Introduction
Lung cancer has been identified as the most lethal
malignancy in the past decades. Among cancers, non-small cell lung
cancer (NSCLC) is considered a complex malignancy owing to its
diverse alterations in molecular and genomic pathways. Early
diagnosis is crucial for increasing the chance for survival;
however, no highly sensitive and effective biomarker for NSCLC has
been validated in clinical applications. Therefore, a potential
biomarker that is specific and early for cancer screening is always
a perpetual goal for scientists.
Human mitochondrial DNA (mtDNA), a 16,569 kb
circular DNA that contains 37 genes, encodes 12S and 16S rRNAs, 22
tRNAs, and 13 polypeptides (1). It
contains a non-coding circular loop (D-loop) with a large number of
unmethylated dinucleotide fragments called CpG islands (2). Since 1973, mtDNA has been suggested to
contribute to carcinogenesis (3).
Structural differences in mitochondria between patients with cancer
and normal controls, observed under electron microscopy, indicated
that mitochondrial instability is cancer-associated (4). For example, deletion and insertions
within the mtDNA D-loop were observed in primary breast cancer
(5). An aberrant mtDNA copy number
was also highly associated with lung cancer in a dose-dependent
manner (6). Moreover, through
comprehensive resequencing microarrays, a wide variety of mutations
in mtDNA have been confirmed to be related to early stages of
cancers which suggests that analysis of mtDNA in clinical samples
may be a potential approach to cancer diagnosis (7).
Recent evidence has revealed that mtDNA participates
in a myriad of immune responses. It serves as an effector of
pattern recognition receptor (PRR) signalling that triggers the
innate immune system to respond to cellular damage, stress, and
infection by pathogens (8). One of
PRRs, Toll-like receptor (TLR), is activated by mtDNA and induces a
signalling cascade that ultimately results in an inflammatory
response involving cytokines and other downstream effectors
(9). Among the TLRs, TLR1, TLR2,
TLR4, TLR5 and TLR6 are activated by bacterial and fungal cell
surface molecules, whereas TLR3, TLR7, TLR8 and TLR9 are triggered
by pathogen-specific nucleic acids (10–12).
While mtDNA shares unmethylated CpG DNA repeats with bacterial DNA,
mtDNA is also a ligand for TLR9 (13,14).
CpG DNA directly binds to TLR9 and induces TLR9 translocation (with
alteration) from the endoplasmic reticulum to early endosomes and
the later lysosomal compartment (15). Intracellular adapter protein myeloid
differentiation factor-88 (MyD88) associated with TLR9 activates
downstream signalling proteins, such as the interleukin-1
receptor-associated kinase (IRAK) family, mitogen-activated kinases
(MAPK), or interferon-regulatory factors (IRFs) (16,17).
Previous studies have indicated that TLR9 is also
widely expressed in various human cancers, including breast,
ovarian, prostate, brain, gastric, renal cell carcinoma, and
oesophageal tumours (18–21). These data indicated that TLR9 serves
a role as a prognosis marker as well as a biomarker for cancer
diagnosis. A CpG island-containing oligodeoxyribonucleotide (ODN)
called ODN-M362 is a synthetic CpG-rich sequence that contains
unmethylated motifs, which is used for effective triggering of the
TLR9-ligand binding-related events, which also mimics mtDNA
function (22). Several preclinical
findings have demonstrated the anticancer activity of CpG ODNs,
which have been developed into TLR9-based agonist treatment
(22,23). In the present study, we quantified
serum mtDNA levels in healthy subjects and NSCLC patients. It was
found that lung cancer patients without metastasis had more mtDNA
in serum as compared to the patients with metastasis. Moreover,
TLR9-associated signalling was demonstrated after treatment with
ODN-M362. In A549 and HCC827 cell lines, TLR9 signalling was
activated and in addition, its adaptor protein, MyD88, was induced
by ODN-M362 in a dose-dependent manner. This finding indicated that
TLR9 may serve as a serological marker for NSCLC identification.
Furthermore, we employed a human cytokine array to evaluate
ODN-M362 stimulation of cytokines secretion. Hopefully, our
findings may identify the role that TLR9 and mtDNA play in lung
cancer progression and metastasis.
Materials and methods
Cell culture
A549 and HCC827 cells were cultured with F12K medium
and RPMI-1640 (both from Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany) at 37°C in a humidified atmosphere (95% air, 5%
CO2), respectively. Fetal bovine serum (FBS; 10%) (JRH
Biosciences, Lenexa, KS, USA), 100 µg/ml penicillin/streptomycin
and 2 mM L-glutamine (Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) were supplemented in culture medium.
