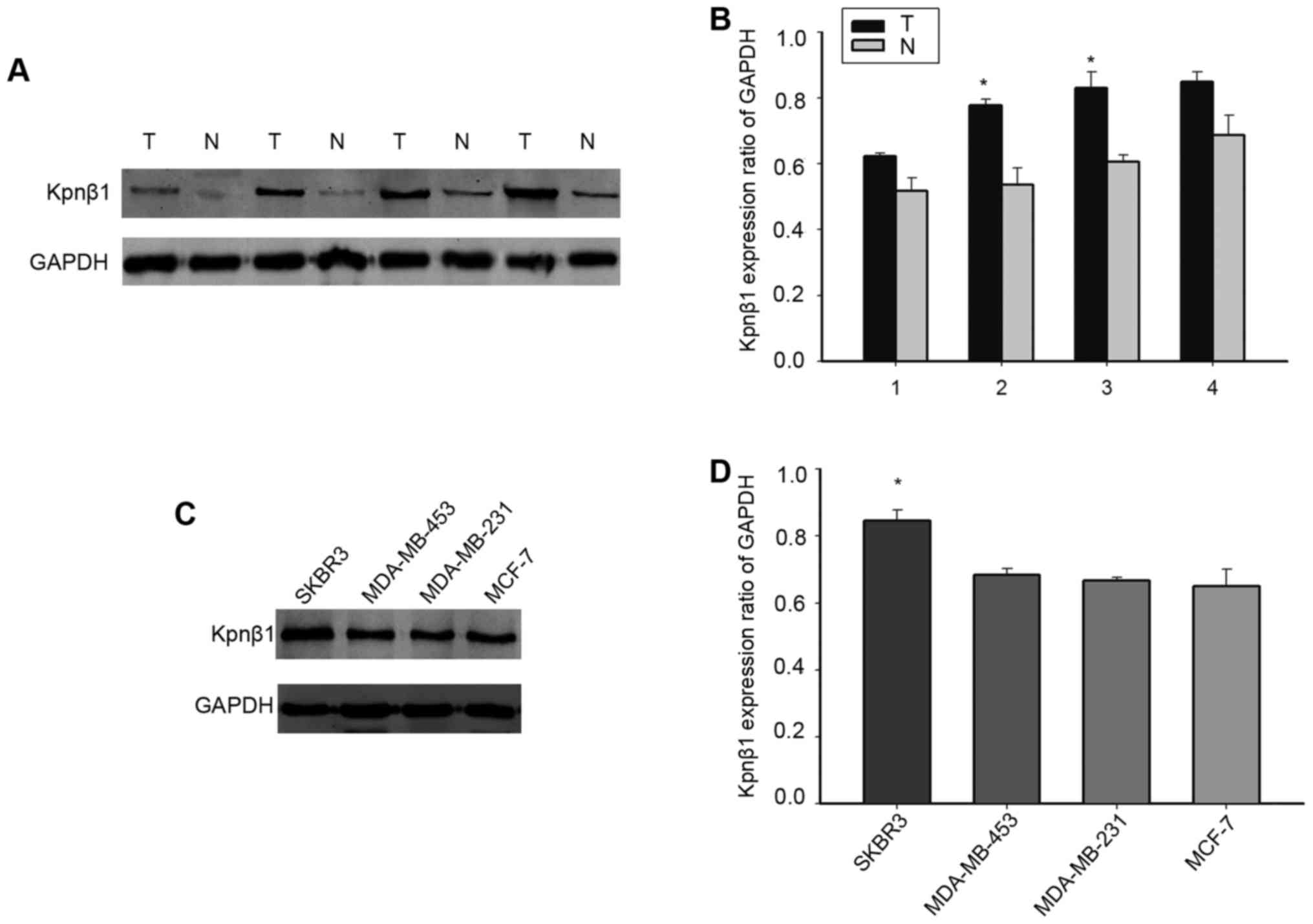
To verify whether Kpnβ1 is highly expressed in human
BC, we further detected the expression of Kpnβ1 by western blot
analysis in four pairs of fresh samples. As shown in Fig. 2A and B, the expression of Kpnβ1 was
much higher in malignant tumors when compared with that noted in
the adjacent normal tissues, consistent with our previous
observation. To further detect the expression of Kpnβ1 in BC cell
lines, we selected SKBR-3, MDA-MB-453, MCF-7, and MDA-MB-231 cells.
Kpnβ1 was expressed to a greater extent in SBKR-3 cells compared
with the other cell lines (Fig. 2C and
D). SKBR-3 cells were therefore used for subsequent
experimentation. We supposed that the expression of Kpnβ1 may be
related to the proliferation of BC based on these results.
