Introduction
The risk of developing breast cancer markedly
increases in individuals carrying a germline mutation in the
BRCA1 or BRCA2 caretaker genes, which are activated
in the DNA damage response machinery and which exert their activity
in homologous recombination (HR). HR is a DNA double-strand break
(DSB) repair pathway, activated in the S and G2 phases of the cell
cycle following exposure to genotoxic agents, such as ionizing
radiation (IR) (1,2).
It has been hypothesized that individuals harboring
a germline mutation in the BRCA1 and BRCA2 genes may
exhibit enhanced radiosensitivity and may thus be exposed to an
increased carcinogenic risk following exposure to IR for
therapeutic or diagnostic purposes (3-19).
The authors have recently demonstrated an increased
radiosensitivity and micronucleus formation in peripheral blood
lymphocytes of healthy women carrying BRCA1 or BRCA2
mutations (18,19). These results, obtained in
peripheral blood lymphocytes, suggested that the deficient repair
of DNA DSBs might also occur in mammary epithelial cells of women
exhibiting a reduced expression of wild-type BRCA proteins.
Hence, the main aim of this in vitro study
was to assess the DNA DSB repair capacity and the formation of
chromosomal abnormalities in irradiated, non-tumorigenic human
mammary epithelial cells exhibiting a decreased expression of
wild-type BRCA1/2 proteins, as may occur in the case of the healthy
mammary tissue of women harboring heterozygous BRCA1/2
mutations. In the MCF10A non-tumorigenic human mammary epithelial
cell line, by lentivirus-mediated RNA interference, the partial
reduction of the expression levels of BRCA1 and BRCA2 was achieved
to levels which may functionally mimic those of mammary cells in
heterozygous BRCA mutation carriers. In these cells, the
repair capacity following irradiation was investigated using the
RAD51 foci assay, which specifically detects DNA DSB repair by the
homologous recombination pathway. A deficient repair capacity was
phenotypically confirmed by analyzing chromosomal abnormalities
with the micronucleus assay and by cell viability testing.
Materials and methods
Cell lines
Mycoplasma-free MCF10A cells (cat. no. CRL-10317,
freshly obtained from ATCC) were cultured in monolayers using equal
volumes of DMEM-glutamax and F12-glutamax (Life Technologies;
Thermo Fisher Scientific) supplemented with 5% fetal calf serum
(Invitrogen; Thermo Fisher Scientific), antibiotics (50 U/ml
penicillin and 50 µg/ml streptomycin, Invitrogen; Thermo Fisher
Scientific), 10 µg/ml insulin (Sigma-Aldrich), 0.5 µg/ml
hydrocortison (Sigma-Aldrich) and 20 ng/ml epidermal growth factor
(Peprotech). Experiments were performed on cells in which BRCA1 and
BRCA2 were knocked down (these cells are referred to as BRCA1i and
BRCA2i cells). RNA interference (RNAi) was achieved by stable
transduction with lentiviral vectors harboring DNA sequences
encoding short hairpin RNAs specific for BRCA1 or BRCA2. In brief,
lentiviral particles were constructed using pLKO.1-puro vectors
(Addgene). The RNAi sequences for BRCA1 and BRCA2
were 5'-GCCCACCTAATTGTACTGAAT-3' and 5'-TACAATGTACACATGTAACAC-3',
respectively. A negative control cell line, transduced with an
empty pLKO.1-puro lentiviral vector (hereafter referred to as the
control) was also established. This is an acknowledged limit of
this study, as scrambled sequence transduction would have been
preferable, though according to previous experience, this procedure
may target unintended mRNAs. Moreover, empty vectors allow for the
determination of the effects of transduction on cell response and
gene expression (20). The
transduction of the MCF10A cells was achieved by the addition of 1
µg/ml DNA, TurboFect (1.5 µg/ml, Thermo Fisher Scientific) and
polybrene (1 µg/ml, Sigma-Aldrich) to a 30% confluent culture.
Cells were grown in puromycin-supplemented DMEM medium (2 µg/ml,
Life Technologies; Thermo Fisher Scientific) for 15 days to obtain
stably transduced cell lines. Notably, stable BRCA1 knockdown by
retrovirus-mediated RNAi does not alter the non-tumorigenic
phenotype of MCF10A cells (21).
According to our personal experience, BRCA1/2 knockdown neither
induces transformation in vitro, nor tumorigenesis in
vivo.
Reverse transcription-quantitative PCR
(RT-qPCR) analysis
BRCA1 and BRCA2 mRNA knockdown was
evaluated by RT-qPCR analysis. Total RNA was extracted using RNeasy
Mini kits (Qiagen Benelux), following the manufacturer's
instructions, without optional DNase treatment. The RNA
concentration and quality were determined using a DropSense96 kit
(Trinean) before removing contaminating DNA with the Heat&Run
gDNA removal kit (Articzymes). Reverse transcription was achieved
with the iScript cDNA synthesis kit, following the manufacturer's
instructions. A total of 5 µl of qPCR reaction contained 10 ng cDNA
(total RNA equivalents), 2.5 µl SsoAdvanced universal SYBR-Green
supermix (Bio-Rad) and 2.5 µM forward and reverse primers. Cycling
and detection was performed on a Lightcycler LC480 (Roche) with 2
min denaturation at 95˚C followed by 45 cycles with 5 sec at 95˚C,
30 sec at 60˚C and 1 sec at 72˚C. Melting curve analysis was
performed to test for non-specific amplification. Three biological
repeats were performed for each amplification. The analysis of
relative gene expression was performed using a qBase+ platform
(Biogazelle), developed by Hellemans et al (22), based on the ΔΔCq method by Livak
and Schmittgen (23).
Western blot analysis
BRCA1 and BRCA2 protein knockdown was evaluated by
western blot analysis. Protein extraction was performed in
subconfluent cultures of the control, BRCA1i and BRCA2i cells using
a tris-EDTA lysis buffer containing 1% NP-40 and 1% protease
inhibitor (Sigma-Aldrich). For each sample, 50 µg protein were
loaded together with 25% SDS sample buffer (Thermo Fisher
Scientific) and 10% dithiothrietol (DDT, Sigma-Aldrich) on a 3-8%
Tris-Acetate gel (Novex; Thermo Fisher Scientific), and run for 5 h
at 25 mA. Proteins were transferred to a methanol-pretreated PVDF
membrane in Tris/glycine blotting buffer enriched with 10% methanol
for 16 h at 30 V. Following standard blocking, the membrane was
incubated overnight at 4˚C with a primary antibody (rabbit
polyclonal anti-BRCA1, cat. no. 07-434, diluted 1:1,000, or
monoclonal anti-BRCA2, cat. no. OP95, diluted 1:500; Millipore),
together with mouse monoclonal α-actinin (cat. no. 05-384, diluted
1:30,000 Millipore). The membranes were washed and incubated for 2
h at room temperature with horseradish peroxidase (HRP)-conjugated
secondary antibodies (goat anti-rabbit-HRP, Perbio, cat. no. 31460,
diluted 1:1,000, or goat anti-mouse-HRP, cat. no. 31432, Thermo
Fisher Scientific, diluted 1:1,000). Visualization was achieved
with a chemoluminescence kit (Thermo Fisher Scientific), and
analysis of relative expression at the protein level was performed
using a shareware ImageJ software (version 1.52q, bundled with Java
1.8.0_172; developer: NIH).
