Esophageal cancer (ESCA) is a solid tumor and
frequently occurs in the digestive tract. It is highly invasive and
has a high incidence rate and mortality (1); its cancer mortality ranks sixth
worldwide and the overall 5-year survival rate is low (2–4); it is
estimated to be ~15–25% (4,5). According to histopathological results,
it can be classified into esophageal adenocarcinoma (EAC) and
esophageal squamous cell carcinoma (ESCC). Due to differences in
etiology and geographical incidence rate, EAC is common in the
Western population, while ESCC is prevalent in the Eastern
population (5–8). The incidence of ESCA is closely
associated with smoking, alcohol consumption, unhealthy dietary
habits and obesity (9). In China,
the incidence rate of ESCC is higher due to its early symptoms not
being obvious (10); therefore, the
majority of the patients have developed late-stage disease when
diagnosed, with low quality of life and poor prognosis (2). At present, the conventional clinical
treatment methods for ESCA are surgery, chemotherapy and
radiotherapy (11); however, with
the complex development of the disease, including invasion and
metastasis, as well as drug resistance and normal cell toxicity,
traditional treatment methods face significant challenges (12). In order to enhance treatment
effectiveness and improve the low survival rate, treatment methods
need to be constantly updated and strengthened, which requires an
urgent proposal of new strategies.
MicroRNAs (miRNAs/miRs) are a class of non-coding
conserved small-molecule RNAs with a length of ~20 nucleotides
(13). Mature RNA binds to the
corresponding mRNA molecule, regulates the relevant gene
expression, participates in the regulation of biological behaviors,
such as cell proliferation, differentiation, apoptosis and
metabolism, and plays a wide range of regulatory roles (13–18).
In cancer, certain miRNAs are often abnormally expressed and become
tumor promoters or suppressors (13,16,19–23).
Signal transduction and transcription activator 3 (STAT3) is a
widely studied crucial component that connects cellular signaling
and transcription (24), controls
multiple signaling pathways and acts as a hub gene (25). Abnormal expression of STAT3 can lead
to cellular dysfunction, development of diseases and various
malignant tumors in humans (26).
In tumor tissues, constitutively activated STAT3 can promote
multiple malignant behaviors of cancer cells (24,25,27–33).
Numerous studies have shown that miRNAs can participate in
regulating the STAT3 signaling pathway; furthermore, STAT3 can
modulate the expression levels of multiple miRNAs. MiRNAs and STAT3
interact via direct or indirect mechanisms, forming signaling
circuits that affect cellular balance (34–37).
With the continuous advancement in molecular biology
research, an accumulating number of studies have indicated that the
occurrence of cancer is related to unusual signaling routes within
cells. Furthermore, targeted therapy has become a method of
precision treatment for tumors (12). The STAT3 signaling pathway mediated
by miRNAs has been widely explored in various cancer types; e.g. in
gastric cancer (38), leukemia
(39), colorectal cancer (26) and ESCA (40). A review was conducted on the miRs
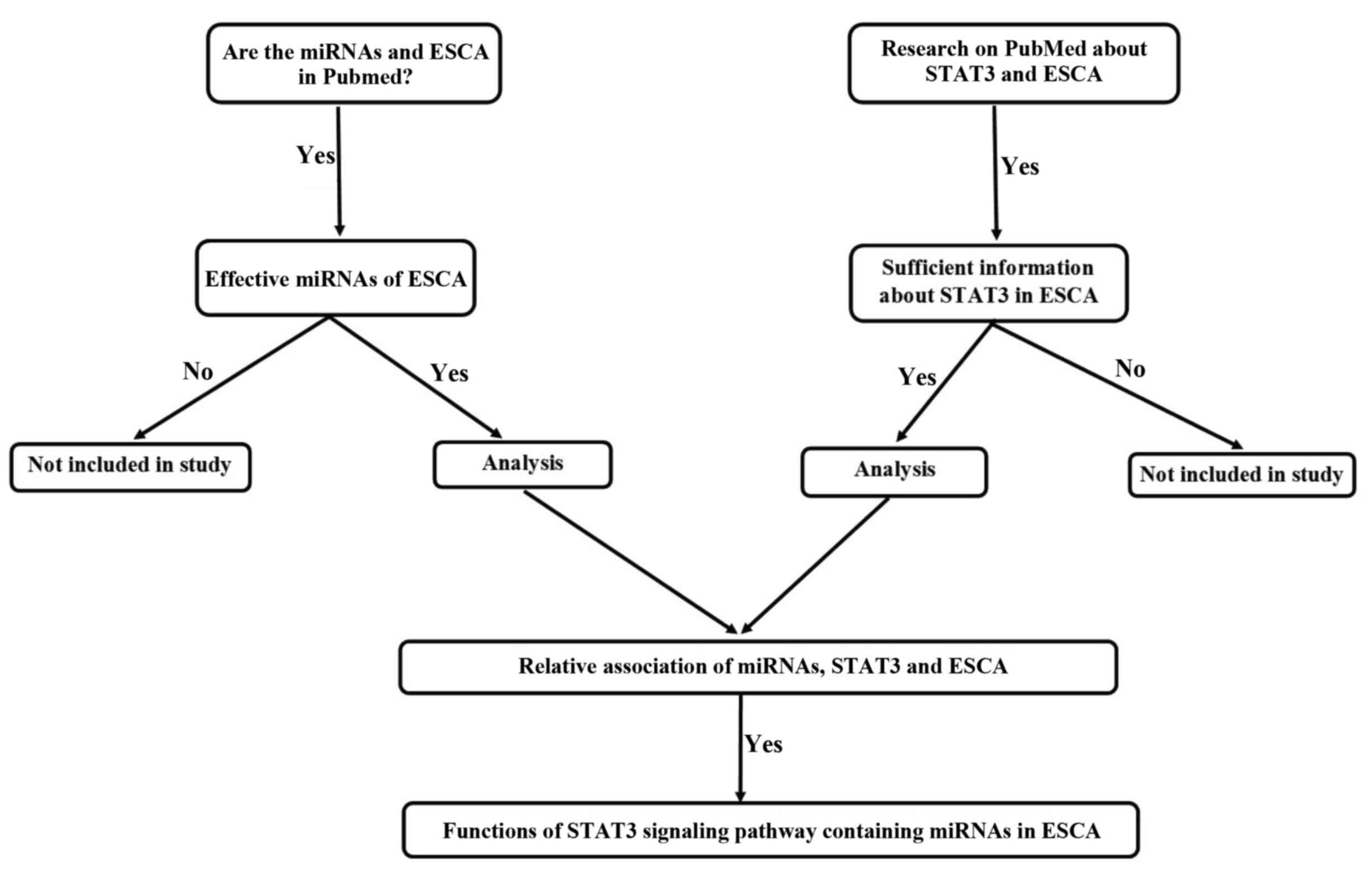
involved in the STAT3 signaling pathway in ESCA following a
literature search in the PubMed database using the key words
‘esophageal cancer’, ‘STAT3’ and ‘miRNA’. The present review
unveiled the complex pathogenesis behind ESCA by analyzing the
interaction between STAT3 and different miRNAs, as well as the
impact of miRNAs on the STAT3 signaling pathway (Fig. 1); it also reviewed the effects of
different miRNAs and the STAT3 signaling pathway on the
proliferation, invasion, metastasis, apoptosis, drug resistance and
immune escape of ESCA. Understanding the molecular mechanisms of
the interaction between miRNAs and STAT3 can provide favorable
conditions to identify potential therapeutic targets for ESCA and
develop novel anticancer drugs, laying a foundation for formulating
new clinical plans. This is of great significance for improving
disease prognosis and provides hope for the survival of patients
with ESCA.