Subjects
Study subjects were recruited during routine health
examinations with signed consents. Twenty-nine study subjects with
varying age and sex were randomly chosen including 16 lung
adenocarcinoma patients and 13 healthy volunteers, enrolling for
analysis. This study was a prospective study that aimed to identify
potential biomarkers to predict cancer progression. The Ethics
Committee of the Taipei Tzu-Chi General Hospital approved the study
(Approval no. TCRD-TPE-100-43; IRB no. 98-IRB-019-X; IRB extension
no. 05-XD55-107). Our study was conducted from January 2010-January
2012 and December 2017-July 2018. The inclusion criteria for lung
adenocarcinoma patients were as follows: The patients were required
to be >18 years of age with the pathology type confirmed by
pathology or cytology. Patients were excluded from the study if
they had received chronic steroid therapy, adjuvant or neoadjuvant
chemotherapy, radiotherapy, or both. Patients with a history of any
other type of cancer were not eligible. The inclusion criteria for
healthy volunteers were as follows: >18 years of age was
required with no evidence of cancers. Healthy volunteers were
excluded if they had inflammatory or infectious conditions
(empyema, interstitial lung disease, or active pneumonitis) or a
history of any other type of cancer.
Sample preparation and DNA
extraction
Blood samples were centrifuged at 200 × g at 8°C for
45 min. Serum was transferred into new polypropylene tubes and
re-centrifuged at 8,000 × g for 20 min. The supernatant was
collected and stored at −80°C. Serum DNA was extracted from 400 µl
sample with QIAmp DNA Blood Mini Kit (Qiagen, Inc., Valencia, CA,
USA).
Quantitative PCR
DNA from patient serum was collected to performed
quantitative (qPCR) on Applied Biosystems 7900 Sequence Detector
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The TaqMan
probe sequence purchased from Applied Biosystems (Thermo Fisher
Scientific, Inc.) was Mit 3153T
(5′-FAM-TTCACAAAGCGCCTTCCCCCGTAAATGA-TAMRA-3′), where FAM was
6-carboxyfluorescein and TAMRA was 6-carboxytetramethylrhodamine.
The thermocycling conditions included an initial denaturation at
50°C for 2 min and 95°C for 10 min, followed by 45 cycles at 95°C
for 15 sec, 60°C for 1 min. The internal control primer sequences
were plasmid forward primer (pls F) (5′-AATACGCAAACCGCCTCTCC-3′)
and plasmid reverse primer (pls R) (5′-ACAACATACGAGCCGGAAGC-3′).
The internal control for the TaqMan probe sequence was plasmid
TaqMan probe (pls T)
(5′-FAM-CGCAACGCAATTAATGTGAGTTAGCTCAC-TAMRA-3′). The method of
quantification used was 2−ΔΔCq (24).
Mitochondrial DNA copy number
calibration
A 172-bp mtDNA from genomic DNA of a healthy
volunteer was cloned and used as a calibrator. Calibrators were
prepared by serial dilution of the stock solution and contained
10–107 mitochondrial DNA copies/µl. The results were
expressed as genome-equivalent (GE)/ml of serum using a conversion
factor of 6.6 pg of DNA/cell. The concentration was calculated
using an equation that was previously described (25). Primers used to amplify Mt3130-3301
were Mit3130F (5′-AGGACAAGAGAAATAAGGCC-3′) and Mit 3301R
(5′-TAAGAAGAGGAATTGAACCTCTGACTGTAA-3′).
Western blotting
Cells were washed twice with ice-cold
phosphate-buffered saline (PBS) (Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA), lysed in 1 ml/plate with ice-cold IP lysis
buffer supplemented with protease inhibitors (Gibco; Thermo Fisher
Scientific, Inc.) and harvested by scraping. Protein concentration
was determined by BCA method. A total of 30 µg of protein sample
was separated by 10% SDS-PAGE, and then transferred onto
polyvinylidene fluoride (PVDF) membranes (Bio-Rad Laboratories,
Inc., Hercules, CA, USA). Membrane was blocked by 5% non-fat dry
milk in TBST with 0.05% Tween-20 for 1 h. Primary antibodies to
TLR9 (cat. no. ab88101; Abcam, Cambridge, MA, USA), MyD88 (cat. no.
GTX112986; GeneTex International Corporation, Hsinchu, Taiwan) and
β-tubulin (cat. no. sc-73242; Santa Cruz Biotechnology, Dallas, TX,
USA) were used at a dilution of 1:1,000 and incubated at 4°C for
overnight. Secondary antibodies to anti-mouse-HRP and
anti-rabbit-HRP (cat. no. 31430 and 31460, Invitrogen; Thermo
Fisher Scientific, Inc.) were used at a dilution of 1:5,000 and
incubated at room temperature for 1 h. Signals were detected with
chemiluminescence (Amersham; GE Healthcare, Chicago, IL, USA) and
exposed to X-ray film (Kodak, Rochester, NY, USA). Signal
quantification was done by using ImageJ.
Cytokine array analysis
Culture medium (400 µl) was harvested after cells
were treated with CpG ODN-M362 for 24 h according to the
manufacturer's instructions (Human Cytokine Array Panel A; R&D
Systems, Inc., Minneapolis, MN, USA). CpG-ODN-M362 sequence,
5′-TCGTCGTCGTTC:GAACGACGTTGAT-3′; and negative control, pCpG was a
synthetic sequence called pCpG Giant that failed to trigger
downstream TLR9 signaling and served as a negative control. pCpG
Giant was purchased from InvivoGen (San Diego, CA, USA) (cat. no.
tlrl-cpgg).
Statistical analysis
All experiments were repeated at least 3 times. Mean
values and standard deviation (SD) were calculated and analyzed
with a one-way analysis of variance (ANOVA) or Student's t-test
with PRISM 5.0 (GraphPad Software, Inc., San Diego, CA, USA).