RAD51 foci assay
The HR pathway is active in the S and G2 phases of
the cell cycle to repair DSBs induced by IR (2,24-26),
and relies on RAD51, whose recruitment to a DSB site is mediated by
BRCA1 and BRCA2. RAD51 recruitment can be typically detected in the
form of nuclear foci upon immunostaining (25,27). The RAD51 irradiation-induced foci
assay has been widely used to detect HR defects both in cancer
biopsy samples and in non-tumor cells of BRCA1/2 breast cancer
patients (28-30).
Sample preparation
The cells were switched to puromycin-free culture
DMEM medium (Life Technologies) at the beginning of each
experiment. Cells (n=200,000) were seeded in 2 ml culture medium in
6-well plates. Approximately 24 h after seeding, the cells were
examined for subconfluency. To achieve a maximum number of cells in
the S and G2 phases of the cell cycle, the cells were synchronized
by the addition of the DNA polymerase inhibitor, aphidicolin (1
µg/ml, Sigma-Aldrich) to the culture medium for 24 h. The cells
were subsequently washed with PBS (1.78 g/l
Na2HPO4; 0.42 g/l
KH2PO4; 7.2 g NaCl, VWR) and incubated at
37˚C for 8 h with fresh culture medium.
Cell cycle analysis
The MCF10A control cells (empty vector-transduced)
were harvested at various time points following synchronization to
evaluate the percentage of cells in each phase of the cell cycle.
Cell permeabilization was achieved by fixation in 95% ethanol at
-20˚C and the DNA was subsequently stained with propidium iodide
(PI, Sigma-Aldrich) in a hypotonic staining buffer containing 0.1%
Sodium citrate, 0.3% Triton X-100, 0.01% PI and 0.002% ribonuclease
A (all reagents were from Sigma-Aldrich). The PI cell content was
analyzed on a FACSCantoTM (BD Biosciences). Cells of
interest were selected based on forward and side scatter area
patterns. A non-synchronized sample was used as reference.
Irradiation and olaparib
treatment
At 3 h following aphidicolin removal, the cells were
irradiated with 2 Gy 220 kV-13 mA X-rays to generate DSBs (SARRP
unit, XSTRAHL Ltd.). The cells were subsequently incubated at 37˚C
for 5 h for optimal RAD51 foci formation. To determine this optimal
time point, different time points varying between 2 and 8 h were
previously tested (data not shown).
Since cells defective in HR are sensitive to
poly(ADP-ribose) polymerase (PARP) inhibitor (PARPi) drugs
(31-33),
the PARPi, olaparib, was used to enhance the number of DSBs in
S-phase cells. To part of the cultures, 5 µM olaparib (Bio-Connect)
was added to the medium 1 h prior to irradiation, in order to block
the repair of radiation-induced single-strand breaks (SSBs). RAD51
foci formation was also evaluated in untreated cell cultures and in
cultures exposed to olaparib alone. In total, 8 repeats were
performed for each of the 4 experimental conditions (irradiated or
sham-irradiated cells, with or without olaparib treatment).
The induction of DNA DSBs by IR was assessed by
gamma-H2AX staining as previously described (34) (Fig.
S1). Inhibition by olaparib was confirmed by the PARP activity
following exposure to H2O2 in the presence of
olaparib, as previously described (35) (Fig.
S2).
RAD51 foci immunostaining
Prior to RAD51 foci staining, the cells were
harvested, cytospinned on polylysine-coated slides (VWR) and fixed
in 3% paraformaldehyde (Sigma-Aldrich) for 20 min. The slides were
washed twice in PBS and antigen retrieval was achieved by
incubation for 20 min in heated (95˚C) citrate buffer (0.02% citric
acid, pH 6, Sigma-Aldrich). The slides were subsequently washed and
incubated with a blocking serum containing 1% BSA (Roche), 5% goat
serum (Dako) and 0.2% Tween-20 (Sigma-Aldrich) in PBS. The slides
were then incubated overnight at 4˚C with a RAD51 H-92 rabbit
primary antibody (dilution: 1:2,000, Santa Cruz Biotechnology, cat.
no. sc-8349), washed with PBS containing 3% Tween-20
(Sigma-Aldrich), and incubated for 30 min at room temperature with
a secondary antibody (goat anti-rabbit; dilution: 1:1,000, Thermo
Fisher Scientific; cat. no. A32731). Finally, the slides were
washed in PBS/3% Tween-20 and mounted with 200 ng/ml DAPI in
fluoromount (Sigma-Aldrich).
The slides were scanned using the Metacyte software
module on a Metafer4 scanning platform (Axio Imager, Metasystems)
at a x63 magnification. This software module enables automatic cell
detection and foci counting according to set parameters, resulting
in an unbiased data acquisition. The number of RAD51 foci was
automatically scored in at least 500 cells for each experimental
condition, and expressed as the number of RAD51 foci per cell
(RAD51 foci/cell).
Micronucleus (MN) assay
Chromosomal damage was assessed with the MN assay as
previously described (36).
Briefly, the cells were seeded in 2 ml culture medium in 6-well
plates (200,000 cells/well) 1 day prior irradiation. Subconfluent
cultures of exponentially dividing cells were irradiated with doses
of 0.2, 0.5, 1, 2 and 4 Gy, and cytochalasin B (2.25 µg/ml;
Sigma-Aldrich) was immediately added to block cytokinesis. The
cells were maintained at 37˚C in a humidified 5% CO2
atmosphere incubator for 16 h. Sham-irradiated cultures were
included in each experiment. The cells were then harvested and
subjected to a cold hypotonic shock with 0.075 M KCl, followed by
overnight fixation in 3/1/4 methanol/acetic acid/ringer solution
(ringer: 9 g/l NaCl, 0.42 g/l Kcl and 0.24 g/l CaCl2).
