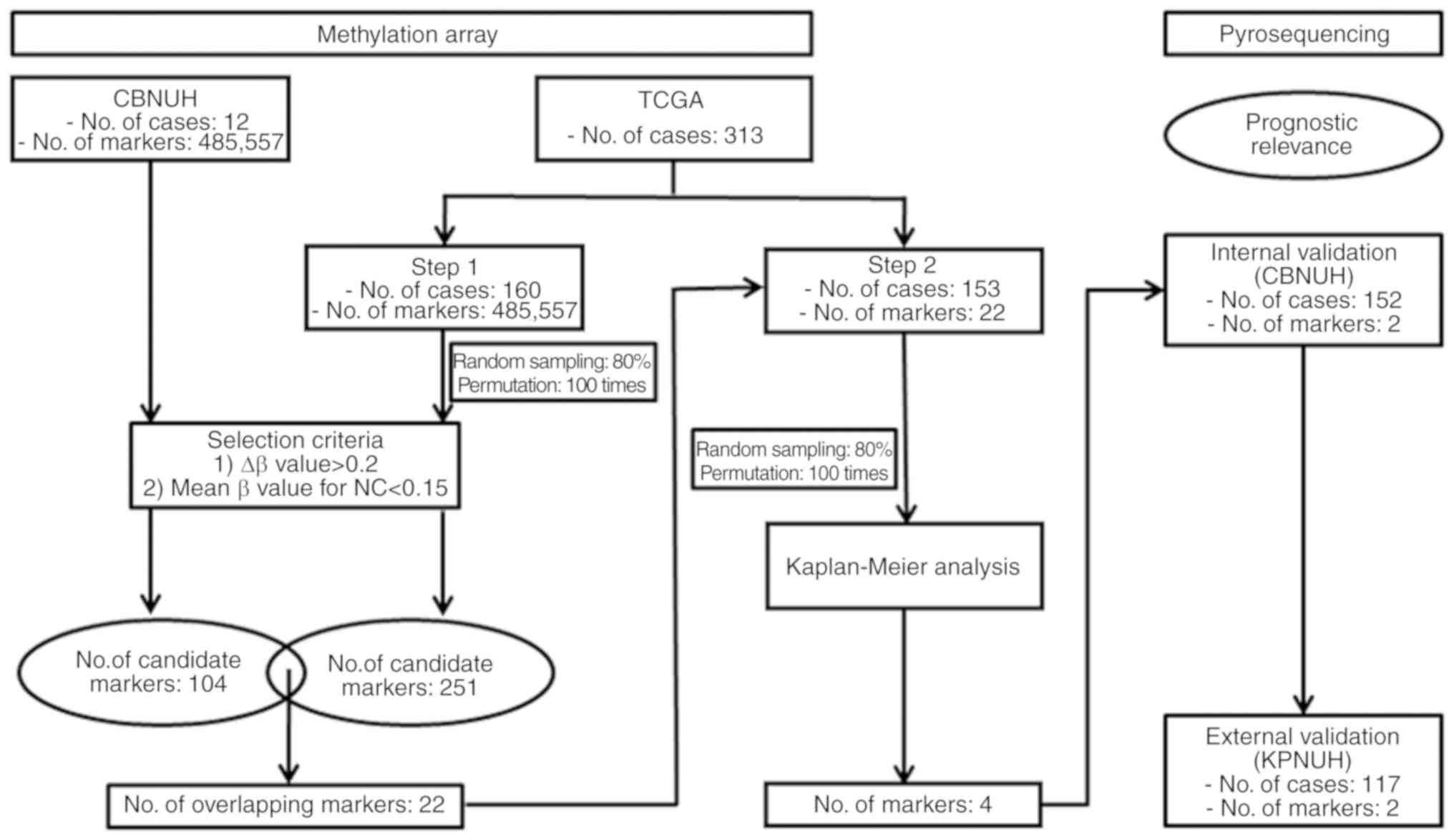
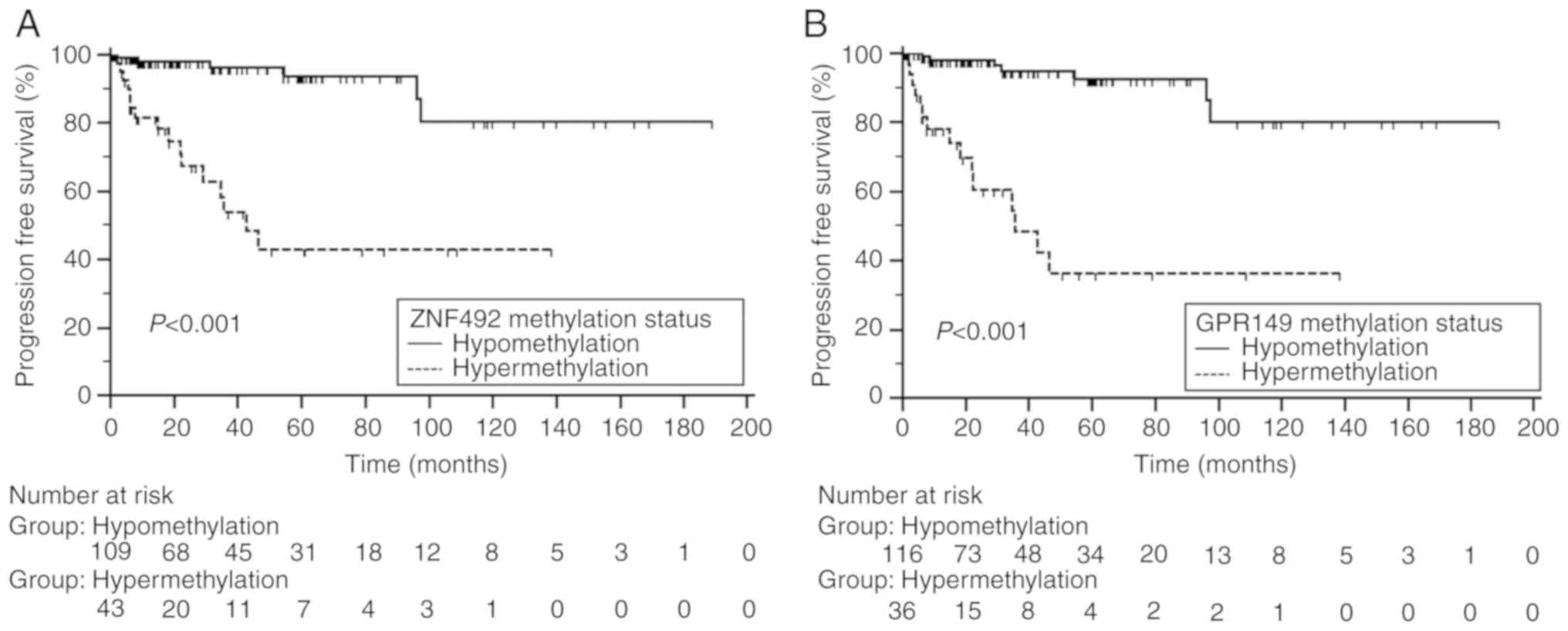
|
1
|
Ljungberg B, Bensalah K, Canfield S,
Dabestani S, Hofmann F, Hora M, Kuczyk MA, Lam T, Marconi L,
Merseburger AS, et al: EAU guidelines on renal cell carcinoma: 2014
update. Eur Urol. 67:913–924. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Srinivasan R, Ricketts CJ, Sourbier C and
Linehan WM: New strategies in renal cell carcinoma: Targeting the
genetic and metabolic basis of disease. Clin Cancer Res. 21:10–17.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Eggers H, Steffens S, Grosshennig A,
Becker JU, Hennenlotter J, Stenzl A, Merseburger AS, Kuczyk MA and
Serth J: Prognostic and diagnostic relevance of hypermethylated in
cancer 1 (HIC1) CpG island methylation in renal cell carcinoma. Int
J Oncol. 40:1650–1658. 2012.PubMed/NCBI
|
|
4
|
Morris MR, Ricketts C, Gentle D,
Abdulrahman M, Clarke N, Brown M, Kishida T, Yao M, Latif F and
Maher ER: Identification of candidate tumour suppressor genes
frequently methylated in renal cell carcinoma. Oncogene.
29:2104–2117. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
van Vlodrop IJ, Baldewijns MM, Smits KM,
Schouten LJ, van Neste L, van Criekinge W, van Poppel H, Lerut E,
Schuebel KE, Ahuja N, et al: Prognostic significance of Gremlin1
(GREM1) promoter CpG island hypermethylation in clear cell renal