Scheffe's post hoc test was used following one-way ANOVA. Data with
P<0.05 was considered to indicate a statistically significant
difference.
Results
MtDNA is cloned from normal serum as a
copy number calibrator
In order to evaluate the CpG level of different
stages of tumorigenesis in lung cancer patients, a 172-bp mtDNA
segment between nucleotide positions 3130 and 3301 was amplified
from genomic DNA of a healthy volunteer with the primers.
Mt3130-3301 was successfully amplified (Fig. 1A). The mtDNA amplicons were then
cloned into the pCRII TOPO TA plasmid (Invitrogen; Thermo Fisher
Scientific, Inc.) and were confirmed with restriction enzyme
digestion and sequencing (Fig. 1B and
C). The cloned mtDNA plasmid was to be used as a calibrator for
the mtDNA copy number.
To calibrate the correlation between threshold cycle
(CT) in a TaqMan assay and the serum mtDNA copy number,
a plot of the CT against the input target quantity, the
plotted on a common log scale, was generated (Fig. 1D). The linearity of the quantitative
assay was assessed with a calibrator, which was 172 bp mtDNA
flanked with a plasmid. The calibrator was serially diluted to
concentrations from 10 to 107 copies/µl before use. The
mtDNA copy number was determined by division of the total DNA
concentration by the weight of each plasmid molecule (25). The assay was sensitive and able to
detect 10 copies of mitochondrial DNA/µl. As the plot indicated,
the threshold cycle (CT) was set to 15 and was
proportional to the target copy number from 10 to 107
mtDNA copies.
Patients without metastasis have
higher serum mitochondrial DNA levels
To investigate the mtDNA copy number at different
malignancy stages of lung cancer, we performed quantitative PCR
analysis to evaluate the levels of mtDNA. Total DNA extraction from
13 healthy volunteers, 5 patients with non-metastatic, and 11
patients with metastatic lung adenocarcinoma (clinical
characteristics of healthy subjects and lung adenocarcinoma
patients are summarised in Table I)
was conducted, and we determined the concentration of serum mtDNA.
Among these patients, 11 had lung adenocarcinoma of stage IV with
metastasis, and 5 had stage II lung adenocarcinoma without
metastasis. Serum mtDNA levels of the M0 (non-metastatic) group
were significantly higher than those of the Meta (metastatic) and