Subsequently, the cells were fixed in a 3:1 methanol/acetic acid
solution. For further analysis, the cells were stained with DAPI
(200 ng/ml; Sigma-Aldrich). The slides were scanned at 10 X
magnification with the MSearch software module of the Metafer 4
scanning system and the MNScore software (Metasystems). The
automated image analysis system selects BN cells and determines the
number of MN for each BN cell. BN cells and MN were manually
examined for false positives and negatives. Two slides for each
culture were automatically scanned and approximately 400 BN cells
were scored in each slide. All experiments were performed in
duplicate. Each experiment was repeated thrice.
Cell viability assay
A protocol described previously was used for this
assay (37,38). Briefly, following irradiation
(doses of 0.5, 1, 2, 3, 4, 6 and 8 Gy), the cultures were further
incubated for 4 days at 37˚C until sham-irradiated plates nearly
reached confluence. The cells were fixed for 10 min in a solution
of buffered formalin (3.7%), washed with PBS (pH 7.3) and stained
with a 0.01% crystal violet solution (Sigma-Aldrich). The stain was
dissolved overnight in 1 ml 10% sodium dodecyl sulfate (SDS). The
optical density of the samples was measured with a
spectrophotometer at 590 nm. All cell viability assays were
performed in quadruplicate. Each experiment was repeated 3
times.
Statistical analysis
Differences in mean RAD51 foci, in micronucleus
counts and in cell viability data were analyzed by one-way analysis
of variance (ANOVA). The Tukey's range test was applied to perform
post-hoc analysis of the significance of comparisons. A 5% alpha
error threshold (P-value <0.05) was applied to all analyses.
Statistical inference was performed using the R software
environment. For the Tukey post-hoc test, the multcompView
package in R was used.
Results
Effects of RNAi
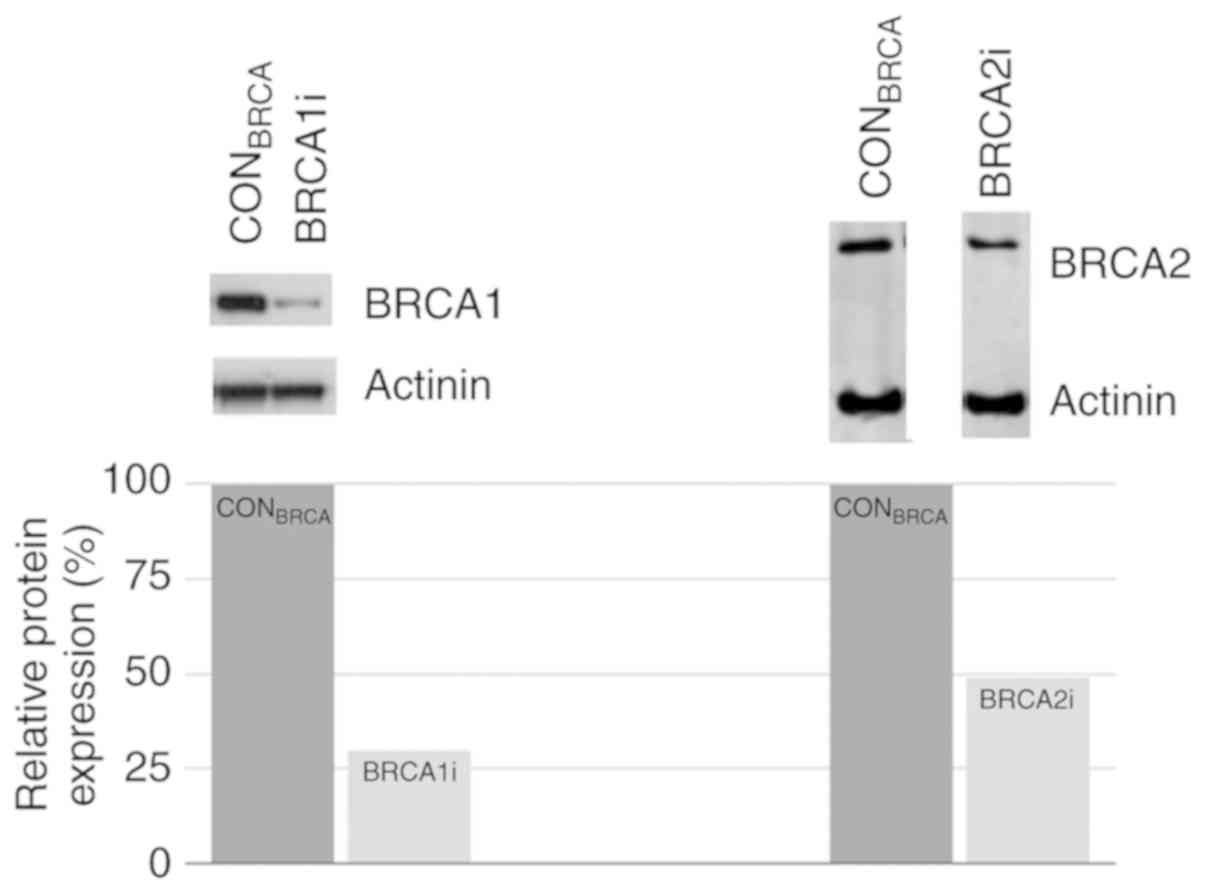
The lentivirus-mediated RNA interference of
BRCA1 and BRCA2 was confirmed at the RNA and protein
levels by RT-qPCR analysis and western blot analysis. Compared to
the control cells, 45 and 35% reductions in the BRCA1 and
BRCA2 mRNA levels were achieved, respectively (Fig. S3). Notably, in experiments
assessing knockout specificity, it was demonstrated that the
knockdown of BRCA1 in the BRCA1i cells did not affect the mRNA
levels of BRCA2 (97% of controls), and the knockdown of
BRCA2 in the BRCA2i cells did not affect the mRNA levels of
BRCA1 (97% of controls). The quantification of the protein
knockdown using ImageJ software revealed an estimated 70% reduction
of BRCA1 in BRCA1i cells, and an estimated 51% BRCA2 reduction in
BRCA2i cells (Fig. 1).