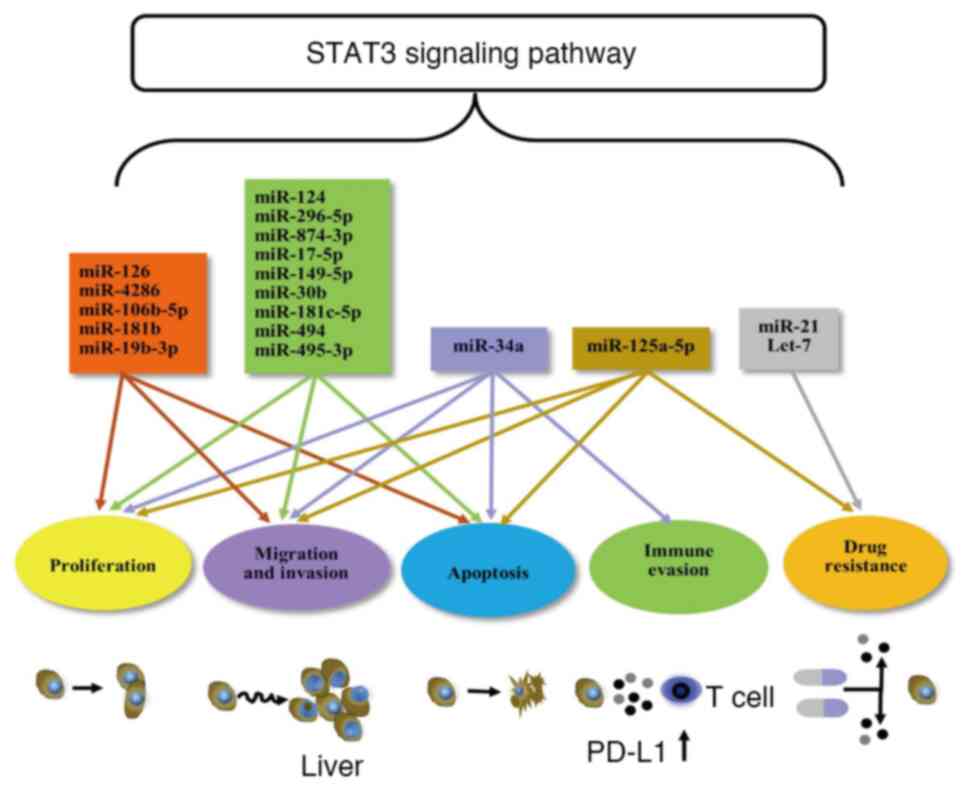
The majority of the miRNAs associated with the STAT3
signaling pathway exert inhibitory effects on tumors, including
miR-124 (41,42), miR-125a-5p (43), miR-296-5p (44), miR-874-3p (45), miR-17-5p (46), miR-149-5p (47), miR-30b (48), miR-181c-5p (49), miR-494 (50), miR-495-3p (51) and miR-34a (52) (Table
I; Figs. 2 and 3).