HS (healthy volunteer) groups (Fig.
2A). In addition, the Meta group had the lowest serum mtDNA
level that was about a half of that of the M0 group. The qPCR
results were then used to calculate mtDNA copy numbers with the
calibrator (Fig. 2B). The mtDNA
copy number was significantly higher in the M0 group, and up to
4×104, ~4 times higher than that in the HS group,
indicating an associtaion between serum mtDNA levels and metastasis
status during lung carcinogenesis.
 | Table I.Clinical characteristics of healthy
and lung adenocarcinoma patients. |
Table I.
Clinical characteristics of healthy
and lung adenocarcinoma patients.
|
Characteristics | Mean, medium
(range) or number (n) |
|---|
| Healthy | n=13 |
|
Sex | 4 males/9
females |
|
Age | 48.2/49.0
(34–69) |
| Non-metastatic | n=5 |
|
Sex | 2 males/2
females |
|
Age | 59.0/60.0
(49–68) |
|
Pathological stage | Stage II |
| Metastatic | n=11 |
|
Sex | 2 males/7
females |
|
Age | 63.0/63.5
(57–67) |
|
Pathological stage (brain,
bone, and lung metastasis) | Stage IV |
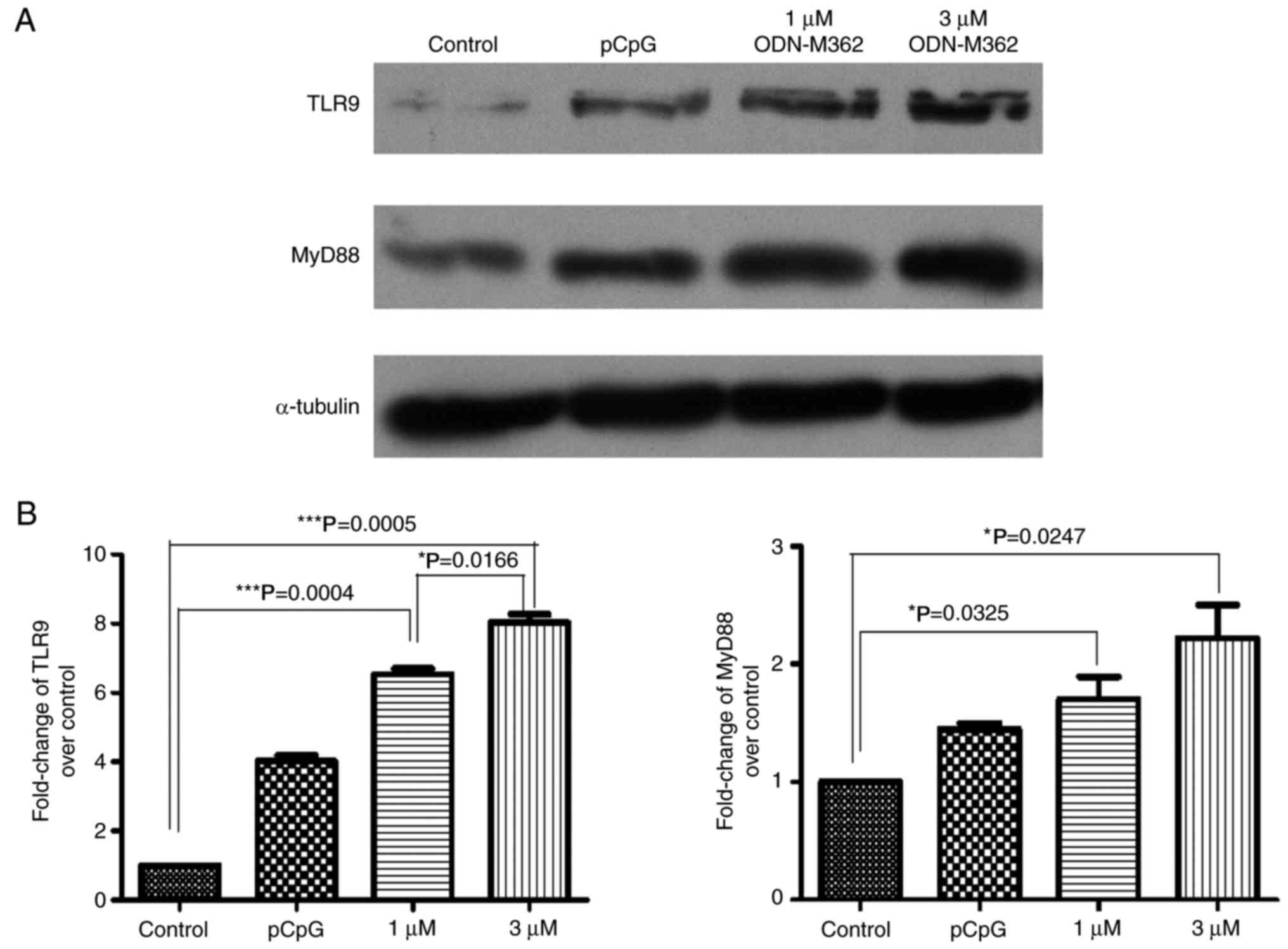
Synthetic CpG ODN, ODN-M362,
stimulates TLR9 and downstream MyD88 expression
To confirm that the high copy number of mtDNA
functionally affects its receptor, TLR9, two human lung
adenocarcinoma cell lines were treated with different
concentrations of a synthetic unmethylated CpG ODN, ODN-M362, which
has been known to serve as an agonist of TLR9, thereby simulating
to the role of mtDNA by triggering TLR9-mediated signalling. A549
cells were treated with 1 or 3 µM ODN-M362 for 24 h before
harvesting. The expression level of TLR9 in A549 was induced by
ODN-M362, and the downstream adaptor protein, MyD88, was also
upregulated (Fig. 3A). In addition,
the HCC827 cell line was also treated with 1, 3, or 5 µM ODN-M362
for 24 h. Compared with the A549 cells, the HCC827 cell line is
also a lung adenocarcinoma cell line however it carries an acquired
mutation in the EGFR tyrosine kinase domain. TLR9 and MyD88 in
HCC827 cells were significantly upregulated by ODN-M362 treatment
in a dose-dependent manner (Fig.
4A). Quantitative analysis of the western blotting was carried
out relative to untreated cells (Figs.
3B and 4B). Thus, ODN-M362 was
able to stimulate the expression of TLR9 in both lung cancer cell
lines, and TLR9 may be regulated by mtDNA copy numbers in A549 and
HCC827 cells.