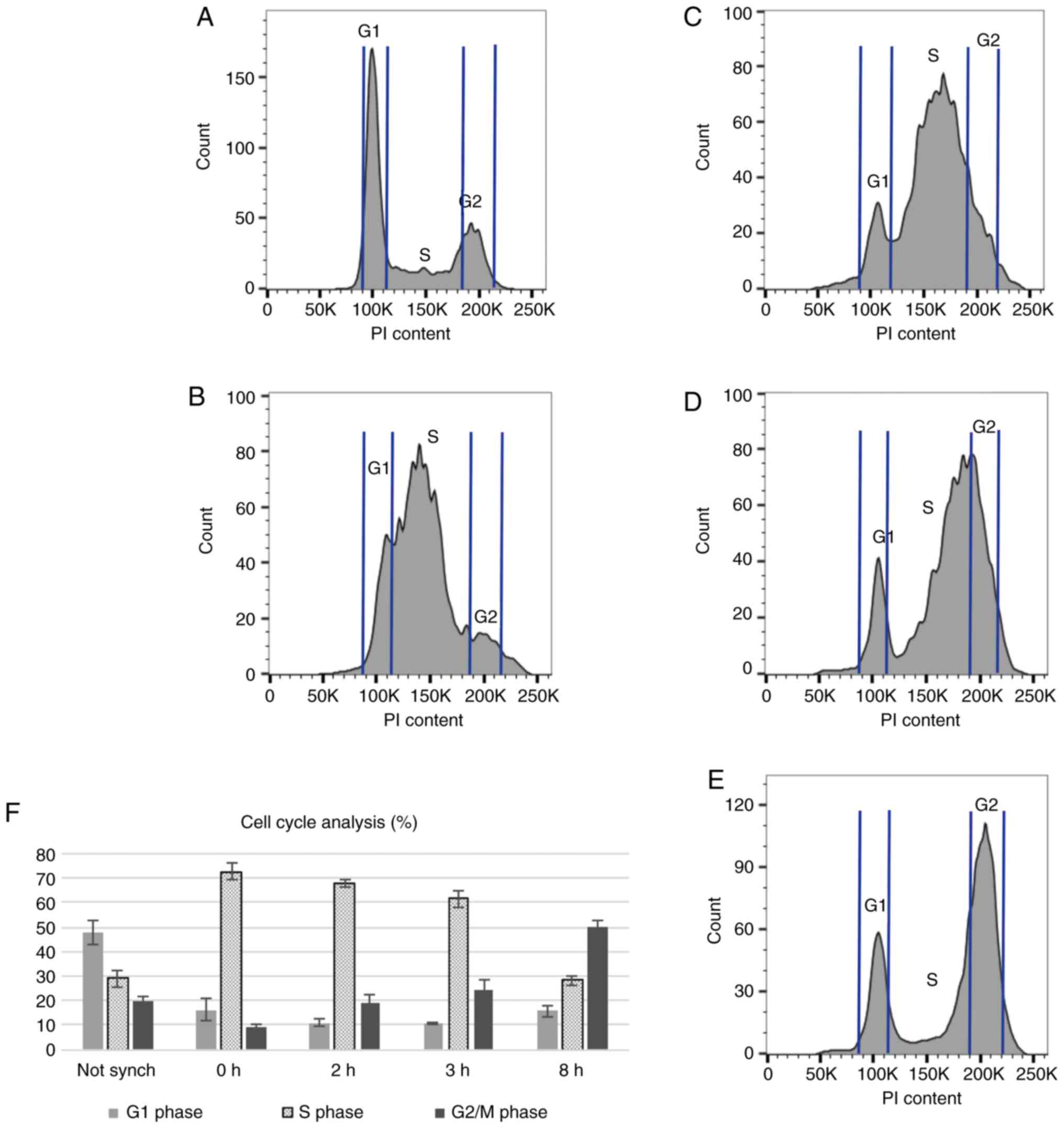
Cell cycle analysis
The results of cell cycle analysis in
non-synchronized cells, and in synchronized cells at various time
points following aphidicolin removal are shown in Fig. 2. As an example, the histogram
charts of all time points for one repeat are illustrated in Fig.
2A-E. The mean percentage (4 repeats) of cells in each phase of the
cell cycle in a non-synchronized sample, and in synchronized cells
at various time points (0, 2, 3 and 8 h) following aphidicolin
removal is shown in Fig. 2F.
Synchronization with aphidicolin increased the
number of cells in the S phase of the cell cycle. Immediately
following aphidicolin synchronization, approximately 70% of the
cells were at the beginning of the S phase, compared to 30% in
non-synchonized cells. At 2 and 3 h following aphidicolin removal,
the lowest number of cells in the G1 phase was achieved, while the
number of cells in the S phase remained between 60 and 70%,
compared to the non-synchronized cultures. At 8 h following
aphidicolin synchronization, the cells shifted towards the G2 and M
phases of the cell cycle. Since the number of cells in the late S
phase peaked at 2 and 3 h following synchronization, these time
points were selected for the addition of olaparib (2 h following
the removal of aphidicolin) and irradiation (3 h following the
removal of aphidicolin) to maximize the effects of these agents on
RAD51 foci formation.
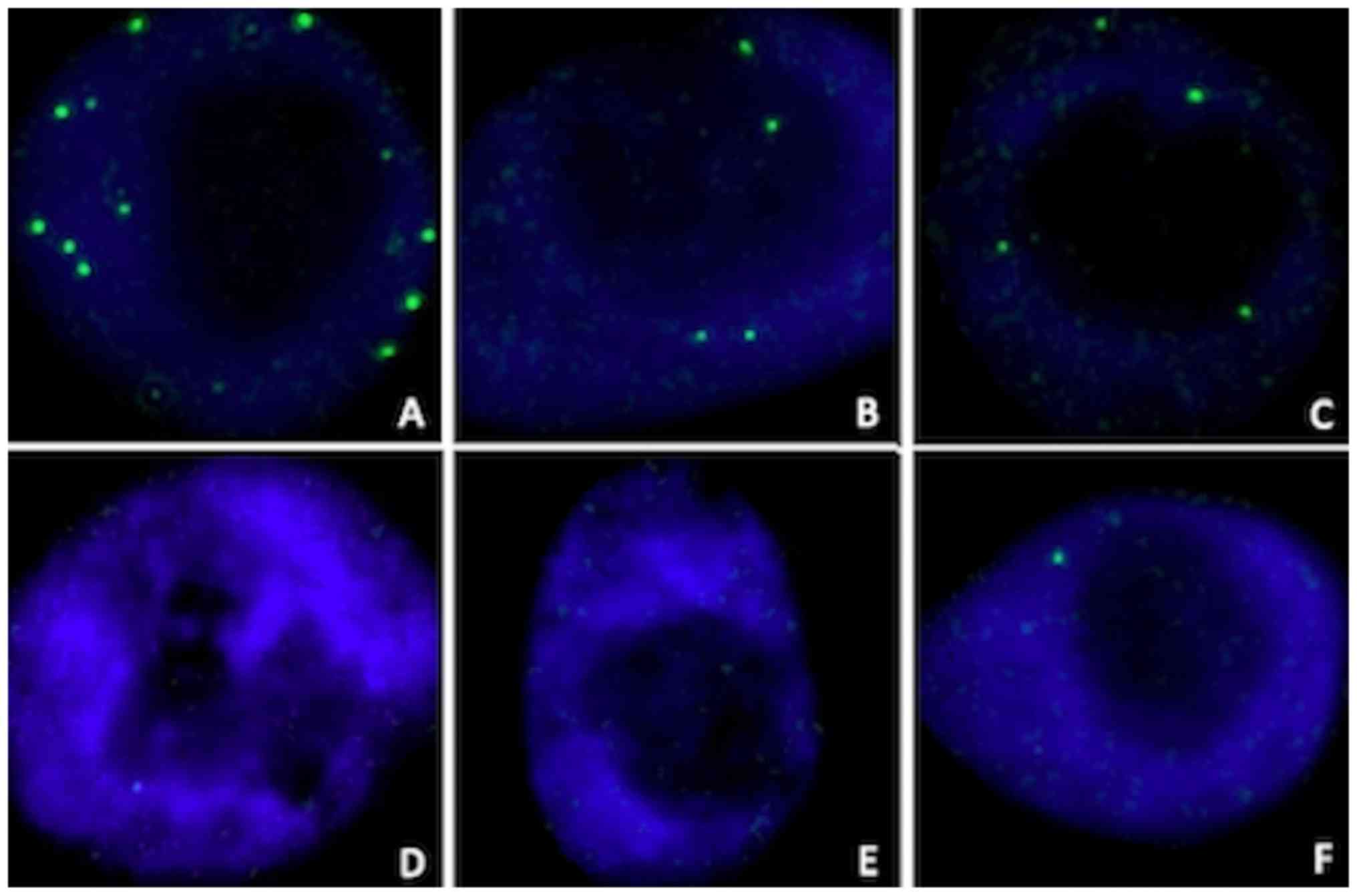
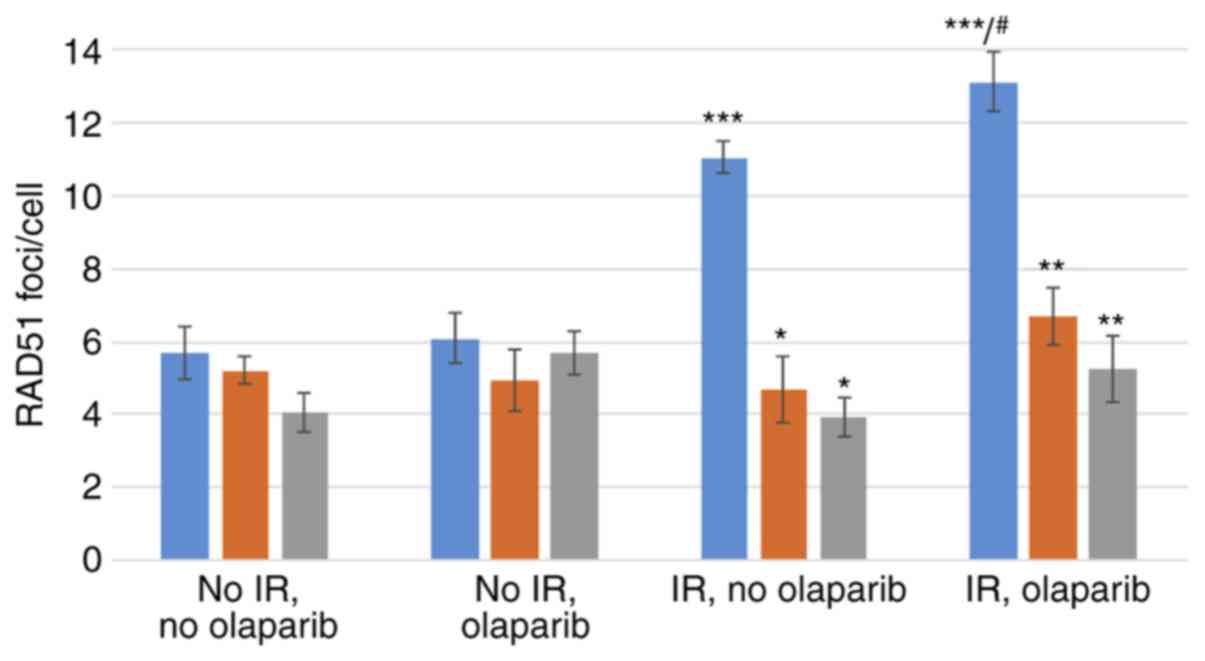
RAD51 foci formation
Representative examples of the induction of RAD51
foci by radiation in the nuclei of MCF10A cells, with or without a
knockdown of BRCA1 or BRCA2 are illustrated in Fig. 3. The mean number of RAD51 foci per