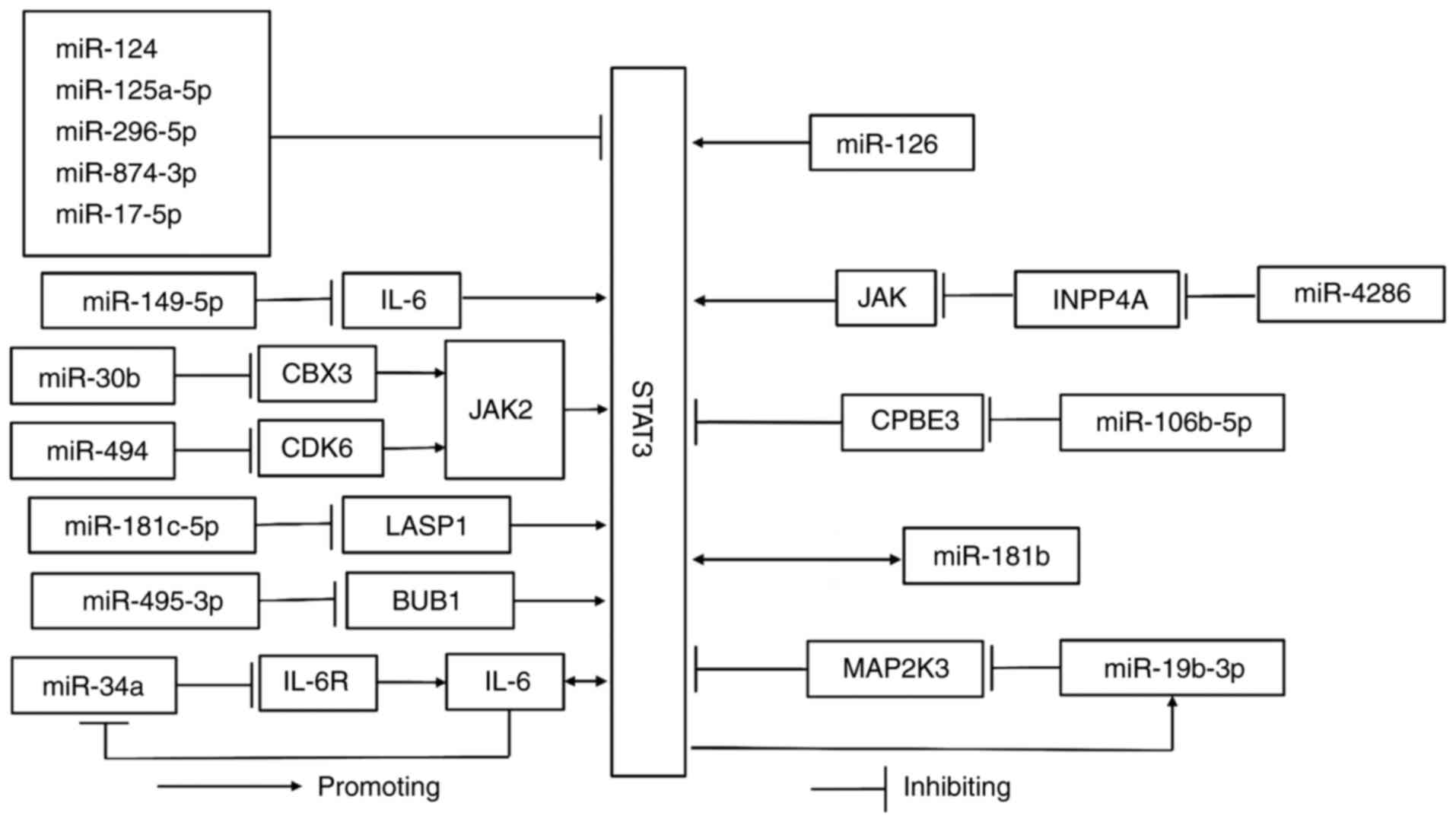
As a highly conserved miRNA, miR-124 is a mediator
of the STAT3 signaling pathway, which inhibits cancer development;
it has been confirmed in various studies. Overexpressed miR-124 can
downregulate STAT3 and further inhibit the growth of bladder cancer
(53), colorectal cancer (54) and hepatocellular carcinoma (55). In ESCA, the expression levels of
miR-124 are significantly reduced, STAT3 is constitutively
activated and its content is negatively associated with miR-124.
Previous research studies have shown that miR-124 can directly
inhibit STAT3 and further hinder cell proliferation, invasiveness
and metastasis and aid the induction of apoptosis (41,42).
Similarly, upregulation of miR-125a-5p levels causes binding to
STAT3 and reduces STAT3 expression, leading to cell-cycle arrest
and prevention of cell growth in ESCC (43); miR-296-5p inhibits the occurrence of
ESCC by downregulating STAT3 levels (44). STAT3 is a direct functional target
of miR-874-3p in ESCC cells; following their combination, they can
form the miR-874-3p/STAT3 signaling axis to prevent the malignant
behaviors of tumor cells (45).
STAT3 is the direct downstream target of miR-17-5p, which binds to
STAT3 to reduce the proliferative, migratory and invasive
capacities of esophageal cancer cell line TE-1 (46).
In addition to the direct binding to STAT3, miRNAs
can indirectly mediate the STAT3 signaling pathway. MiR-149-5p
inhibits the expression of interleukin-6 (IL-6) at the
transcriptional and translational level. IL-6 is a general STAT3
activator (29,56) and overexpression of miR-149-5p can
suppress the abnormal proliferation, migration and invasion of ESCC
cells via the miR-149-5p/IL-6/STAT3 signaling pathway (47). Chromobox 3 (CBX3) is a member of the
chromobox protein family, which promotes the development of various
cancer types, including gastric cancer (57), breast (58) and pancreatic cancer (59). The Janus kinase (JAK)2/STAT3 is a
common signaling pathway found in various tumors (24,31,60–62).
CBX3 is a downstream target of miR-30b expressed in ESCC cells.
When the levels of miR-30b are reduced, the expression levels of
CBX3, phosphorylated (p-)JAK2 and p-STAT3 are markedly increased
and the JAK2/STAT3 signaling pathway is apparently active.
Therefore, upregulation of miR-30b expression can inhibit cancer
development via the miR-30b/CBX3/JAK2/STAT3 signaling axis
(48). MiR-181c-5p indirectly
controls ESCC by binding to LIM and the SH3 protein 1 (LASP1),
which can activate STAT3. MiR-181c-5p decreases STAT3 expression
and hinders cell proliferation, invasiveness and migration by
interacting with LASP1 (49).
MiR-494 can negatively regulate the expression of cyclin-dependent
kinase (CDK)6, which promotes cell-cycle progression (63). Upregulated miR-494 binds to CDK6 to
inhibit the downstream JAK2/STAT3 signaling pathway, resulting in
an antitumor effect (50). By
targeting Budding uninhibited by benzimidazoles 1 (BUB1), which can
directly upregulate the phosphorylation levels of STAT3, miR-495-3p
can also hinder the growth of ESCA via the miR-495-3p/BUB1/STAT3
signaling pathway (51). The
aforementioned miRNAs are upstream molecules of STAT3 and miR-34a,
an effective tumor suppressor, which can form a feedback loop with
STAT3 to inhibit ESCA. Programmed cell death-ligand 1 (PD-L1) and
IL-6 receptor (IL-6R) are targets in the loop and miR-34a
competitively blocks the binding of IL-6 to its receptor to inhibit
STAT3 activation. However, overactivated STAT3 can induce IL-6.
Ultimately, they can form the miR-34a/STAT3/IL-6R feedback loop and
increase miR-34a expression, which can hinder the progression of
ESCC (52). In addition, miR-34a
targets the E2F transcription factor 5, which can affect epithelial
mesenchymal transition (EMT) and inhibit migration of ESCC
(64). By inhibiting the
PI3K/AKT/mTOR signaling pathway, miR-34a can reverse radiation
resistance of ESCC and improve radiotherapy efficacy (65). Overexpressed miR-34a levels can also
control the expression of matrix metalloproteinase (MMP)-2 and
MMP-9 or bind to Yinyang-1 to inhibit ESCC metastasis and invasion
(66,67).