The relevance of Kpnβ1 expression with
clinicopathological variables and BC patient survival
To further examine the pathophysiological
significance of Kpnβ1 with respect to tumor characteristics and
behavior, the clinicopathological data were summarized and are
shown in Table I. We found that
Kpnβ1 expression was markedly correlated with grade (P=0.048),
tumor size (P=0.007), histology (P=0.032), and Ki-67 expression
(P=0.015). Whereas there was no association between Kpnβ1 and age
(P=0.975), estrogen receptor (ER) (P=0.500), progesterone receptor
(PR) (P=0.866), Her2 expression (P=0.129), axillary lymph node
status (P=0.060), nerve invasion and metastasis (P=0.282) and
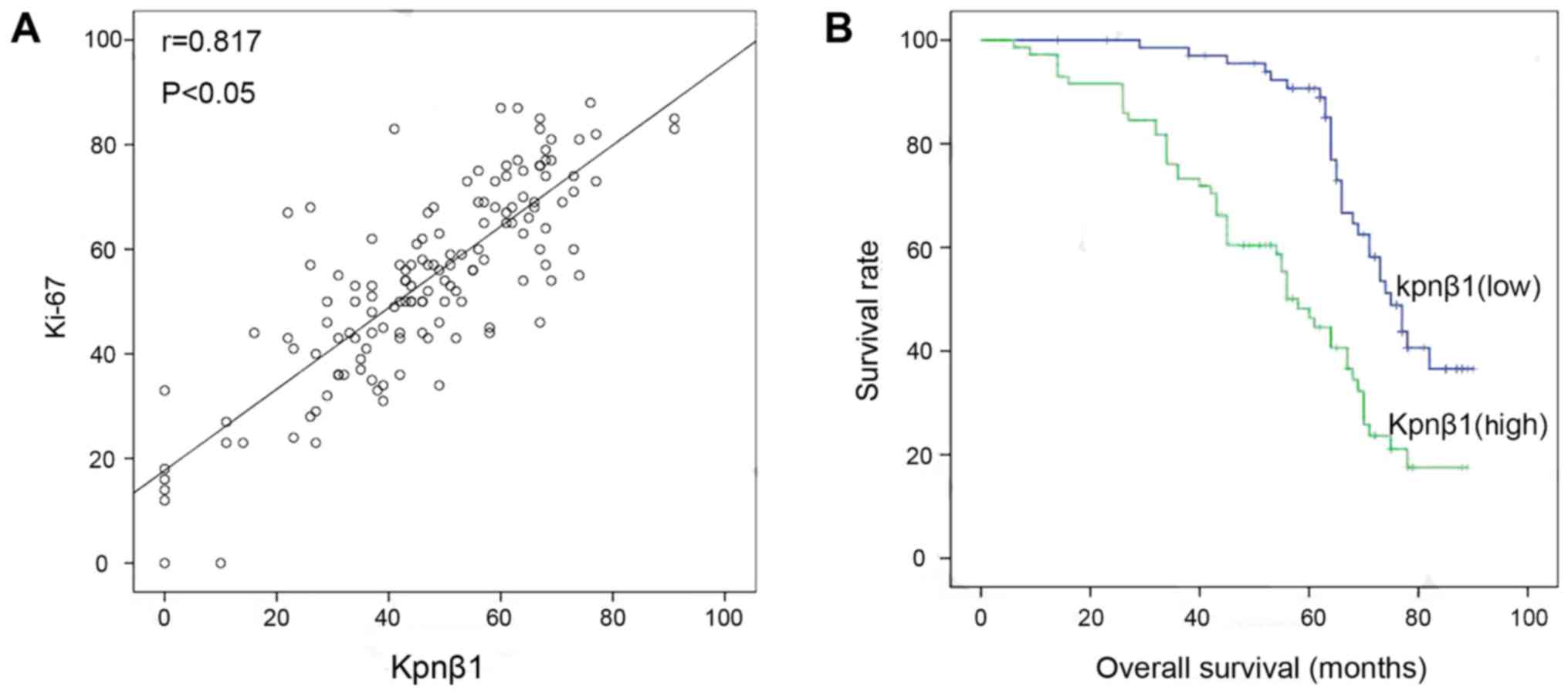
vascular metastasis (P=0.088). Furthermore, the relationship
between the Ki-67 proliferation index and Kpnβ1 expression in
breast carcinoma tissues revealed a significant correlation
according to Spearmans correlation coefficient (Fig. 3A). We next sought to understand the
correlation between Kpnβ1 expression level and patient survival
using Kaplan-Meier analysis. At the end of the clinical follow-up,
survival analysis was restricted to 140 patients with complete
follow-up data and results for Kpnβ1 expression by
immunohistochemistry. Kaplan-Meier survival curves revealed that
high expression of Kpnβ1 was significantly associated with poor
overall survival (Fig. 3B). In
addition, in the univariate analysis, when all variables were
compared separately to survival status, tumor grade (P=0.004),
axillary lymph node status (P=0.000), nerve invasion and metastasis
(P=0.002), vascular metastasis (P=0.000), Ki-67 expression
(P=0.017), and Kpnβ1 expression (P=0.002) were prognostic factors
for overall survival (Table II).
Multivariate analysis using the Coxs proportional hazards model
indicated that Kpnβ1 expression (P=0.001), grade (P=0.001), Her2
expression (P=0.035), axillary lymph node status (P=0.001), and
vascular metastasis (P=0.029) were independent prognostic
indicators of overall survival (Table
III).
 | Table II.Survival status and
clinicopathological parameters of the 140 breast carcinomas
specimens. |
Table II.
Survival status and
clinicopathological parameters of the 140 breast carcinomas
specimens.