cell in synchronized cell cultures, assessed in the 4 tested
conditions (irradiated or sham-irradiated, with or without
olaparib) is shown in Fig. 4. The
significance values for all comparisons are listed in Table SI.
Control MCF10A cells
Compared to the cells not irradiated and not exposed
to olaparib, exposure to IR induced marked and significant
increases in the mean RAD51 foci/cell, both in the presence
(~2.3-fold) or absence (~1.9-fold) of olaparib (Fig. 4, blue bars and Table SI).
The exposure of sham-irradiated control cells to
olaparib did not significantly increase the number of foci/cell.
Conversely, in the irradiated control cells, exposure to olaparib
caused a small, yet significant increase in RAD51 foci/cell
(~1.2-fold; Fig. 4, blue bars and
Table SI).
BRCA1i and BRCA2i cells
Compared to their respective controls, a
significantly lower yield of RAD51 foci was observed in the
irradiated BRCA1i and BRCA2i cells, exposed (BRCA1i cells, 49%
reduction; BRCA2i cells, 60% reduction) or not (BRCA1i cells, 58%
reduction; BRCA2i cells, 64% reduction) to olaparib (Fig. 4 and Table SI).
Compared to the BRCA1i or BRCA2i cells not
irradiated and not exposed to olaparib, exposure to IR did not
induce statistically significant increases in the mean RAD51
foci/cell, both in the presence or absence of olaparib (Fig. 4 and Table SI).
Micronucleus assay
The spontaneous MN yield (mean number of micronuclei
per 1,000 sham-irradiated binucleated cells ± SD) for the control,
BRCA1i and BRCA2i cells was 28.1±2.5; 31.7±4.2 and 26.0±1.7,
respectively. There was no significant difference in the
spontaneous MN values between the BRCA1i or BRCA2i cell lines and
control MCF10A cells.