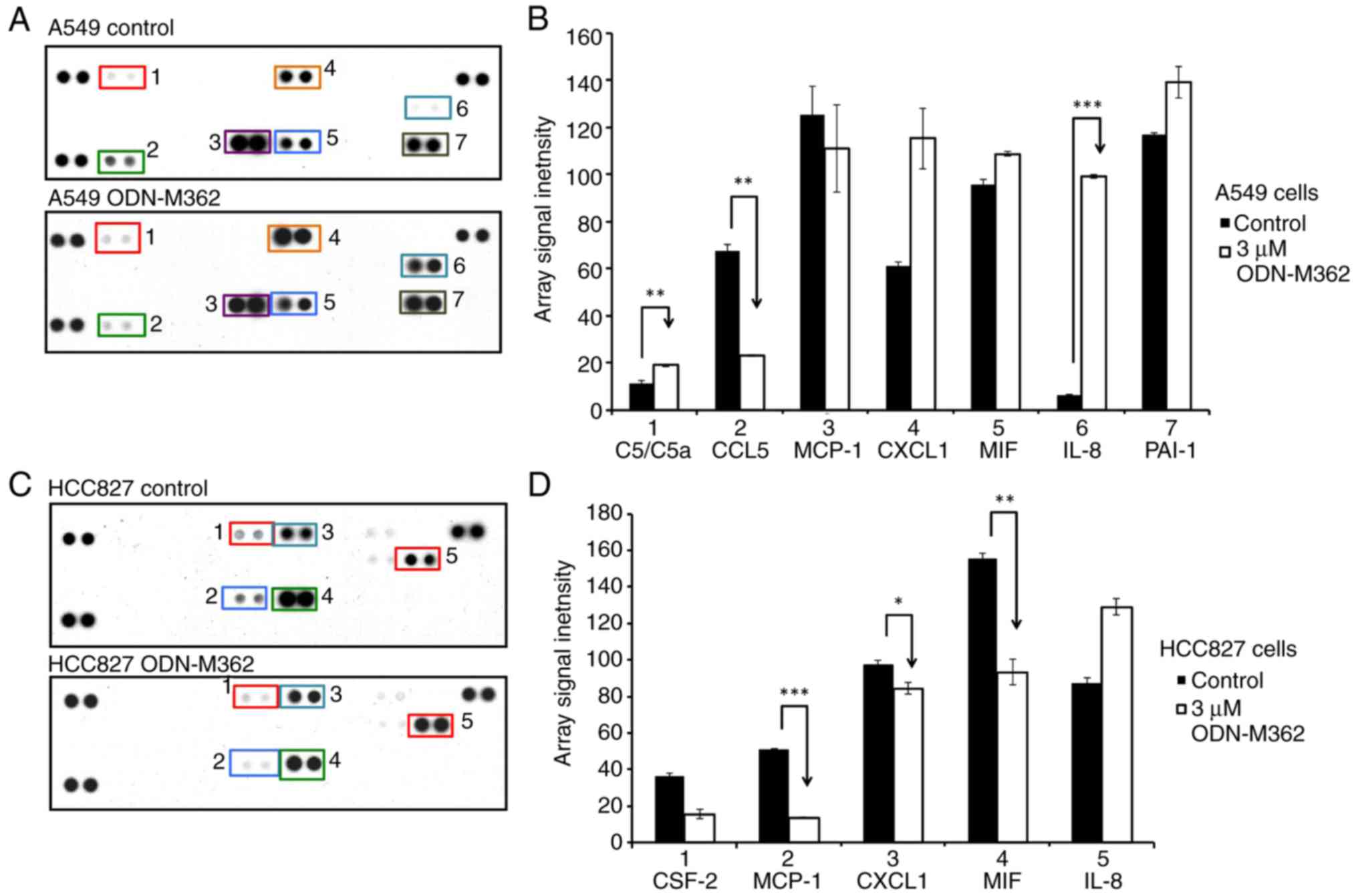
Cytokine expression levels are altered
by ODN-M362-mediated TLR9 signalling
We hypothesised that induction of TLR9 in lung
adenocarcinoma cell lines activated downstream signalling through
ODN-M362. Therefore, we performed a cytokine array analysis to
assess the expression levels of 36 human cytokines with/without
ODN-M362 treatment of A549 and HCC827 cell lines (Fig. 5A and C). Among the changes of
cytokine expression, ODN-M362-treated A549 cells secreted IL-8
>15-fold compared to the untreated cells, while C5/C5a was
induced 1.6-fold in ODN-M362-treated A549 cells (Fig. 5B). It is known that C5/C5a is a
direct inducer of interleukin-8 (IL-8) secretion (26). Therefore, marked increase of IL-8
supports the previous finding and may be due to serial activation
from the C5/C5a-mediated complement cascade. IL-8, an
inflammation-associated chemokine, is also likely involved in
cancer-associated processes, such as angiogenesis, proliferation,
migration, and invasion (27).
Moreover, IL-8 has been identified as an immunosuppressive cytokine
that promotes carcinogenesis (28).
Therefore, the ODN-M362-mediated induction of TLR9 signalling may
cause inflammation-induced carcinogenesis through the IL-8
abnormality. Conversely, CCL5 was downregulated in ODN-M362-treated
A549 cells (Fig. 5B). Although CCL5
has been reported as a protein that promotes cancer cell
proliferation and migration (29),
CCL5 also induces proper immune responses to tumour formation, and
the mechanism is still unclear (28).
Both A549 and HCC827 are NSCLC cell lines; however,
HCC827 cells are EGFR-mutated and highly sensitive to EGFR
inhibitors with deletions from E746 to A750, whereas A549 cells
encode wild-type EGFR, and these cell lines may induce different
immune responses (2,30). We found that after HCC827 cells were
treated with ODN-M362, monocyte chemoattractant protein-1 (MCP-1),
CXCL1, and macrophage migration inhibitory factor (MIF) were
significantly downregulated (Fig. 5B
and D). These results indicated that impaired EGFR signalling
not only affects TLR9-mediated cytokine expression levels but also
fails to induce chemotaxis for monocytes, neutrophils, and
macrophages when TLR9 signalling is activated.
Discussion
It has been well established that mtDNA is involved
in immune responses via TLR signalling. However, the precise role
of mtDNA-mediated TLR signalling in different stages of human lung
cancer has remained unclear. In the present study, the expression
levels of serum mtDNA of healthy people and patients with NSCLC
with or without metastasis were quantified. High levels of serum
mtDNA in patients with non-metastatic lung adenocarcinoma as
compared to healthy volunteers were determined; in contrast, NSCLC
patients with metastasis displayed the lowest level of serum mtDNA.