|
|
| Survival
status |
|
|
|---|
|
|
|
|
|
|
|---|
| Parameters | Total | Alive | Dead |
P-valuea | χ2 |
|---|
| Age (years) |
|
|
|
|
|
|
≤50 | 57 | 22 | 35 | 0.325 | 0.968 |
|
>50 | 83 | 39 | 44 |
|
|
| Grade |
|
|
|
|
|
| I | 17 | 13 | 4 | 0.004a | 11.310 |
| II | 60 | 28 | 32 |
|
|
|
III | 63 | 20 | 43 |
|
|
| ER |
|
|
|
|
|
|
Negative | 69 | 35 | 34 | 0.092 | 2.832 |
|
Positive | 71 | 26 | 45 |
|
|
| PR |
|
|
|
|
|
|
Negative | 70 | 30 | 40 | 0.865 | 0.029 |
|
Positive | 70 | 31 | 39 |
|
|
| Her2 |
|
|
|
|
|
|
Negative | 68 | 28 | 40 | 0.579 | 0.308 |
|
Positive | 72 | 33 | 39 |
|
|
| Tumor size |
|
|
|
|
|
|
≤2×2×2 | 77 | 33 | 34 | 0.194 | 1.687 |
|
>2×2×2 | 63 | 28 | 45 |
|
|
| Axillary lymph node
status |
|
|
|
|
|
| N0 | 52 | 34 | 18 | 0.000a | 16.010 |
| Nx | 88 | 27 | 61 |
|
|
| Nerve invasion and
metastasis |
|
|
|
|
|
|
Negative | 85 | 46 | 39 | 0.002a | 9.788 |
|
Positive | 55 | 15 | 40 |
|
|
| Vascular
metastasis |
|
|
|
|
|
|
Negative | 75 | 48 | 27 | 0.000a | 27.419 |
|
Positive | 65 | 13 | 52 |
|
|
| Histology |
|
|
|
|
|
|
Ductal | 109 | 50 | 59 | 0.303 | 1.059 |
|
Others | 31 | 11 | 20 |
|
|
| Ki-67 |
|
|
|
|
|
|
Low | 49 | 28 | 21 | 0.017a | 5.647 |
|
High | 91 | 33 | 58 |
|
|
| Kpnβ1 |
|
|
|
|
|
|
Low | 69 | 39 | 30 | 0.002a | 9.281 |
|
High | 71 | 22 | 49 |
|
|
 | Table III.Contribution of various potential
prognostic factors to survival by Cox regression analysis in 140
breast carcinoma specimens. |
Table III.
Contribution of various potential
prognostic factors to survival by Cox regression analysis in 140
breast carcinoma specimens.
| Parameters | Hazard radio | 95.0% CI | P-value |
|---|
| Kpnβ1 | 2.925 | 1.716–4.984 | 0.001a |
| Ki-67 | 0.861 | 0.496–1.495 | 0.596 |
| Age (years) | 0.919 | 0.569–1.483 | 0.729 |
| Histology | 0.698 | 0.402–1.210 | 0.200 |
| Grade | 2.121 | 1.384–3.251 | 0.001a |
| ER | 0.992 | 0.541–1.821 | 0.980 |
| PR | 1.207 | 0.653–2.230 | 0.548 |
| Her2 | 0.554 | 0.320–0.959 | 0.035a |
| Tumor size | 0.792 | 0.487–1.288 | 0.348 |
| Axillary lymph node
status | 4.065 | 2.212–7.469 | 0.001a |
| Nerve invasion and
metastasis | 1.078 | 0.635–1.829 | 0.780 |
| Vascular
metastasis | 1.830 | 1.064–3.147 | 0.029a |
Kpnβ1 expression is positively
correlated with cell proliferation and its expression is cell-cycle
dependent
Since the expression of Kpnβ1 was significantly
correlated with Ki-67 expression, a cellular marker for
proliferation in BC specimens, we hypothesized that Kpnβ1 may play