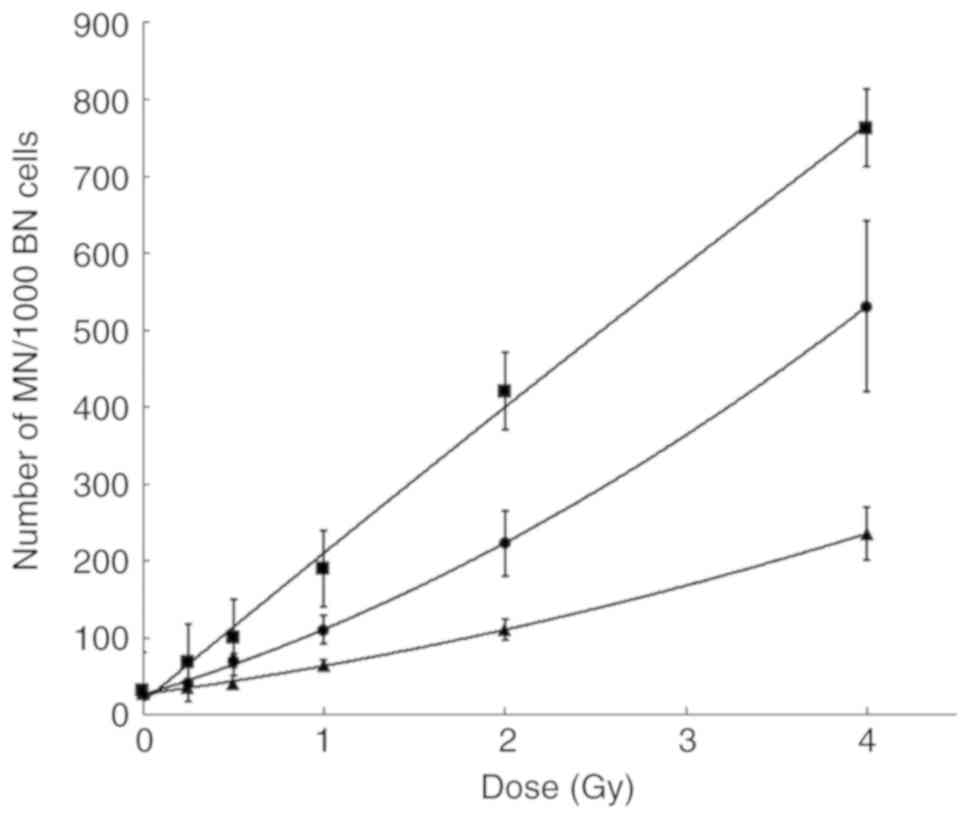
The MN dose-response curves in the control cells,
and in the BRCA1i and BRCA2i cells are shown in Fig. 5. Compared to the irradiated
controls, significantly higher MN yields were obtained in both the
BRCA1i and BRCA2i irradiated cell lines. The BRCA1i cells were the
most radiosensitive, resulting in a steeper, quasi-linear
dose-response curve. The P-values for all the comparisons are
listed in Table SI.
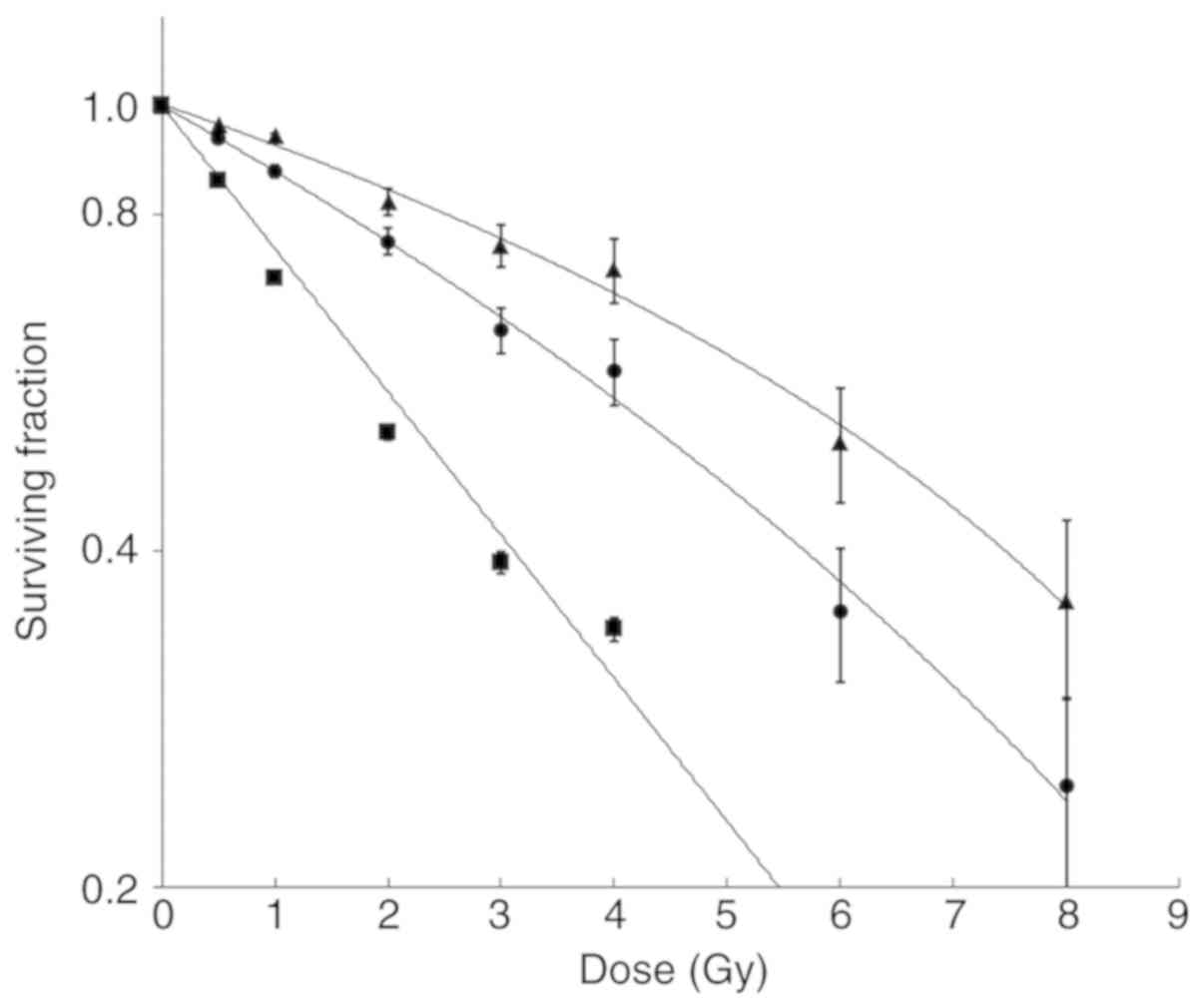
Cell viability assay
Cell viability/survival curves of knockdown cell
lines together with the control cell line are shown in Fig. 6. The BRCA1i and BRCA2i irradiated
cells exhibited a dose-dependent decrease in survival, when
compared to the control cells. The P-values for all the comparisons
are listed in Table SI.