As an important coordinating factor, miR-126 binding
to different targets exerts a certain influence on ESCA. A previous
study indicated that overexpressed miR-126 can downregulate the
content of microtubule-associated protein 1 light chain 3β and
sequestosome 1 (p62) related to autophagy. STAT3 is a direct target
of miR-126 and its silencing leads to a decrease in autophagy;
therefore, miR-126 can bind to STAT3 and may activate it, which
further inhibits apoptosis and autophagy of the esophageal squamous
cell and promotes the development of ESCC (68). However, miR-126 has an inhibitory
effect on ESCC via integrating with vascular endothelial growth
factor-A (VEGF-A) (73) or insulin
receptor substrate-1 (IRS-1) and Golgi phosphoprotein 3 (GOLPH3)
(74) and it can also mediate the
PI3K/AKT (75) or the ADAM
metallopeptidase domain 9 (ADAM-9)/EGFR/AKT (76) signaling pathway to suppress the
progression of ESCC. In addition, miR-126-5p can influence the
proliferation and invasion of ESCC by negatively regulating CDK
(77). The different signaling
pathways involved in miR-126 have the opposite effect; therefore,
the definite role of miR-126 requires further exploration under
specific conditions in ESCA. Unlike miR-126, miR-4286 and
miR-106b-5p bind to downstream molecules to indirectly induce STAT3
activation. MiR-4286 targets inositol polyphosphate-4-phophatase
type IA and negatively regulates it to evoke the JAK/STAT3 pathway;
in this way, it indirectly facilitates ESCA (69). Furthermore, miR-4286 can be a member
of the miR spectrum that distinguishes ECA from Barrett's esophagus
(78). Overexpression of
cytoplasmic polyadenylation element binding protein 3 (CPBE3) can
hinder tumor migration, invasion, angiogenesis and STAT3 activity
and suppress EMT in ESCA. MiR-106b-5p can negatively regulate
CPBE3, thereby enhancing the activation of STAT3 and facilitating
EMT; this results in the formation of the miR-106b-5p/CPBE3/STAT3
signaling axis, which aids tumor growth (70).
MiR-181b and miR-19b-3p promote the development of
ESCA via feedback loops. In ESCA stem cells, the combination of
STAT3 and miR-181b promoter induces miR-181b expression;
furthermore, upregulated miR-181b can increase p-STAT3 levels,
resulting in the formation of a feedback loop that leads to
anti-apoptotic effects and the formation of cell colonies (71). In addition, the interaction of
miR-181b-1, cylindromatosis and STAT3 can establish a link between
inflammation and tumor formation (79). By analogy, miR-19b-3p can inhibit
mitogen-activated protein kinase kinase 3 (MAP2K3) that can bind to
STAT3 and facilitate its degradation. STAT3 is a transcription
driver of miR-19b-3p and the three components form a positive
circuit to induce ESCC progression (72). Furthermore, miR-19b-3p targeting
phosphatase and tensin homolog can induce the occurrence of ESCA
(80), which is of great
significance for miR biomarker studies performed in ECA (81).
Cisplatin is a conventional first-line
chemotherapeutic drug used for the treatment of ESCA and resistance
to this drug limits its clinical application (82). Tumor cells treated with cisplatin
can release IL-6 and induce the phosphorylation of STAT3 in ESCA.
The release of IL-6 can evoke the survival of the IL-6/STAT3
signaling pathway, finally protecting cancer cells from cisplatin.
Low expression of Let-7 is associated with low chemotherapy
sensitivity. In contrast to these observations, upregulation of
Let-7 can decrease p-STAT3 levels to counteract the activation of
the IL-6/STAT3 pathway by cisplatin and reduce the resistance to
this drug (83). The cisplatin
resistance mechanism of miR-125a-5p is similar to that of Let-7,
which is in parallel to the deactivation of the STAT3 signaling
pathway. The ability of miR-125a-5p to regulate EMT is also
relevant to the chemical sensitivity of cisplatin. By contrast,
STAT3 activated by miR-21 can increase the production of monocytic
myeloid-derived suppressor cells, which can protect tumor cells
against the effects of cisplatin. MiR-21 can increase cisplatin
resistance via the STAT3 signaling pathway in ESCC (84) (Table
I, Fig. 2).
MiR-34a mediates the miR-34a/STAT3/IL-6R feedback
loop. PD-L1 and IL-6R are downstream targets of miR-34a present in
ESCA cells. MiR-34a can inhibit them via binding to PD-L1 and
IL-6R, while IL-6R can promote STAT3 expression, which can inhibit
miR-34a. Based on the aforementioned mechanism, PD-L1 can
participate in the miR-34a/IL-6R/STAT3 feedback loop. Furthermore,
low levels of PD-L1 expression can boost the immune system. When
miR-34a levels are reduced, loss of PD-L1 expression causes an
upstream gene constraint, further promoting immune escape. As
miR-34a levels are upregulated, PD-L1 expression is restricted by
upstream molecular pathways to reverse tumor immune escape and
strengthen T-cell immune response (52,85)
(Table I; Fig. 2).
It has been shown that downregulation of the
expression levels of CPBE3 and MAP2K3 is linked to low clinical
survival and poor prognosis in ESCA, whereas miR-106b-5p and
miR-19b-3p respectively inhibit their expression levels (70,72).
It has been suggested that certain miRNAs are associated via STAT3
signaling with low survival in patients. MiR-21, which can promote
cisplatin resistance, originates from cancer-associated fibroblasts
(CAFs). The highly infiltrated CAFs are associated with a low
survival rate and can increase miR-21; therefore, overexpressed
miR-21 levels affect patient survival in ESCC (84). A reduction in the expression levels
of miR-125a-5p and miR-874-3p is often associated with a lower rate
of survival and their expression levels can also aid the
determination of the clinical stage of ESCC (43,45).
Upregulation of Let-7 expression frequently suggests improved
prognosis for patients with this disease (83) (Table
I).