a role in the cell cycle progression of BC cells. We demonstrated
that the expression of Kpnβ1 was high in BC cells, especially in
SKBR-3 cells (Fig. 2C and D). To
ascertain that Kpnβ1 was invovled in the cell cycle, SKBR-3 cells
were subjected to serum starvation and re-feeding. Flow cytometric
analysis revealed that after 48 h serum deprivation, SKBR-3 cells
were arrested in the G1 phase (86.56%). Upon re-feeding, the cells
excited at the G1 phase and gradually entered S phase over time
(Fig. 4A and B). We then employed
western blotting to examine whether the expression of Kpnβ1 was
cell-cycle dependent. Kpnβ1 expression was found to increase
gradually after serum stimulation with increased time, consistent
with the expression of the proliferation marker PCNA (Fig. 4C and D). These findings indicated
that Kpnβ1 may play an important role in cell proliferation of
SKBR-3 cells via involvement in the cell cycle.
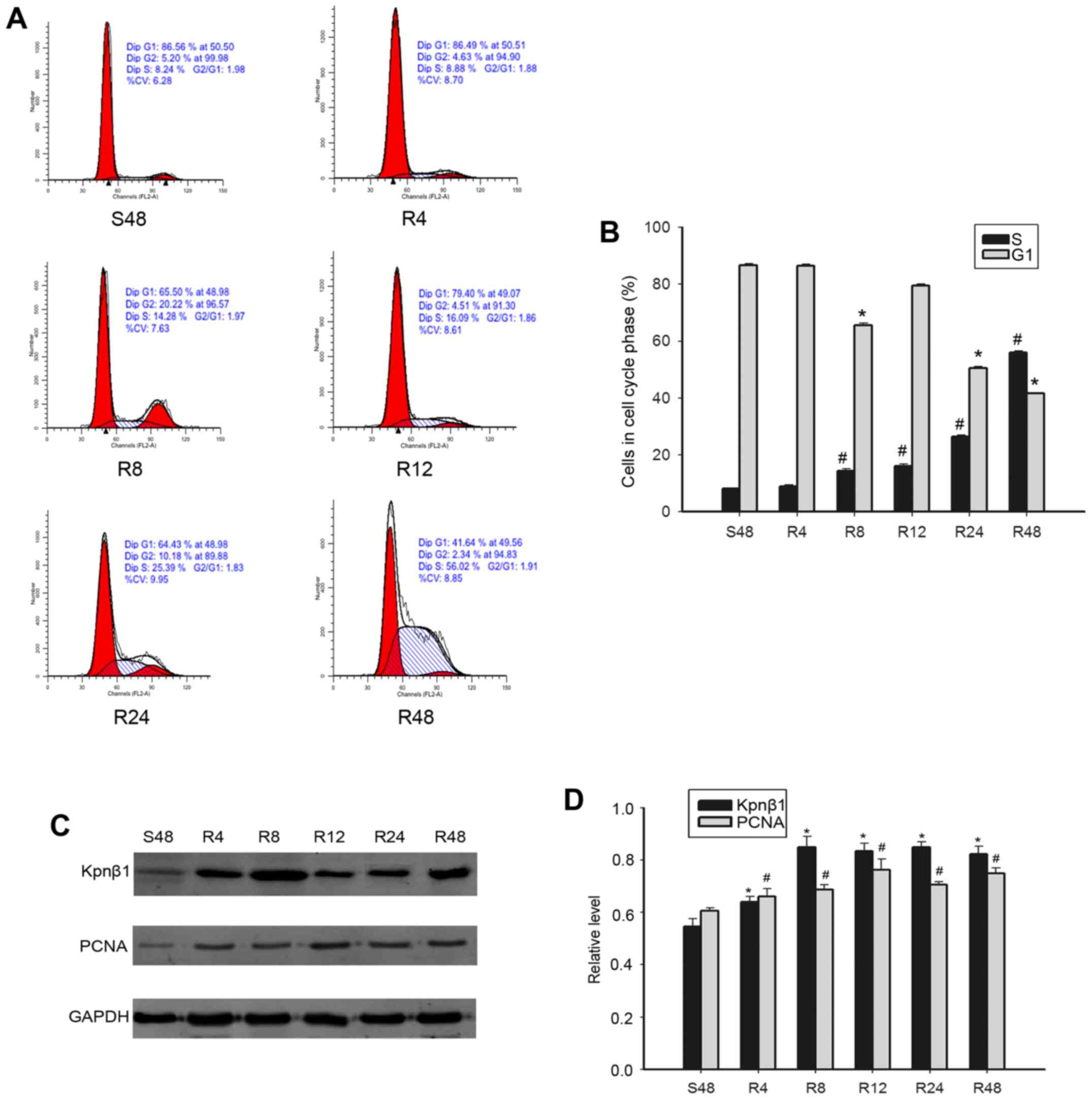 | Figure 4.Expression of Kpnβ1 and cell
cycle-related molecules detected in proliferating MDA-MB-231 cells
by flow cytometry. (A and B) Cells synchronized at the phase G1
progressed into the cell cycle when serum was added for S48h, R4h,
R8h, R12h, R24h, and R48h. (C and D) S48h MDA-MB-231 cells were
released by re-feeding with serum and cell lysates were prepared
and analyzed by western blotting using antibodies against Kpnβ1,
PCNA, and GAPDH (loading control). Bar charts revealing the ratio
of Kpnβ1, PCNA, and GAPDH by densitometry. Data are expressed or
presented as the means ± SEM. *,#P<0.05, compared
with the control cells that were serum starved for 48 h (S48h). SEM
denotes the standard error of the mean. S denotes serum starvation.
R denotes serum release. Kpnβ1, karyopherin β-1; PCNA,
proliferating cell nuclear antigen. |
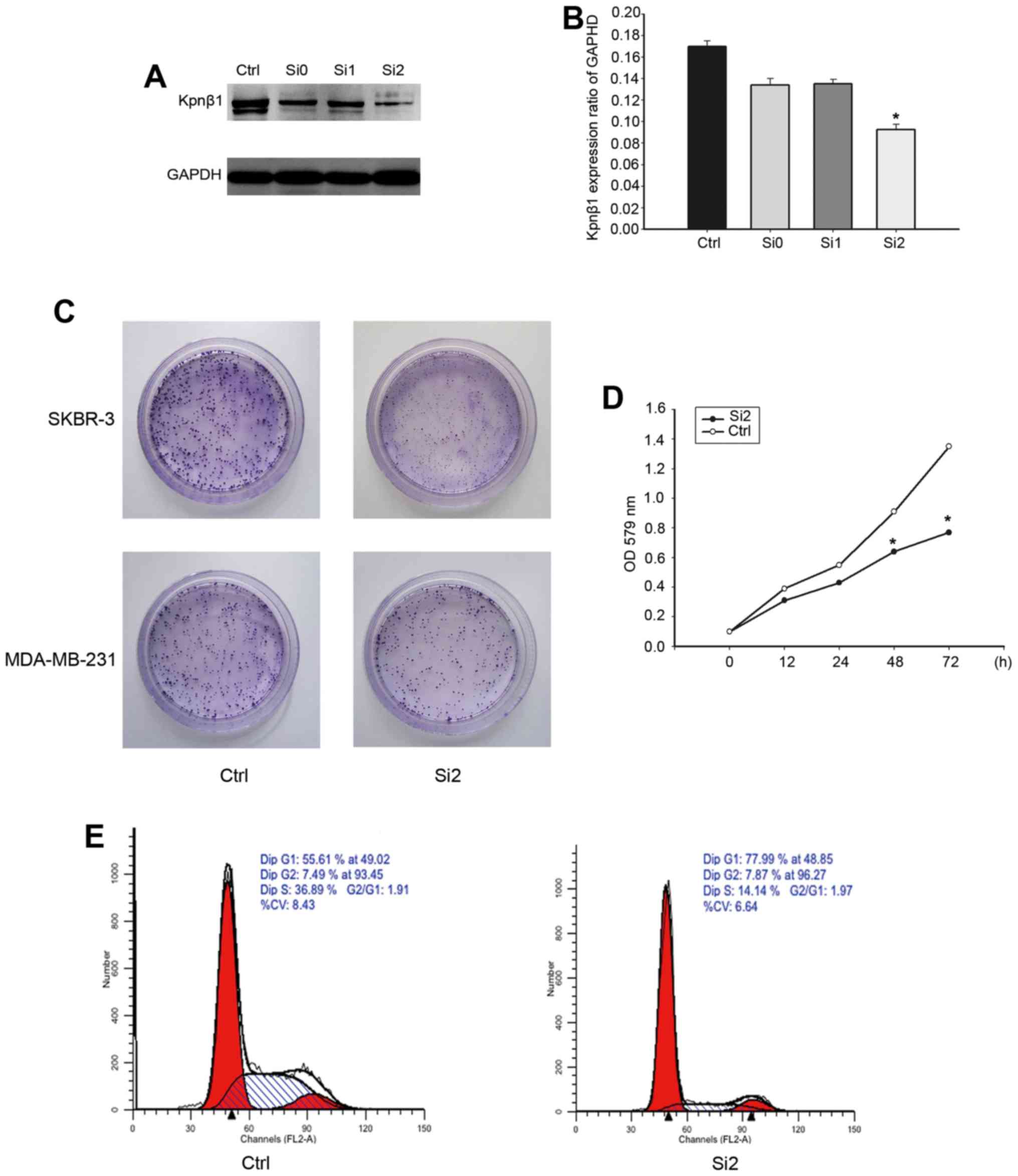
Kpnβ1 knockdown inhibits cellular
proliferation and promotes cell cycle arrest
To further verify the effect of Kpnβ1 on BC cell
proliferation, chemically synthesized siRNA was employed to knock
down endogenous Kpnβ1 in SKBR-3 cells. SKBR-3 cells were
transfected with Kpnβ1-siRNA (siRNA-0, siRNA-1, siRNA-2) and
negative control siRNA, and then the expression of Kpnβ1 was
assessed by western blotting 48 h post-transfection (Fig. 5A). The results revealed that Kpnβ1
expression levels were decreased in the SKBR-3 cells transfected
with Kpnβ1-siRNA compared with cells transfected with the negative
control siRNA, with siRNA-2 achieving the most marked knockdown
efficiency (Fig. 5A and B).