cell carcinoma. Am J Pathol. 176:575–584. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kawai Y, Sakano S, Suehiro Y, Okada T,
Korenaga Y, Hara T, Naito K, Matsuyama H and Hinoda Y: Methylation
level of the RASSF1A promoter is an independent prognostic factor
for clear-cell renal cell carcinoma. Ann Oncol. 21:1612–1617. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lasseigne BN, Burwell TC, Patil MA, Absher
DM, Brooks JD and Myers RM: DNA methylation profiling reveals novel
diagnostic biomarkers in renal cell carcinoma. BMC Med. 12:2352014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Arai E, Ushijima S, Tsuda H, Fujimoto H,
Hosoda F, Shibata T, Kondo T, Imoto I, Inazawa J, Hirohashi S and
Kanai Y: Genetic clustering of clear cell renal cell carcinoma
based on array-comparative genomic hybridization: Its association
with DNA methylation alteration and patient outcome. Clin Cancer
Res. 14:5531–5539. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ricketts CJ, Morris MR, Gentle D, Shuib S,
Brown M, Clarke N, Wei W, Nathan P, Latif F and Maher ER:
Methylation profiling and evaluation of demethylating therapy in
renal cell carcinoma. Clin Epigenetics. 5:162013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Slater AA, Alokail M, Gentle D, Yao M,
Kovacs G, Maher ER and Latif F: DNA methylation profiling
distinguishes histological subtypes of renal cell carcinoma.