ESCA is a familiar malignant tumor with a low early
diagnostic rate and late cure rate. It is necessary to positively
explore the pathogenic mechanism for formulating novel treatment
strategies. It has been found that the STAT3 signaling pathway
involving miRNAs can play a crucial role in the occurrence and
development of ESCA. Upregulation of miR-124, miR-125a-5p,
miR-296-5p, miR-874-3p, miR-17-5p, miR-149-5p, miR-30b,
miR-181c-5p, miR-494, miR-495-3p and miR-34a can form signaling
regulatory axes with STAT3 to inhibit proliferation, invasiveness
and metastasis and promote apoptosis in ESCA. However, miR-126,
miR-4286, miR-106b-5p, miR-181b and miR-19b-3p positively regulate
ESCA growth and foster malignant behaviors by interacting with
STAT3. These roles of the miRNA/STAT3 signaling axis may provide
novel opportunities for the use of miRNAs as molecular targets for
treating ESCA. Perhaps targeted promotion or inhibition of the
aforementioned miRNAs via various means can be used as a method to
treat ESCA. The STAT3 signaling pathways mediated by Let-7 and
miR-21 can influence ESCA resistance to cisplatin and further
affect drug efficacy. Increasing Let-7 levels or downregulating
miR-21 levels can promote the potential of drug therapy for ESCA.
It is worth noting that certain signaling pathways can regulate
multiple functions. MiR-125a-5p inhibits the progression of tumors
and lowers resistance to cisplatin; miR-34a not only hinders cancer
development but also increases immunity, weakening the escape of
cancer cells. The higher survival rate and improved prognosis of
patients are associated with increased expression levels of
miR-125a-5p, miR-874-3p and Let-7, while overexpression of
miR-19b-3p, miR-106b-5p and miR-21 are related to poor prognosis
and a lower survival rate. Of note, when miR-126 targets VEGF-A,
IRS-1, GOLPH3 or CDK6 and mediates the PI3K/AKT or ADAM-9/EGFR/AKT
signaling axis, it has an inhibitory effect on ESCA. As it combines
with STAT3, its effect is inverse. The same signaling molecules
have completely opposite effects on different signaling pathways.
Therefore, it is important to accurately determine the role of each
signaling pathway.
In conclusion, the complex signaling network formed
by miRNAs and STAT3 affects multiple pathophysiological processes
in ESCA. Therefore, further research is required to assess the
detailed mechanisms by which miRNAs and STAT3 signaling pathways
control ESCA. These results can provide new insights into the
clinical treatment required for various diseases and improve
patient quality of life.
Not applicable.
This work was financially supported by Sichuan Science and
Technology Program (grant no. 2022YFS0636) and Luzhou Science and
Technology Program (grant no. 2024JYJ061).
Not applicable.
XY made major contributions to the data analysis and
manuscript writing. LYF collected data and figures. YZH and HCG
participated in writing and revising the manuscript. All authors
discussed, carefully read and approved the final manuscript. Data
authentication is not applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Obermannová R, Alsina M, Cervantes A,
Leong T, Lordick F, Nilsson M, van Grieken NCT, Vogel A and Smyth
EC; ESMO Guidelines Committee. Electronic address, : simpleclinicalguidelines@esmo.org:
Oesophageal cancer: ESMO clinical practice guideline for diagnosis,
treatment and follow-up. Ann Oncol. 33:992–1004. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zeng H, Zhang F, Sun Y, Li S and Zhang W:
Treatment options for neoadjuvant strategies of esophageal squamous
cell carcinoma (Review). Mol Clin Oncol. 20:42024. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pennathur A, Gibson MK, Jobe BA and
Luketich JD: Oesophageal carcinoma. Lancet. 381:400–412. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chung CS, Lee YC, Wang CP, Ko JY, Wang WL,
Wu MS and Wang HP: Secondary prevention of esophageal squamous cell
carcinoma in areas where smoking, alcohol, and betel quid chewing
are prevalent. J Formos Med Assoc. 109:408–421. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Watanabe M, Otake R, Kozuki R, Toihata T,
Takahashi K, Okamura A and Imamura Y: Correction to: Recent
progress in multidisciplinary treatment for patients with
esophageal cancer. Surg Today. 50:4252020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Uhlenhopp DJ, Then EO, Sunkara T and
Gaduputi V: Epidemiology of esophageal cancer: Update in global
trends, etiology and risk factors. Clin J Gastroenterol.
13:1010–1021. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Abnet CC, Arnold M and Wei WQ:
Epidemiology of esophageal squamous cell carcinoma.
Gastroenterology. 154:360–373. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chu LY, Peng YH, Weng XF, Xie JJ and Xu
YW: Blood-based biomarkers for early detection of esophageal
squamous cell carcinoma. World J Gastroenterol. 26:1708–1725. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu CQ, Ma YL, Qin Q, Wang PH, Luo Y, Xu
PF and Cui Y: Epidemiology of esophageal cancer in 2020 and
projections to 2030 and 2040. Thorac Cancer. 14:3–11. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lv H, He Z, Wang H, Du T and Pang Z:
Differential expression of miR-21 and miR-75 in esophageal
carcinoma patients and its clinical implication. Am J Transl Res.
8:3288–3298. 2016.PubMed/NCBI
|
|
11
|
Rui W, Li C, Da Q, Yue Y, Jing L, Ruirui
G, Youbin C, Lu T and Li B: Analysis of the influencing factors in
the long-term survival of esophageal cancer. Front Oncol.
13:12740142023. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ren G, Wang Z, Tian Y, Li J, Ma Y, Zhou L,
Zhang C, Guo L, Diao H, Li L, et al: Targeted chemo-photodynamic
therapy toward esophageal cancer by GSH-sensitive theranostic
nanoplatform. Biomed Pharmacother. 153:1135062022. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Del Valle-Morales D, Le P, Saviana M,
Romano G, Nigita G, Nana-Sinkam P and Acunzo M: The
Epitranscriptome in miRNAs: Crosstalk, detection, and function in
cancer. Genes (Basel). 13:12892022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shi Y, Liu Z, Lin Q, Luo Q, Cen Y, Li J,
Fang X and Gong C: MiRNAs and cancer: Key link in diagnosis and
therapy. Genes (Basel). 12:12892021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zarrilli G, Galuppini F, Angerilli V,
Munari G, Sabbadin M, Lazzarin V, Nicolè L, Biancotti R and Fassan
M: miRNAs involved in esophageal carcinogenesis and miRNA-Related
therapeutic perspectives in esophageal carcinoma. Int J Mol Sci.