In the colony formation assay, we observed that
SKBR-3 and MDA-MB-231 cell proliferation was significantly
inhibited in the cells treated with siRNA-2, and was more
significant in the SKBR-3 cells (Fig.
5C). This indicated that in SKBR-3 cells, the effect of Kpnβ1
on cell proliferation was more significant. Furthermore, the CCK-8
assay found that the rate of SKBR-3 cell proliferation after
treatment with siRNA-2 exhibited a significant decline compared
with the negative control siRNA (Fig.
5D). Additionally, flow cytometric analyses of the cell cycle
in SKBR-3 cells transfected with different treatments revealed a
significant increase in the G1 phase and a marked decrease in the S
phase, suggesting that downregulation of Kpnβ1 arrested the cell
cycle (Fig. 5E). Based on these
findings, Kpnβ1-knockdown exhibited a specific inhibitory effect on
cell proliferation associated with cell cycle arrest in the SKBR-3
cells.
Kpnβ1 interacts with Her2 and
suppression of Kpnβ1 expression abrogates Her2 nuclear
transport
Despite a lack of statistical correlation between
the expression of Kpnβ1 and Her2 detected in our 140 BC tissues
(P=0.129), we investigated the association between Kpnβ1 and Her2.
We hypothesized that Kpnβ1-knockdown may suppress BC cell
proliferation by blocking Her2 nuclear entry, and we assessed this
using the SKBR-3 cell line (Her2-overexpressing cells) The results
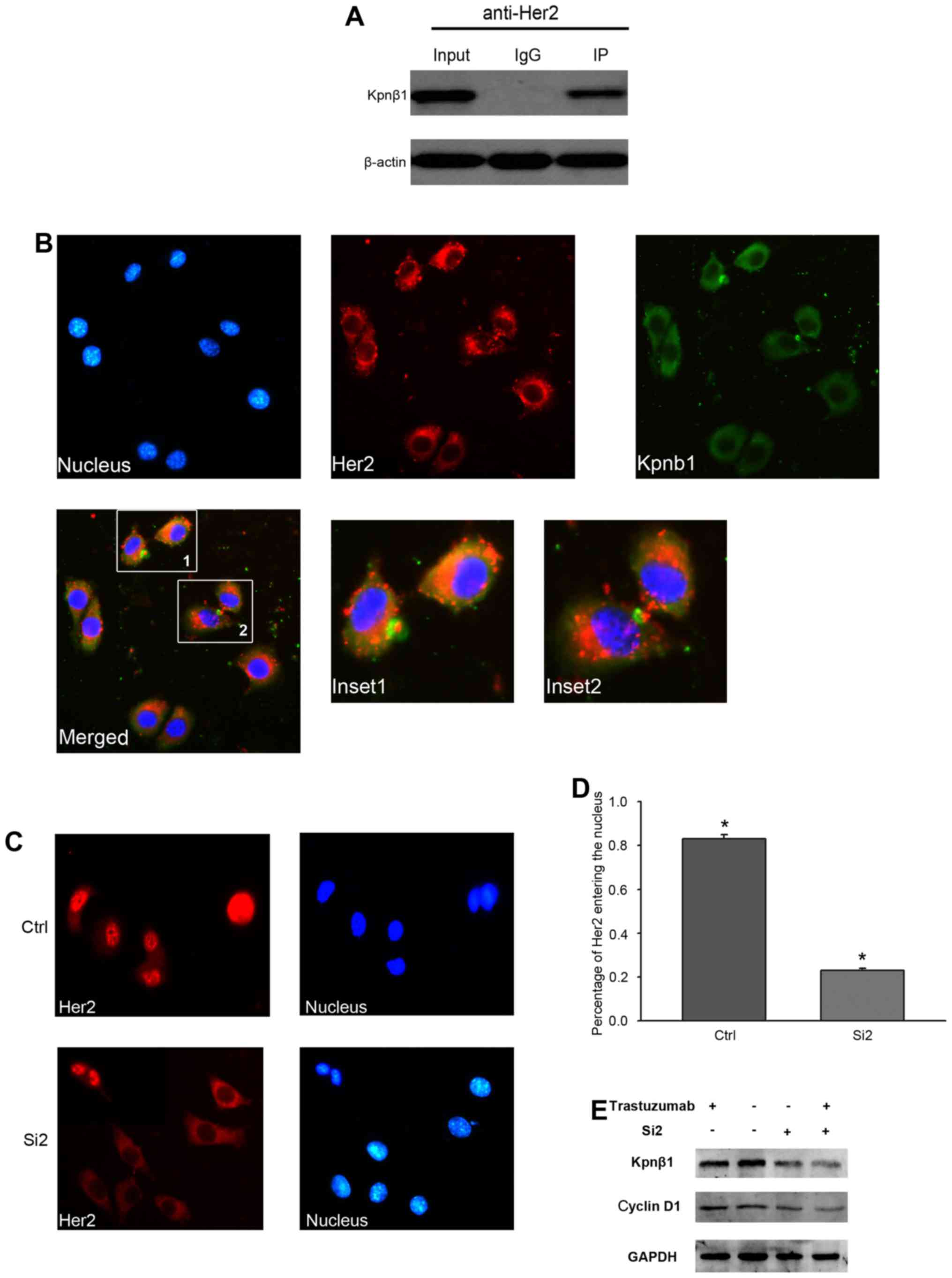
confirmed the effect of Kpnβ1 on Her2 in SKBR-3 cells (Fig. 6A). Furthermore, immmunofluorescence
detected co-localization of Kpnβ1 and Her2 in the SKBR-3 cell
cytoplasm (Fig. 6B).