Epigenetics. 8:252–267. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wei JH, Haddad A, Wu KJ, Zhao HW, Kapur P,
Zhang ZL, Zhao LY, Chen ZH, Zhou YY, Zhou JC, et al: A
CpG-methylation-based assay to predict survival in clear cell renal
cell carcinoma. Nat Commun. 6:86992015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Arai E, Kanai Y, Ushijima S, Fujimoto H,
Mukai K and Hirohashi S: Regional DNA hypermethylation and DNA
methyltransferase (DNMT) 1 protein overexpression in both renal
tumors and corresponding nontumorous renal tissues. Int J Cancer.
119:288–296. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ellinger J, Kahl P, Mertens C, Rogenhofer
S, Hauser S, Hartmann W, Bastian PJ, Büttner R, Müller SC and von
Ruecker A: Prognostic relevance of global histone H3 lysine 4
(H3K4) methylation in renal cell carcinoma. Int J Cancer.
127:2360–2366. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Greene FL: The American Joint Committee on
Cancer: Updating the strategies in cancer staging. Bull Am Coll
Surg. 87:13–15. 2002.PubMed/NCBI
|
|
15
|
Lopez-Beltran A, Scarpelli M, Montironi R
and Kirkali Z: 2004 WHO classification of the renal tumors of the
adults. Eur Urol. 49:798–805. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fuhrman SA, Lasky LC and Limas C:
Prognostic significance of morphologic parameters in renal cell
carcinoma. Am J Surg Pathol. 6:655–663. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Baylin SB and Jones PA: A decade of
exploring the cancer epigenome-biological and translational
implications. Nat Rev Cancer. 11:726–734. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ushijima T: Detection and interpretation
of altered methylation patterns in cancer cells. Nat Rev Cancer.
5:223–231. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Krishna SS, Majumdar I and Grishin NV:
Structural classification of zinc fingers: Survey and summary.
Nucleic Acids Res. 31:532–550. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jen J and Wang YC: Zinc finger proteins in
cancer progression. J Biomed Sci. 23:532016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cheng Y, Liang P, Geng H, Wang Z, Li L,
Cheng SH, Ying J, Su X, Ng KM, Ng MH, et al: A novel 19q13
nucleolar zinc finger protein suppresses tumor cell growth through
inhibiting ribosome biogenesis and inducing apoptosis but is
frequently silenced in multiple carcinomas. Mol Cancer Res.
10:925–936. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Deng J, Liang H, Ying G, Dong Q, Zhang R,
Yu J, Fan D and Hao X: Poor survival is associated with the
methylated degree of zinc-finger protein 545 (ZNF545) DNA promoter
in gastric cancer. Oncotarget. 6:4482–4495. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jen J, Lin LL, Chen HT, Liao SY, Lo FY,
Tang YA, Su WC, Salgia R, Hsu CL, Huang HC, et al: Oncoprotein
ZNF322A transcriptionally deregulates alpha-adducin, cyclin D1 and
p53 to promote tumor growth and metastasis in lung cancer.
Oncogene. 35:2357–2369. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang L, Hamilton SR, Sood A, Kuwai T,
Ellis L, Sanguino A, Lopez-Berestein G and Boyd DD: The previously
undescribed ZKSCAN3 (ZNF306) is a novel ‘driver’ of colorectal
cancer progression. Cancer Res. 68:4321–4330. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bar-Shavit R, Maoz M, Kancharla A, Nag JK,
Agranovich D, Grisaru-Granovsky S and Uziely B: G protein-coupled
receptors in cancer. Int J Mol Sci. 17:E13202016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lappano R and Maggiolini M: G
protein-coupled receptors: Novel targets for drug discovery in
cancer. Nat Rev Drug Discov. 10:47–60. 2011. View Article : Google Scholar : PubMed/NCBI
|