Discussion
The main aim of this in vitro study was to
assess homologous recombination repair and radiosensitivity in
human mammary cells showing reduced protein levels of wild-type
BRCA proteins, a condition that may model the reduced protein
expression observed in heterozygous cells of women carriers of
BRCA1/BRCA2 mutations. A reduced BRCA expression, and
in turn, an impaired DNA repair capacity may point to an increased
risk of developing radiation-induced carcinogenesis in such women.
One of the authors' concerns was that a BRCA mutation
carrier receiving, for example, repeated mammography screens each
year, or adjuvant radiotherapy following surgical removal of a
primary lesion, may be at increased risk of developing secondary,
radiation-induced neoplasia. Thus, in this study, a dose of
radiation was selected which is commonly administered to women
receiving a single session of radiotherapy (2 Gy). In the
experiments in this study, the non-tumorigenic and
non-mammosphere-forming MCF10A cell line was selected, which is
often used as in vitro model, together, with its in
vitro-transformed derivative cell lines-to investigate the
biology of non-neoplastic human mammary cells (39). These diploid cells have a
relatively stable karyotype and form in vitro acinar
structures that recapitulate many aspects of mammary gland
architecture.
The cells were synchronized in the S and G2 phases
for an optimal evaluation of the DNA repair capacity by homologous
recombination, a pathway which involves both BRCA1 and BRCA2. DSBs
were induced by exposure to ionizing radiation. To increase the
number of DSBs, olaparib was also used, which is a PARPi that
transforms radiation-induced single-strand breaks into DSBs during
the S phase of the cell cycle.
As expected, HR was involved in the repair of
radiation-induced DSBs in synchronized MCF10A cells, as shown by
the highly significant increase of RAD51 foci yields in irradiated
control cells, compared to their sham-irradiated counterparts.
Conversely, exposure to IR did not increase the yield of RAD51
foci, either in BRCA1i or in BRCA2i cells. In fact, significantly
lower yields of RAD51 foci were observed in the BRCA1i and BRCA2i
cell lines compared to the irradiated control cells. The lack of
induction of RAD51 foci in the BRCA1i and BRCA2i cells may suggest
that the knockdown may phenotypically mimic BRCA
haploinsufficiency in breast epithelial cells, where the presence
of a single wild-type allele may result in increased DNA damage
resulting from deficient HR repair, as suggested by Sedic and
Kuperwasser (40).
Studies investigating the influence of BRCA1 and
BRCA2 on RAD51 foci formation and HR function have been previously
performed using non-human, non-mammary cell lines such as CHO, DT40
and mouse embryonic stem cells. All studies have demonstrated an
impaired function of HR pathways, resulting in a reduction in RAD51
foci formation in heterozygous cells (30,41-44).
The results of studies focusing on the effects of
BRCA1 on both RAD51 foci formation and HR have been less univocal
(43,44). In a study using lymphoblastoid
cell lines of heterozygous BRCA1 mutation carriers, Vaclová
et al could not directly demonstrate a decrease in RAD51
foci formation, compared to the controls, at 4 h following exposure
to 10 Gy IR. However, they did observe a significant increase in
staining intensity of yH2AX foci in the same heterozygous
BRCA1 cells compared to controls 4 h following irradiation,
thus suggesting an increase in the number of DSBs. They also argued
that this result implies impaired HR in mutation carriers (29). Pathania et al did not
detect a reduction in radiation-induced RAD51 foci in human mammary
epithelial cells containing a BRCA1 mutation exposed to 10
Gy compared to control cells. However, the combined exposure to UV
and IR did yield a significant reduction of RAD51 foci (45).
Notably, with one exception, these studies were
performed in non-mammary cells. The authors of this study believe
that experiments performed herein modeled more closely the cellular
makeup of human breast cells.
Moreover, none of these studies considered the
variation of HR-based DSB repair throughout the different phases of
the cell cycle. It was demonstrated that 48% of non-synchronized
cycling MCF10A cells were in the G1 phase of the cell cycle, a
phase during which HR cannot be activated due to the absence of the
homologous sister chromatid. In the synchronized cell cultures, at
the moment of irradiation approximately 90% of cells were in the S
or G2 phases of the cell cycle, during which HR activation is
maximal for repair of DSBs (2,46).
In order to activate the HR pathway more
extensively, olaparib was added to the cultures prior to
irradiation. A small, yet significant increase was observed in the
number of RAD51 foci in irradiated, olaparib-treated control cells
compared to cells not exposed to olaparib. However, PARP inhibition
did not significantly increase the yield of RAD51 foci in
irradiated BRCA1i or BRCA2i cells. The limited effect of olaparib
treatment in the experiments in this study may be due to an
exhaustion of HR capacity by aphidicolin synchronization and
exposure to IR. Furthermore, it should be considered that PARP
fulfills a number of different functions in the DNA damage
response, including detection and signaling of DSBs and
stabilization of stalled replication forks (47,48). It was hypothesized that in the
experiments herein, PARP trapping, initiated by olaparib at the SSB
site (47,49), may have partly impaired HR
activation once the SSB is transformed in a DSB due to the
replication fork collapse.
To investigate whether the impairment of HR shown by
a decrease in RAD51 foci would result in increased chromosomal
abnormalities, we have performed the micronucleus assay. Compared
to controls, a marked and dose-dependent increase of micronucleus
formation was observed in irradiated cells harboring a BRCA1 or
BRCA2 knockdown. This effect was more pronounced in BRCA1i cells
likely because BRCA1 plays a broader role in DNA DSB repair, which
includes the non-homologous end-joining pathway (1,2).
This pathway is active also in the G1 phase of the cell cycle, and
the micronucleus assay was performed in cycling MCF10A cells, of
which a fraction of >40% was in the G1 phase.
The impaired DNA repair capacity in BRCA1i and
BRCA2i cells also resulted in decreased cell survival compared to
controls. In addition, in this case, the BRCA1i cells were affected
by radiation to a greater extent, when compared to the BRCA2i
cells.
In conclusion, this study demonstrated that in cells
containing a knockdown for either BRCA1 or BRCA2, the HR pathway is
impaired, resulting in a >50% reduction of RAD51 foci, in
significant increases in MN yields and in decreased cell viability
following exposure to ionizing radiation, compared to the cells not
subjected to knockdown. As in both BRCA1i and BRCA2i cell lines
<50% of the protein is retained after lentiviral knockdown, the
results obtained in the MCF10A cell line may mimic the situation in
healthy carriers of a germline BRCA1/2 mutation. Therefore,
the assessment of RAD51 foci in heterozygous BRCA1 and
BRCA2 mutation carriers may be a useful strategy to measure
radiosensitivity and HR capacity in these subjects. However, as the
results of this study are preliminary, this hypothesis is currently
being tested in breast epithelial cells and lymphocytes of
BRCA1/BRCA2 women who may present impaired expression of
BRCA due to the presence of germline heterozygous mutations in
different functional domains of both genes. Information from these
investigations will also be crucial to direct further research,
ultimately aiming at improving protection strategies in subjects
showing increased risk for radiation-induced carcinogenesis.