We next found that the localization of Her2 was
affected by Kpnβ1-knockdown. When SKBR-3 cells were transfected
with Kpnβ1-siRNA2, confocal microscopy demonstrated the distinct
localization of Her2 in the cytoplasm and at the cell surface, but
not in the nucleus as observed in the control siRNA-treated cells
(Fig. 6C and D). These results
indicated that Kpnβ1-knockdown abrogated nuclear transport of Her2.
From the above mentioned results, we speculated that Kpnβ1
stimulated cell proliferation by promoting the nuclear
translocation of Her2. However, it was not determined whether cell
proliferation depended on Her2 status. Thus, we used trastuzumab to
inhibit the activity of the cell membrane surface Her2. The results
revealed the Her2 activities of different cases. Kpnβ1 knockdown
also inhibited cellular proliferation (Fig. 6E). Therefore, Kpnβ1 interacted with
Her2, then promoted nuclear transport of Her2, which was not
dependent on the state of Her2.
Discussion
BC is the leading form of cancer in women, and
although substantial progress has been made in its screening and
management, globally it has the highest mortality rate (23). Although BC is presently incurable,
it has the potential to become curable, or at least have improved
prognostics when detected at an early stage. Thus a deeper
understanding of the molecular events associated with BC is
essential to develop novel treatments. In this study, we identified
and characterized Kpnβ1 as an important player in BC progression.
Kpnβ1 appears to be involved in the process of BC cell
proliferation, partially by participating in the nuclear transport
of Her2.
Kpnβ1 functions as a transportation cargo protein
into and out of the cell nucleus during a selective, multistep
process (24). Kpnβ1-mediated
nuclear transportation is involved in multiple biological processes
such as insulin resistance, circadian rhythm and viral infections.
Kpnβ1 was found to promote palmitate-induced insulin resistance via
NF-κB signaling in hepatocytes (25), and mediate PER/CRY nuclear
translocation and therefore circadian clock function (26). Upregulation of Kpnβ1 was effective
in nuclear localization signaling for the minor capsid proteins,
VP2 and VP3, during BKPyV nuclear entry (27). Moreover, Kpnβ1 likely plays a key
role in the inflammation process (28).
Studies on the effect of Kpnβ1 expression on tumors
have been recently reported. There is a consensus that increased
expression of certain Kpnβ proteins in cancer cells results in
increased nuclear transport efficiency, thus facilitating increased
oncogenic signaling and promoting the cancer phenotype (19). Increased expression of Kpnβ1 was
recognized in malignant BC cells and Kpnβ1 knockdown has been shown
to decrease nuclear import efficiency in malignant but not in
non-transformed cells (29).
Furthermore, previous studies have revealed that Kpnβ1 is
overexpressed in colon, breast, lung, ovarian, and cervical cancer
specimens when compared with normal tissues (17).
Many articles exist on the close relationship
between Kpnβ1 expression and tumor proliferation, and Kpnβ1
inhibition has been shown to prolong mitotic arrest and apoptosis
in cervical cancer cells (30).
Furthermore, Kpnβ1 knockdown abrogated nuclear transport of DR5 and
increased its cell surface expression. Kpnβ1-mediated nuclear
localization of DR5 limited DR5/TRAIL-induced cell death of human
tumor cells (31), suggesting that
the molecular mechanism of Kpnβ1 and tumor proliferation was
related to its mediated nuclear entry efficiency. Therefore,
inhibition of Kpnβ1 represents a novel therapeutic approach for the
treatment of cancer (32).
A previous study reported that the regulation of
Kpnβ1 expression in cancer cells was related to the level of
EZH2/miR-30a. Inhibition of E2F in cancer cells caused increased
activation of Kpnβ1 promoters, leading to elevated levels of Kpnβ1
proteins, and ultimately impacting the phenotype in cervical
carcinoma (33). In malignant
peripheral nerve sheath tumor (MPNST) cells, EZH2 expression was
significantly upregulated and the miR-30a level was significantly
increased in EZH2-knockdown cells, which may inhibit the expression
of Kpnβ1. Based on this, EZH2 regulated miR-30a targeted Kpnβ1 in
MPNST cells (34). However, limited
studies exist describing the overexpression of Kpnβ1 in BC.