22:36402021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Inoue J and Inazawa J: Cancer-associated
miRNAs and their therapeutic potential. J Hum Genet. 66:937–945.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Saliminejad K, Khorram Khorshid HR,
Soleymani Fard S and Ghaffari SH: An overview of microRNAs:
Biology, functions, therapeutics, and analysis methods. J Cell
Physiol. 234:5451–5465. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
He B, Zhao Z, Cai Q, Zhang Y, Zhang P, Shi
S, Xie H, Peng X, Yin W, Tao Y and Wang X: miRNA-based biomarkers,
therapies, and resistance in Cancer. Int J Biol Sci. 16:2628–2647.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ali Syeda Z, Langden SSS, Munkhzul C, Lee
M and Song SJ: Regulatory mechanism of MicroRNA expression in
cancer. Int J Mol Sci. 21:17232020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kousar K, Ahmad T, Abduh MS, Kanwal B,
Shah SS, Naseer F and Anjum S: miRNAs in regulation of tumor
microenvironment, chemotherapy resistance, immunotherapy modulation
and miRNA therapeutics in cancer. Int J Mol Sci. 23:138222022.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Popov A and Mandys V:
Senescence-associated miRNAs and their role in pancreatic cancer.
Pathol Oncol Res. 28:16101562022. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chakrabortty A, Patton DJ, Smith BF and
Agarwal P: miRNAs: Potential as biomarkers and therapeutic targets
for cancer. Genes (Basel). 14:13752023. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hussen BM, Hidayat HJ, Salihi A, Sabir DK,
Taheri M and Ghafouri-Fard S: MicroRNA: A signature for cancer
progression. Biomed Pharmacother. 138:1115282021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Haura EB, Turkson J and Jove R: Mechanisms
of disease: Insights into the emerging role of signal transducers
and activators of transcription in cancer. Nat Clin Pract Oncol.
2:315–324. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Furtek SL, Backos DS, Matheson CJ and
Reigan P: Strategies and approaches of targeting STAT3 for cancer
treatment. ACS Chem Biol. 11:308–318. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rahbar Farzam O, Najafi S, Amini M, Rahimi
Z, Dabbaghipour R, Zohdi O, Asemani Shahgoli G, Baradaran B and
Akbari B: Interplay of miRNAs and lncRNAs in STAT3 signaling
pathway in colorectal cancer progression. Cancer Cell Int.
24:162024. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kamran MZ, Patil P and Gude RP: Role of
STAT3 in cancer metastasis and translational advances. Biomed Res
Int. 2013:4218212013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang Z, Zhu S, Shen M, Liu J, Wang M, Li
C, Wang Y, Deng A and Mei Q: STAT3 is involved in esophageal
carcinogenesis through regulation of Oct-1. Carcinogenesis.
34:678–688. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yuan J, Zhang F and Niu R: Multiple
regulation pathways and pivotal biological functions of STAT3 in
cancer. Sci Rep. 5:176632015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Siveen KS, Sikka S, Surana R, Dai X, Zhang
J, Kumar AP, Tan BK, Sethi G and Bishayee A: Targeting the STAT3
signaling pathway in cancer: Role of synthetic and natural
inhibitors. Biochim Biophys Acta. 1845:136–154. 2014.PubMed/NCBI
|
|
31
|
Frank DA: STAT3 as a central mediator of
neoplastic cellular transformation. Cancer Lett. 251:199–210. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Germain D and Frank DA: Targeting the
cytoplasmic and nuclear functions of signal transducers and
activators of transcription 3 for cancer therapy. Clin Cancer Res.
13:5665–5669. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Turkson J and Jove R: STAT proteins: Novel
molecular targets for cancer drug discovery. Oncogene.
19:6613–6626. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cao Q, Li YY, He WF, Zhang ZZ, Zhou Q, Liu
X, Shen Y and Huang TT: Interplay between microRNAs and the STAT3
signaling pathway in human cancers. Physiol Genomics. 45:1206–1214.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu S, Li W, Liang L, Zhou Y and Li Y: The
regulatory relationship between transcription factor STAT3 and
noncoding RNA. Cell Mol Biol Lett. 29:42024. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yu H, Lee H, Herrmann A, Buettner R and
Jove R: Revisiting STAT3 signalling in cancer: New and unexpected
biological functions. Nat Rev Cancer. 14:736–746. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kohanbash G and Okada H: MicroRNAs and
STAT interplay. Semin Cancer Biol. 22:70–75. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xiang X, Ma HZ, Chen YQ, Zhang DZ, Ma SX,
Wang HJ, Liu DM, Yuan Y and Cai H: GM-CSF-miRNA-Jak2/Stat3
signaling mediates chemotherapy-induced cancer cell stemness in
gastric cancer. Front Pharmacol. 13:8553512022. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sajjadi-Dokht M, Merza Mohamad TA,
Sulaiman Rahman H, Suliman Maashi M, Danshina S, Shomali N, Solali
S, Marofi F, Zeinalzadeh E, Akbari M, et al: MicroRNAs and
JAK/STAT3 signaling: A new promising therapeutic axis in blood
cancers. Genes Dis. 9:849–867. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ma RJ, Ma C, Hu K, Zhao MM, Zhang N and
Sun ZG: Molecular mechanism, regulation, and therapeutic targeting
of the STAT3 signaling pathway in esophageal cancer (Review). Int J
Oncol. 61:1052022. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li Z, Qin X, Bian W, Li Y, Shan B, Yao Z
and Li S: Exosomal lncRNA ZFAS1 regulates esophageal squamous cell
carcinoma cell proliferation, invasion, migration and apoptosis via
microRNA-124/STAT3 axis. J Exp Clin Cancer Res. 38:4772019.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Cheng Y, Li Y, Nian Y, Liu D, Dai F and
Zhang J: STAT3 is involved in miR-124-mediated suppressive effects
on esophageal cancer cells. BMC Cancer. 15:3062015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhao Y, Ma K, Yang S, Zhang X, Wang F,
Zhang X, Liu H and Fan Q: MicroRNA-125a-5p enhances the sensitivity
of esophageal squamous cell carcinoma cells to cisplatin by
suppressing the activation of the STAT3 signaling pathway. Int J
Oncol. 53:644–658. 2018.PubMed/NCBI
|
|
44
|
Wang ZZ, Luo YR, Du J, Yu Y, Yang XZ, Cui
YJ and Jin XF: MiR-296-5p inhibits cell invasion and migration of
esophageal squamous cell carcinoma by downregulating STAT3
signaling. Eur Rev Med Pharmacol Sci. 23:5206–5214. 2019.PubMed/NCBI
|
|
45
|
Yuan RB, Zhang SH, He Y, Zhang XY and
Zhang YB: MiR-874-3p is an independent prognostic factor and
functions as an anti-oncomir in esophageal squamous cell carcinoma
via targeting STAT3. Eur Rev Med Pharmacol Sci. 22:7265–7273.