Although it has been confirmed that the copy number of mtDNA in
gastric cancer is higher than that in benign tissue, the
correlation between the copy number of mtDNA and clinical
characteristics of cancer progression is still ambiguous (31). In spite of the controversy regarding
the status of a prognostic biomarker, if we take the tumour node
metastasis stage and the mtDNA copy number into consideration,
there is still a significant synergistic effect on prognosis of
colorectal cancer (32). In the
present study, it was concluded that the serum mtDNA level is lower
in patients with metastatic lung cancer, in agreement with a
previous study on breast cancer (33). Although 3 years were spent
collecting samples from NSCLC patients with or without metastasis;
it was generally found that patients had less awareness before
cancer metastasis to seek diagnosis, particularly for lung cancer.
Therefore, this increased our challenge in obtaining enough samples
for NSCLC without metastasis (M0). Nonetheless, according to the
association between serum mtDNA copy number and metastasis status,
the potential of detection of mtDNA levels in patients with or
without metastasis as a pre-clinical diagnosis for identification
of cancer progression is suggested.
Next, a TLR9 agonist, ODN-M362 was used, to mimic
the serum mtDNA effect and examine the expression level of TLR9 and
its downstream targets in two different human lung cancer cell
lines. It was observed that the expression of TLR9 was upregulated
in both A549 and HCC827 cells after ODN-M362 treatment.
Nonetheless, one of the downstream targets of TLR9 signalling,
MyD88, was more significantly induced in HCC827 cells. While A549
is an EGFR wild-type cell line, HCC827 cells carry an oncogenic
EGFR exon 19 deletion, rendering these cells sensitive to EGFR
tyrosine kinase inhibitors (TKIs) (30). This finding indicated that ODN-M362
triggered much more effective TLR9 signalling in EGFR-mutated
cells.
In order to clarify the effects of mtDNA-induced
TLR9 signalling, we performed cytokine array analysis to identify
the potential responsive elements. In A549 cells, activated TLR9
expression led to cancer progression and metastasis by increasing
secretion of C5/C5a, IL-8, and CXCL1. When A549 cells were treated
with ODN-M362, they may have transformed into cells with
inflammation-induced malignancy through IL-8 aberrancy. In
parallel, increasing TLR9 expression in HCC827 cells downregulated
MCP-1, CXCL1, MIF and CSF-2. CpG DNAs have long been recognised as
a therapeutic strategy with an immunostimulatory antitumour
activity (22). However, the
antitumour characteristic of ODN-M362 may be diminished by cell
properties. When ODN-M362 was introduced into HCC827 cells, TLR9
signalling was increased, which revealed an mtDNA expression
phenomenon similar to that in non-metastatic cancer patients. This
result was consistent with the findings that patients with
colorectal cancer had higher mtDNA expression and exhibited higher
expression of immunosuppressive cytokines, such as IL-2 and TGF-β1
(32).
Therefore, we deduced that the antitumour
characteristic of ODN-M362 may have an improved therapeutic effect
in NSCLC patients without metastasis. Moreover, HCC827 cells had
more in common in terms of biological features with NSCLC patients
without metastasis, and may provide a model for mtDNA-related
research in the future. In addition, the cytokine expression
diversity in A549 and HCC827 cell lines may serve as a referral
index for identification of the corresponding cancer stages.
As for A549 and HCC827 cells, these two cell lines
exhibited innate discrepancies in drug responses, including
gefitinib and erlotinib. Although many well-known TKIs have been
investigated regarding NSCLC treatment, EGFR appears to be the key
element that determines drug sensitivity (34). Therefore, it is believed that EGFR
mutation is the pivot that causes differential responses in A549
and HCC827 cells to ODN-M362 treatment; however, other factors may
be involved and need to be clarified in the future.
In line with a previous finding concerning EGFR
mutations associated with NSCLC, one upstream protein, CMTM7 was
found to be responsible for a reduction in EGFR/AKT signalling in
the HCC827 cell line. In addition, when the expression of CMTM7 was
knocked down, it promoted NSCLC cell proliferation and metastasis
(35). This result not only
indicated that the intrinsic characteristic of HCC827 cells is less
or no metastasis, but also indicated that the application of
ODN-M362 to NSCLC therapy holds promise.
The ODN-M362-based therapy has been applied to
numerous cancers, particularly lung cancer. It was proposed that
TLR9 expression in malignant tumour cells may affect treatment
approaches using a TLR9 agonist, such as CpG ODN. Combined with our
findings, these data indicated that the NSCLC patients without
metastasis have more serum mtDNA. This notion indicated that the
endogenous high mtDNA level revealed suppression of tumour
metastasis. Therefore, we concluded that serological mtDNA testing
may serve as a diagnostic marker, and ODN-M362 therapy may be more
effective before metastasis (36).