Consequently the molecular mechanism of Kpnβ1 remains unclear.
In our study, the expression of Kpnβ1 in SKBR-3
cells was higher than that in MDA-MB-231 cells (Fig. 2C). Although siRNA induced inhibition
of Kpnβ1 in both cell lines, the effect on SKBR-3 cell
proliferation was greater than that in MDA-MB-231 cells (Fig. 5C). Considering SKBR-3 cells are
Her2-overexpressing BC cells and MDA-MB-231 cells are
triple-negative BC cells, we hypothesized that Kpnβ1 is closely
related to Her2. Previous studies have highlighted Kpnβ1-mediated
Her2 nuclear entry in MCF-7/HER18 BC cells (5).
A study on the association between tumors and Her2
nuclear entry have recently increased. After Her2 nuclear entry,
the transcription and expression of the ribosomal RNA (rRNA)
transcription factor is accelerated, thus fostering BC protein
translation and stimulating tumor proliferation and development
(22). In another study, inhibition
of Her2 nuclear entry reduced the expression of cyclin D1, thus
decreasing the growth of BC cells (35). Therefore, we hypothesized that Kpnβ1
overexpression could increase Her2 nuclear entry, thus promoting BC
proliferation, while Kpnβ1-knockdown could decrease this
proliferation by blocking Her2 nuclear entry.
In this study, we identified that high expression of
Kpnβ1 in BC often leads to poor prognosis and that Kpnβ1-knockdown
markedly reduced SKBR-3 cell proliferation. Based on these
findings, we believe that Kpnβ1 is related to Her2 and its
knockdown reduces Her2 nuclear entry. To our knowledge, this is the
first study to report the expression of Kpnβ1 and relevant
pathological parameters in BC, and the effect on its prognosis. Our
results revealed that Kpnβ1 knockdown reduced cell proliferation in
SKBR-3 cell lines, and elucidated the relationship between Kpnβ1
and Her2 and its effect on nuclear entry. We identified the
potential mechanism involved in the effect of Kpnβ1 on Her2
overexpression in BC cell proliferation.
One study limitation was the statistical parameters
used in this study. Atypical statistical parameters should be used
in future studies. Furthermore, experiments on Kpnβ1-knockdown in
affected BC cells after blocking Her2 nuclear entry have not been
fully demonstrated and therefore warrant further study to further
uncover the mechanism.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (no. 81672596), Nantong Science and
Technology Project (no. MS22015058) and Nantong University
Innovation Project (YKC15085).
References
|
1
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nakielny S and Dreyfuss G: Transport of
proteins and RNAs in and out of the nucleus. Cell. 99:677–690.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chook YM and Blobel G: Karyopherins and
nuclear import. Curr Opin Struct Biol. 11:703–715. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Giri DK, Ali-Seyed M, Li LY, Lee DF, Ling
P, Bartholomeusz G, Wang SC and Hung MC: Endosomal transport of
ErbB-2: Mechanism for nuclear entry of the cell surface receptor.
Mol Cell Biol. 25:11005–11018. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lo HW, Ali-Seyed M, Wu Y, Bartholomeusz G,
Hsu SC and Hung MC: Nuclear-cytoplasmic transport of EGFR involves
receptor endocytosis, importin beta1 and CRM1. J Cell Biochem.
98:1570–1583. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhen Y, Sørensen V, Skjerpen CS, Haugsten
EM, Jin Y, Wälchli S, Olsnes S and Wiedlocha A: Nuclear import of
exogenous FGF1 requires the ER-protein LRRC59 and the importins
Kpnα1 and Kpnβ1. Traffic. 13:650–664. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mosammaparast N and Pemberton LF:
Karyopherins: From nuclear-transport mediators to nuclear-function
regulators. Trends Cell Biol. 14:547–556. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Altan B, Yokobori T, Mochiki E, Ohno T,
Ogata K, Ogawa A, Yanai M, Kobayashi T, Luvsandagva B, Asao T, et
al: Nuclear karyopherin-α2 expression in primary lesions and
metastatic lymph nodes was associated with poor prognosis and
progression in gastric cancer. Carcinogenesis. 34:2314–2321. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Grupp K, Habermann M, Sirma H, Simon R,
Steurer S, Hube-Magg C, Prien K, Burkhardt L, Jedrzejewska K,
Salomon G, et al: High nuclear karyopherin α 2 expression is a
strong and independent predictor of biochemical recurrence in
prostate cancer patients treated by radical prostatectomy. Mod
Pathol. 27:96–106. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huang L, Wang HY, Li JD, Wang JH, Zhou Y,
Luo RZ, Yun JP, Zhang Y, Jia WH and Zheng M: KPNA2 promotes cell
proliferation and tumorigenicity in epithelial ovarian carcinoma
through upregulation of c-Myc and downregulation of FOXO3a. Cell
Death Dis. 4:e7452013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pavlou MP, Dimitromanolakis A,
Martinez-Morillo E, Smid M, Foekens JA and Diamandis EP:
Integrating meta-analysis of microarray data and targeted
proteomics for biomarker identification: Application in breast
cancer. J Proteome Res. 13:2897–2909. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ikenberg K, Valtcheva N, Brandt S, Zhong
Q, Wong CE, Noske A, Rechsteiner M, Rueschoff JH, Caduff R, Dellas
A, et al: KPNA2 is overexpressed in human and mouse endometrial
cancers and promotes cellular proliferation. J Pathol. 234:239–252.