2018.PubMed/NCBI
|
|
46
|
Zang HL, Ji FJ, Ju HY and Tian XF:
Circular RNA AKT3 governs malignant behaviors of esophageal cancer
cells by sponging miR-17-5p. World J Gastroenterol. 27:240–254.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Xu Z, Tie X, Li N, Yi Z, Shen F and Zhang
Y: Circular RNA hsa_circ_0000654 promotes esophageal squamous cell
carcinoma progression by regulating the miR-149-5p/IL-6/STAT3
pathway. IUBMB Life. 72:426–439. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Meng L, Wang F, Sun S, Zheng Y, Ding Z,
Sun Y, Li B, Meng Q and Xu M: MicroRNA-30b targets CBX3 and
regulates cell proliferation, apoptosis, and migration in
esophageal squamous cell carcinoma via the JAK2/STAT3 signaling
pathway. Int J Clin Exp Pathol. 10:11828–11837. 2017.PubMed/NCBI
|
|
49
|
Ke S, Fang M, Li R, Wang J and Lu J:
Downregulation of long noncoding RNA breast cancer anti-estrogen
resistance 4 inhibits cell proliferation, invasion, and migration
in esophageal squamous cell carcinoma by regulating the
microRNA-181c-5p/LIM and SH3 protein 1 axis. Bioengineered.
13:12998–13010. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Chen Z, Hu X, Wu Y, Cong L, He X, Lu J,
Feng J and Liu D: Long non-coding RNA XIST promotes the development
of esophageal cancer by sponging miR-494 to regulate CDK6
expression. Biomed Pharmacother. 109:2228–2236. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yang H, Chen XW, Song XJ, Du HY and Si FC:
Baitouweng decoction suppresses growth of esophageal carcinoma
cells through miR-495-3p/BUB1/STAT3 axis. World J Gastrointest
Oncol. 16:3193–3210. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Han Y, Fan X, Fan L, Wu Y, Zhou Z, Wang G,
Guo L, Gao W, Chen Y and Gao Q: Liujunzi decoction exerts potent
antitumor activity in oesophageal squamous cell carcinoma by
inhibiting miR-34a/STAT3/IL-6R feedback loop, and modifies
antitumor immunity. Phytomedicine. 111:1546722023. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wang S, Wu G and Han Y, Li X, Feng X, Wang
H, Zhang L, Lin M, Cai Y and Han Y: miR-124 regulates
STAT3-mediated cell proliferation, migration and apoptosis in
bladder cancer. Oncol Lett. 16:5875–5881. 2018.PubMed/NCBI
|
|
54
|
Zhang J, Lu Y, Yue X, Li H, Luo X, Wang Y,
Wang K and Wan J: MiR-124 suppresses growth of human colorectal
cancer by inhibiting STAT3. PLoS One. 8:e703002013. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Lu Y, Yue X, Cui Y, Zhang J and Wang K:
MicroRNA-124 suppresses growth of human hepatocellular carcinoma by
targeting STAT3. Biochem Biophys Res Commun. 441:873–879. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Chen M, Ye X, Wang R and Poon K: Research
progress of cancer stem cells and IL-6/STAT3 signaling pathway in
esophageal adenocarcinoma. Transl Cancer Res. 9:363–371. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Lin H, Lian J, Xia L, Guan G and You J:
CBX3 promotes gastric cancer progression and affects factors
related to immunotherapeutic responses. Cancer Manag Res.
12:10113–10125. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Yang K, Wang YL, Zhu Z, Cao XH, Liang MX,
Xu D, Fei YJ, Yang SY, Zhou HL and Tang JH: CBX3 promotes breast
cancer progression and high level of CBX3 predicts poor prognosis
in patients. Neoplasma. 70:71–81. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Chen LY, Cheng CS, Qu C, Wang P, Chen H,
Meng ZQ and Chen Z: CBX3 promotes proliferation and regulates
glycolysis via suppressing FBP1 in pancreatic cancer. Biochem
Biophys Res Commun. 500:691–697. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zhou C, Fan N, Liu F, Fang N, Plum PS,
Thieme R, Gockel I, Gromnitza S, Hillmer AM, Chon SH, et al:
Linking cancer stem cell plasticity to therapeutic
resistance-mechanism and novel therapeutic strategies in esophageal
cancer. Cells. 9:14812020. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Huang B, Lang X and Li X: The role of
IL-6/JAK2/STAT3 signaling pathway in cancers. Front Oncol.
12:10231772022. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Mengie Ayele T, Tilahun Muche Z, Behaile
Teklemariam A, Bogale Kassie A and Chekol Abebe E: Role of
JAK2/STAT3 signaling pathway in the tumorigenesis, chemotherapy
resistance, and treatment of solid tumors: A systemic review. J
Inflamm Res. 15:1349–1364. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Fassl A, Geng Y and Sicinski P: CDK4 and
CDK6 kinases: From basic science to cancer therapy. Science.
375:eabc14952022. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Jiang H, Guo Y, Huang K, Lu R, Peng X and
Lin S: MicroRNA-34a inhibits esophageal squamous cell carcinoma
progression by targeting E2F5. J Buon. 24:2514–2522.