One research group reported that a novel
immunomodulatory TLR9 agonist (IMO) inhibited the expression of
EGFR, pMAPK, pAkt, TNF-α, and Bcl-2 and had antitumour effects on
human colon cancer xenografts. In addition, combined with an EGFR
TKI, IMO-mediated inhibition of EGFR signalling was synergistically
enhanced (37). More than 40% of
lung cancers exhibit EGFR overexpression or mutations, and
EGFR-targeted treatment has become one of the most common
molecularly targeted therapies for lung cancer (38). In conclusion, although ODN-M362
caused significant induction of TLR9 signalling in the HCC827 cell
line, we hypothesised that TLR9 agonists may fail to induce
chemotaxis for leukocytes and may have a potential
immunotherapeutic value in EGFR-mutated human lung adenocarcinoma.
However, the biological role of TLR9 expression activated by
ODN-M362 in advanced cancer and metastasis is still unclear and
needs to be further evaluated.
In conclusion, serum mtDNA may serve as a diagnostic
biomarker for identifying NSCLC with or without metastasis.
Moreover, the different expression of TLR9-induced cytokines could
also be an index for stage classification and may help diagnose
progression and provide a prognosis in human lung cancer.
Acknowledgements
The authors thank the technical support from the
Taipei Tzu Chi Hospital, Core Laboratory.
Funding
The present study was supported by the Buddhist Tzu
Chi General Hospital grants: TCRD-TPE-95-25, TCRD-TPE-96-29 and
TCRD-TPE-97-13 to SW.
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
SW and YPC conceived and designed the study. HYL,
SW, CYH and YHL performed the experiments. YHL and SW wrote the
paper. CYH, YHL and SW reviewed and edited the manuscript. All
authors read and approved the manuscript and agree to be
accountable for all aspects of the research in ensuring that the
accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
The Ethics Committee of the Taipei Tzu-Chi General
Hospital approved the study (Approval no. TCRD-TPE-100-43; IRB no.
98-IRB-019-X; IRB extension no. 05-XD55-107). Patient written
informed consent was obtained.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wallace DC: Mitochondrial DNA sequence
variation in human evolution and disease. Proc Natl Acad Sci USA.
91:8739–8746. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Liu B, Du Q, Chen L, Fu G, Li S, Fu L,
Zhang X, Ma C and Bin C: CpG methylation patterns of human
mitochondrial DNA. Sci Rep. 6:234212016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Michl L and Shumacher HB Jr: Pulmonary
valvular incompetence in growing animals. Surgery. 73:412–415.
1973.PubMed/NCBI
|
|
4
|
Cavalli LR and Liang BC: Mutagenesis,
tumorigenicity, and apoptosis: Are the mitochondria involved? Mutat
Res. 398:19–26. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Parrella P, Xiao Y, Fliss M,
Sanchez-Cespedes M, Mazzarelli P, Rinaldi M, Nicol T, Gabrielson E,
Cuomo C, Cohen D, et al: Detection of mitochondrial DNA mutations
in primary breast cancer and fine-needle aspirates. Cancer Res.
61:7623–7626. 2001.PubMed/NCBI
|
|
6
|
Hosgood HD III, Liu CS, Rothman N,
Weinstein SJ, Bonner MR, Shen M, Lim U, Virtamo J, Cheng WL,
Albanes D, et al: Mitochondrial DNA copy number and lung cancer
risk in a prospective cohort study. Carcinogenesis. 31:847–849.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jakupciak JP, Maragh S, Markowitz ME,
Greenberg AK, Hoque MO, Maitra A, Barker PE, Wagner PD, Rom WN,
Srivastava S, et al: Performance of mitochondrial DNA mutations
detecting early stage cancer. BMC Cancer. 8:2852008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Galluzzi L, Kepp O and Kroemer G:
Mitochondria: Master regulators of danger signalling. Nat Rev Mol
Cell Biol. 13:780–788. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Goulopoulou S, Matsumoto T, Bomfim GF and
Webb RC: Toll-like receptor 9 activation: A novel mechanism linking
placenta-derived mitochondrial DNA and vascular dysfunction in
pre-eclampsia. Clin Sci. 123:429–435. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Underhill DM, Ozinsky A, Smith KD and
Aderem A: Toll-like receptor-2 mediates mycobacteria-induced
proinflammatory signaling in macrophages. Proc Natl Acad Sci USA.
96:14459–14463. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Latz E, Visintin A, Espevik T and
Golenbock DT: Mechanisms of TLR9 activation. J Endotoxin Res.
10:406–412. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nishiya T, Kajita E, Miwa S and Defranco
AL: TLR3 and TLR7 are targeted to the same intracellular
compartments by distinct regulatory elements. J Biol Chem.
280:37107–37117. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang Q, Raoof M, Chen Y, Sumi Y, Sursal
T, Junger W, Brohi K, Itagaki K and Hauser CJ: Circulating
mitochondrial DAMPs cause inflammatory responses to injury. Nature.
464:104–107. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hemmi H, Takeuchi O, Kawai T, Kaisho T,
Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, et al:
A Toll-like receptor recognizes bacterial DNA. Nature. 408:740–745.
2000. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Latz E, Schoenemeyer A, Visintin A,
Fitzgerald KA, Monks BG, Knetter CF, Lien E, Nilsen NJ, Espevik T
and Golenbock DT: TLR9 signals after translocating from the ER to
CpG DNA in the lysosome. Nat Immunol. 5:190–198. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Akira S and Hoshino K: Myeloid
differentiation factor 88-dependent and -independent pathways in
toll-like receptor signaling. J Infect Dis. 187 Suppl 2:S356–S363.