2014.PubMed/NCBI
|
|
14
|
Hu ZY, Yuan SX, Yang Y, Zhou WP and Jiang
H: Pleomorphic adenoma gene 1 mediates the role of karyopherin
alpha 2 and has prognostic significance in hepatocellular
carcinoma. J Exp Clin Cancer Res. 33:612014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ma S and Zhao X: KPNA2 is a promising
biomarker candidate for esophageal squamous cell carcinoma and
correlates with cell proliferation. Oncol Rep. 32:1631–1637. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
van der Watt PJ, Maske CP, Hendricks DT,
Parker MI, Denny L, Govender D, Birrer MJ and Leaner VD: The
karyopherin proteins, Crm1 and karyopherin beta1, are overexpressed
in cervical cancer and are critical for cancer cell survival and
proliferation. Int J Cancer. 124:1829–1840. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang P, Garnett J, Creighton CJ, Al
Sannaa GA, Igram DR, Lazar A, Liu X, Liu C and Pollock RE:
EZH2-miR-30d-KPNB1 pathway regulates malignant peripheral nerve
sheath tumour cell survival and tumourigenesis. J Pathol.
232:308–318. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Martens-de Kemp SR, Nagel R, Stigter-van
Walsum M, van der Meulen IH, van Beusechem VW, Braakhuis BJ and
Brakenhoff RH: Functional genetic screens identify genes essential
for tumor cell survival in head and neck and lung cancer. Clin
Cancer Res. 19:1994–2003. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
van der Watt PJ, Stowell CL and Leaner VD:
The nuclear import receptor Kpnβ1 and its potential as an
anticancer therapeutic target. Crit Rev Eukaryot Gene Expr.
23:1–10. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nordgard SH, Johansen FE, Alnaes GI,
Bucher E, Syvänen AC, Naume B, Børresen-Dale AL and Kristensen VN:
Genome-wide analysis identifies 16q deletion associated with
survival, molecular subtypes, mRNA expression, and germline
haplotypes in breast cancer patients. Genes Chromosomes Cancer.
47:680–696. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ross JS and Fletcher JA: The HER-2/neu
oncogene in breast cancer: Prognostic factor, predictive factor,
and target for therapy. Oncologist. 3:237–252. 1998.PubMed/NCBI
|
|
22
|
Li LY, Chen H, Hsieh YH, Wang YN, Chu HJ,
Chen YH, Chen HY, Chien PJ, Ma HT, Tsai HC, et al: Nuclear ErbB2
enhances translation and cell growth by activating transcription of
ribosomal RNA genes. Cancer Res. 71:4269–4279. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gucalp A, Gupta GP, Pilewskie ML, Sutton
EJ and Norton L: Advances in managing breast cancer: A clinical
update. F1000Prime Rep. 6:662014. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lu T, Bao Z, Wang Y, Yang L, Lu B, Yan K,
Wang S, Wei H, Zhang Z and Cui G: Karyopherin β1 regulates
proliferation of human glioma cells via Wnt/β-catenin pathway.
Biochem Biophys Res Commun. 478:1189–1197. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang S, Zhao Y, Xia N, Zhang W, Tang Z,
Wang C, Zhu X and Cui S: KPNβ1 promotes palmitate-induced insulin
resistance via NF-κB signaling in hepatocytes. J Physiol Biochem.
71:763–772. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lee Y, Jang AR, Francey LJ, Sehgal A and
Hogenesch JB: KPNB1 mediates PER/CRY nuclear translocation and
circadian clock function. eLife. 4:e086472015. View Article : Google Scholar :
|
|
27
|
Bennett SM, Zhao L, Bosard C and Imperiale
MJ: Role of a nuclear localization signal on the minor capsid
proteins VP2 and VP3 in BKPyV nuclear entry. Virology. 474:110–116.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sun C, Yu Z, Wang Y and Tao T: The
importin protein karyopherin-β1 regulates the mice fibroblast-like
synoviocytes inflammation via facilitating nucleus transportation
of STAT3 transcription factor. Biochem Biophys Res Commun.
471:553–559. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kuusisto HV and Jans DA: Hyper-dependence
of breast cancer cell types on the nuclear transporter importin β1.
Biochim Biophys Acta. 1853:1870–1878. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Angus L, van der Watt PJ and Leaner VD:
Inhibition of the nuclear transporter, Kpnβ1, results in prolonged
mitotic arrest and activation of the intrinsic apoptotic pathway in
cervical cancer cells. Carcinogenesis. 35:1121–1131. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kojima Y, Nakayama M, Nishina T, Nakano H,
Koyanagi M, Takeda K, Okumura K and Yagita H: Importin β1
protein-mediated nuclear localization of death receptor 5 (DR5)
limits DR5/tumor necrosis factor (TNF)-related apoptosis-inducing
ligand (TRAIL)-induced cell death of human tumor cells. J Biol
Chem. 286:43383–43393. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kim YH, Ha S, Kim J and Ham SW:
Identification of KPNB1 as a cellular target of aminothiazole
derivatives with anticancer activity. ChemMedChem. 11:1406–1409.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
van der Watt PJ, Ngarande E and Leaner VD:
Overexpression of Kpnβ1 and Kpnα2 importin proteins in cancer
derives from deregulated E2F activity. PLoS One. 6:e277232011.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang P, Yang X, Ma X, Ingram DR, Lazar
AJ, Torres KE and Pollock RE: Antitumor effects of pharmacological
EZH2 inhibition on malignant peripheral nerve sheath tumor through
the miR-30a and KPNB1 pathway. Mol Cancer. 14:552015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cordo Russo RI, Béguelin W, Díaz Flaqué
MC, Proietti CJ, Venturutti L, Galigniana N, Tkach M, Guzmán P, Roa
JC, O'Brien NA, et al: Targeting ErbB-2 nuclear localization and
function inhibits breast cancer growth and overcomes trastuzumab
resistance. Oncogene. 34:3413–3428. 2015. View Article : Google Scholar : PubMed/NCBI
|