2019.PubMed/NCBI
|
|
65
|
Ye Z, Xie T, Yan F, Wang L, Fang J, Wang
Z, Hu F, Wang F and Fu Z: MiR-34a reverses radiation resistance on
ECA-109 cells by inhibiting PI3K/AKT/mTOR signal pathway through
downregulating the expression of SIRT1. Int J Radiat Biol.
97:452–463. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Yang L, Song X, Zhu J, Li M, Ji Y, Wu F,
Chen Y, Cui X, Hu J, Wang L, et al: Tumor suppressor microRNA-34a
inhibits cell migration and invasion by targeting
MMP-2/MMP-9/FNDC3B in esophageal squamous cell carcinoma. Int J
Oncol. 51:378–388. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Nie J, Ge X, Geng Y, Cao H, Zhu W, Jiao Y,
Wu J, Zhou J and Cao J: miR-34a inhibits the migration and invasion
of esophageal squamous cell carcinoma by targeting Yin Yang-1.
Oncol Rep. 34:311–317. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Li M, Meng X and Li M: MiR-126 promotes
esophageal squamous cell carcinoma via inhibition of apoptosis and
autophagy. Aging (Albany NY). 12:12107–12118. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Zhang M, Tian H, Li R, Yan W and Xu R:
MicroRNA-4286 promotes esophageal carcinoma development by
targeting INPP4A to evoke the JAK2/STAT3 pathway activation.
Pharmazie. 73:342–348. 2018.PubMed/NCBI
|
|
70
|
Wang H, Peng D, Gan M, He Z and Kuang Y:
CPEB3 overexpression caused by miR-106b-5p inhibition inhibits
esophageal carcinoma in-vitro progression and metastasis.
Anticancer Drugs. 33:335–351. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Xu DD, Zhou PJ, Wang Y, Zhang L, Fu WY,
Ruan BB, Xu HP, Hu CZ, Tian L, Qin JH, et al: Reciprocal activation
between STAT3 and miR-181b regulates the proliferation of
esophageal cancer stem-like cells via the CYLD pathway. Mol Cancer.
15:402016. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Zhang Y, Lu W, Chen Y, Lin Y, Yang X, Wang
H and Liu Z: The miR-19b-3p-MAP2K3-STAT3 feedback loop regulates
cell proliferation and invasion in esophageal squamous cell
carcinoma. Mol Oncol. 15:1566–1583. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Kong R, Ma Y, Feng J, Li S, Zhang W, Jiang
J, Zhang J, Qiao Z, Yang X and Zhou B: The crucial role of miR-126
on suppressing progression of esophageal cancer by targeting
VEGF-A. Cell Mol Biol Lett. 21:32016. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Li H, Meng F, Ma J, Yu Y, Hua X, Qin J and
Li Y: Insulin receptor substrate-1 and Golgi phosphoprotein 3 are
downstream targets of miR-126 in esophageal squamous cell
carcinoma. Oncol Rep. 32:1225–1233. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Nie ZC, Weng WH, Shang YS, Long Y, Li J,
Xu YT and Li Z: MicroRNA-126 is down-regulated in human esophageal
squamous cell carcinoma and inhibits the proliferation and
migration in EC109 cell via PI3K/AKT signaling pathway. Int J Clin
Exp Pathol. 8:4745–4754. 2015.PubMed/NCBI
|
|
76
|
Liu R, Gu J, Jiang P, Zheng Y, Liu X,
Jiang X, Huang E, Xiong S, Xu F, Liu G, et al: DNMT1-microRNA126
epigenetic circuit contributes to esophageal squamous cell
carcinoma growth via ADAM9-EGFR-AKT signaling. Clin Cancer Res.
21:854–863. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Gu JF, Liu SG, Pan Q, Qin F and Li YY:
Negative regulation of CDK6 expression by microRNA-126-5p and its
influence on the proliferation and invasion of esophageal cancer
cells. Anat Rec (Hoboken). 303:2811–2820. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Drahos J, Schwameis K, Orzolek LD, Hao H,
Birner P, Taylor PR, Pfeiffer RM, Schoppmann SF and Cook MB:
MicroRNA profiles of Barrett's esophagus and esophageal
adenocarcinoma: Differences in glandular non-native epithelium.
Cancer Epidemiol Biomarkers Prev. 25:429–437. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk
ML and Struhl K: STAT3 activation of miR-21 and miR-181b-1 via PTEN
and CYLD are part of the epigenetic switch linking inflammation to
cancer. Mol Cell. 39:493–506. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Zeng Q, Zhu Z, Song L and He Z:
Transferred by exosomes-derived MiR-19b-3p targets PTEN to regulate
esophageal cancer cell apoptosis, migration and invasion. Biosci
Rep. 40:BSR202018582020. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Chiam K, Mayne GC, Wang T, Watson DI,
Irvine TS, Bright T, Smith LT, Ball IA, Bowen JM, Keefe DM, et al:
Serum outperforms plasma in small extracellular vesicle microRNA
biomarker studies of adenocarcinoma of the esophagus. World J
Gastroenterol. 26:2570–2583. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Pandey P, Suyal G, Aprajita Pasbola K and
Sharma R: NGS-based profiling identifies miRNAs and pathways
dysregulated in cisplatin-resistant esophageal cancer cells. Funct
Integr Genomics. 23:1112023. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Sugimura K, Miyata H, Tanaka K, Hamano R,
Takahashi T, Kurokawa Y, Yamasaki M, Nakajima K, Takiguchi S, Mori
M and Doki Y: Let-7 expression is a significant determinant of
response to chemotherapy through the regulation of IL-6/STAT3
pathway in esophageal squamous cell carcinoma. Clin Cancer Res.
18:5144–5153. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Zhao Q, Huang L, Qin G, Qiao Y, Ren F,
Shen C, Wang S, Liu S, Lian J, Wang D, et al: Cancer-associated
fibroblasts induce monocytic myeloid-derived suppressor cell
generation via IL-6/exosomal miR-21-activated STAT3 signaling to
promote cisplatin resistance in esophageal squamous cell carcinoma.
Cancer Lett. 518:35–48. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Chen K, Wang X, Yang L and Chen Z: The
Anti-PD-1/PD-L1 immunotherapy for gastric esophageal cancer: A
systematic review and meta-analysis and literature review. Cancer
Control. 28:10732748219974302021. View Article : Google Scholar : PubMed/NCBI
|