2003. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Vollmer J: TLR9 in health and disease. Int
Rev Immunol. 25:155–181. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Berger R, Fiegl H, Goebel G, Obexer P,
Ausserlechner M, Doppler W, Hauser-Kronberger C, Reitsamer R, Egle
D, Reimer D, et al: Toll-like receptor 9 expression in breast and
ovarian cancer is associated with poorly differentiated tumors.
Cancer Sci. 101:1059–1066. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Väisänen MR, Väisänen T, Jukkola-Vuorinen
A, Vuopala KS, Desmond R, Selander KS and Vaarala MH: Expression of
toll-like receptor-9 is increased in poorly differentiated prostate
tumors. Prostate. 70:817–824. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kauppila JH, Takala H, Selander KS,
Lehenkari PP, Saarnio J and Karttunen TJ: Increased Toll-like
receptor 9 expression indicates adverse prognosis in oesophageal
adenocarcinoma. Histopathology. 59:643–649. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sandholm J and Selander KS: Toll-like
receptor 9 in breast cancer. Front Immunol. 5:3302014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Krieg AM: Toll-like receptor 9 (TLR9)
agonists in the treatment of cancer. Oncogene. 27:161–167. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wooldridge JE and Weiner GJ: CpG DNA and
cancer immunotherapy: Orchestrating the antitumor immune response.
Curr Opin Oncol. 15:440–445. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2ΔΔCT method. Methods.
25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lo YM, Tein MS, Lau TK, Haines CJ, Leung
TN, Poon PM, Wainscoat JS, Johnson PJ, Chang AM and Hjelm NM:
Quantitative analysis of fetal DNA in maternal plasma and serum:
Implications for noninvasive prenatal diagnosis. Am J Hum Genet.
62:768–775. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Vecchiarelli A, Retini C, Casadevall A,
Monari C, Pietrella D and Kozel TR: Involvement of C3a and C5a in
interleukin-8 secretion by human polymorphonuclear cells in
response to capsular material of Cryptococcus neoformans. Infect
Immun. 66:4324–4330. 1998.PubMed/NCBI
|
|
27
|
Waugh DJ and Wilson C: The interleukin-8
pathway in cancer. Clin Cancer Res. 14:6735–6741. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Arango Duque G and Descoteaux A:
Macrophage cytokines: Involvement in immunity and infectious
diseases. Front Immunol. 5:4912014.PubMed/NCBI
|
|
29
|
Aldinucci D and Colombatti A: The
inflammatory chemokine CCL5 and cancer progression. Mediators
Inflamm. 2014:2923762014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lee MS, Kim HP, Kim TY and Lee JW:
Gefitinib resistance of cancer cells correlated with
TM4SF5-mediated epithelial-mesenchymal transition. Biochim Biophys
Acta. 1823:514–523. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lee H, Lee JH, Kim DC, Hwang I, Kang YN,
Gwon GJ, Choi IJ and Kim S: Is mitochondrial DNA copy number
associated with clinical characteristics and prognosis in gastric
cancer? Asian Pac J Cancer Prev. 16:87–90. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Qu F, Chen Y, Wang X, He X, Ren T, Huang
Q, Zhang J, Liu X, Guo X, Gu J and Xing J: Leukocyte mitochondrial
DNA content: A novel biomarker associated with prognosis and
therapeutic outcome in colorectal cancer. Carcinogenesis.
36:543–552. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Weerts MJ, Sieuwerts AM, Smid M, Look MP,
Foekens JA, Sleijfer S and Martens JW: Mitochondrial DNA content in
breast cancer: Impact on in vitro and in vivo phenotype and patient
prognosis. Oncotarget. 7:29166–29176. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pao W and Chmielecki J: Rational,
biologically based treatment of EGFR-mutant non-small-cell
lung cancer. Nat Rev Cancer. 10:760–774. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu B, Su Y, Li T, Yuan W, Mo X, Li H, He
Q, Ma D and Han W: CMTM7 knockdown increases tumorigenicity of
human non-small cell lung cancer cells and EGFR-AKT signaling by
reducing Rab5 activation. Oncotarget. 6:41092–41107. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Droemann D, Albrecht D, Gerdes J, Ulmer
AJ, Branscheid D, Vollmer E, Dalhoff K, Zabel P and Goldmann T:
Human lung cancer cells express functionally active Toll-like
receptor 9. Respir Res. 6:12005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Damiano V, Caputo R, Bianco R, D'Armiento
FP, Leonardi A, De Placido S, Bianco AR, Agrawal S, Ciardiello F
and Tortora G: Novel toll-like receptor 9 agonist induces epidermal
growth factor receptor (EGFR) inhibition and synergistic antitumor
activity with EGFR inhibitors. Clin Cancer Res. 12:577–583. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Han R, Wang X, Zhong D, Zhao J, Chen Z,
Sun L, Wang J and Zhang J: Molecular mechanism of erlotinib
resistance in epidermal growth factor receptor mutant non-small
cell lung cancer cell line H1650. Zhongguo Fei Ai Za Zhi.
15:689–693. 2012.(In Chinese). PubMed/NCBI
|