Supplementary Material
yH2AX foci staining following the
exposure of control MCF10A cells to ionizing radiation. Induction
of yH2AX foci was assessed 30 min following exposure to ionizing
radiation (2 Gy) or after mock irradiation (0 Gy). Immunostaining
was performed by means of a monoclonal anti-yH2AX antibody, as
previously described (34). This image gallery shows 4 cells for
each experimental condition taken with Metafer 4
(Metasystems).
Evaluation of PARP inhibition by
olaparib. Immunodetection for PARP activity was performed as
previously described by Barazzuol et al (35). PARP
activation was induced by H2O2 treatment (20
mM, 10 min, left panel). Incubation with olaparib (5 μM) 1 h
prior to and during exposure to H2O2,
resulted in the absence of PARP activity (right panel). Images were
obtained with Metafer 4 (Metasystems). PARP, poly(ADP-ribose)
polymerase.
RT-qPCR analysis of BRCA1 and
BRCA2 mRNA levels (± standard deviation) in control, BRCA1i
and BRCA2i cell lines. For an easy comparison, the relative
expression in relation to the control sample is shown for each
knockdown cell line (n=3). The relative mRNA expression of
BRCA1, but not that of BRCA2, was significantly
decreased in the BRCA1i cells, and the relative mRNA expression of
BRCA2, but not that of BRCA1, was significantly
decreased in the BRCA2i cells. Statistical analysis was carried out
using one-way ANOVA with Tukey's post-hoc test and the detailed
results are shown in Table
SI.
Results of statistical analysis with
one-way ANOVA and Tukey's test for post hoc significance of
multiple comparisons.
Acknowledgements
The authors would like to thank Ms. Leen Pieters,
Ms. Greet De Smet and Ms. Johanna Aernoudt for providing technical
assistance.
Funding
This research was funded by the Belgian Foundation
Against Cancer/Stichting Tegen Kanker, grant number 2012-216. The
funders had no role in the design of the study; in the collection,
analyses, or interpretation of data; in the writing of the
manuscript, or in the decision to publish the results.
Availability of data and materials
Data and materials are available to colleagues who
shall make written request of them. The source of such data and
material should be acknowledged in published articles.
Authors' contributions
AV and AB were involved in the conceptualization of
the study. AB, MFP, JD, MVH, BV, JV, KBMC and AS were involved in
the investigative aspects of the study. JP and AV were involved in
the study methodology. AB, MVH and GP were involved in data
analysis. AB was involved in the writing of the original draft. AV,
KBMC and GP were involved in the writing, reviewing and editing of
the manuscript. AV and KBMC were involved in manuscript
supervision. AV was involved in project administration and funding
acquisition.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Roy R, Chun J and Powell SN: BRCA1 and
BRCA2: Different roles in a common pathway of genome protection.
Nat Rev Cancer. 12:68–78. 2011.PubMed/NCBI View
Article : Google Scholar
|
|
2
|
Mao Z, Bozzella M, Seluanov A and
Gorbunova V: DNA repair by nonhomologous end joining and homologous
recombination during cell cycle in human cells. Cell Cycle.
7:2902–2906. 2008.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Pijpe A, Andrieu N, Easton DF, Kesminiene
A, Cardis E, Noguès C, Gauthier-Villars M, Lasset C, Fricker JP,
Peock S, et al: Exposure to diagnostic radiation and risk of breast
cancer among carriers of BRCA1/2 mutations: Retrospective cohort
study (GENE-RAD-RISK). BMJ. 345(e5660)2012.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Andrieu N, Easton DF, Chang-Claude J,
Rookus MA, Brohet R, Cardis E, Antoniou AC, Wagner T, Simard J,
Evans G, et al: Effect of chest X-rays on the risk of breast cancer
among BRCA1/2 mutation carriers in the international BRCA1/2
carrier cohort study: A report from the EMBRACE, GENEPSO,
GEO-HEBON, and IBCCS Collaborators' Group. J Clin Oncol.
24:3361–3366. 2006.PubMed/NCBI View Article : Google Scholar
|
|
5
|
John EM, McGuire V, Thomas D, Haile R,
Ozcelik H, Milne RL, Felberg A, West DW, Miron A, Knight JA, et al:
Diagnostic chest X-rays and breast cancer risk before age 50 years
for BRCA1 and BRCA2 mutation carriers. Cancer Epidemiol Biomarkers
Prev. 22:1547–1556. 2013.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Narod SA, Lubinski J, Ghadirian P, Lynch
HT, Moller P, Foulkes WD, Rosen B, Kim-Sing C, Isaacs C, Domchek S,
et al: Screening mammography and risk of breast cancer in BRCA1 and
BRCA2 mutation carriers: A case-control study. Lancet Oncol.
7:402–406. 2006.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Giannakeas V, Lubinski J, Gronwald J,
Moller P, Armel S, Lynch HT, Foulkes WD, Kim-Sing C, Singer C,
Neuhausen SL, et al: Mammography screening and the risk of breast
cancer in BRCA1 and BRCA2 mutation carriers: A prospective study.
Breast Cancer Res Treat. 147:113–118. 2014.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Bernstein JL, Thomas DC, Shore RE, Robson
M, Boice JD Jr, Stovall M, Andersson M, Bernstein L, Malone KE,
Reiner AS, et al: Contralateral breast cancer after radiotherapy
among BRCA1 and BRCA2 mutation carriers: A WECARE study report. Eur
J Cancer. 49:2979–2985. 2013.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Broeks A, Braaf LM, Huseinovic A, Nooijen
A, Urbanus J, Hogervorst FB, Schmidt MK, Klijn JG, Russell NS, Van
Leeuwen FE and Van 't Veer LJ: Identification of women with an
increased risk of developing radiation-induced breast cancer: A
case only study. Breast Cancer Res. 9(R26)2007.PubMed/NCBI View
Article : Google Scholar
|
|
10
|
Baeyens A, Thierens H, Claes K, Poppe B,
de Ridder L and Vral A: Chromosomal radiosensitivity in BRCA1 and
BRCA2 mutation carriers. Int J Radiat Biol. 80:745–756.
2004.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Gutiérrez-Enríquez S, Ramón Y, Cajal T,
Alonso C, Corral A, Carrasco P, Cornet M, Sanz J, Ribas M, Baiget M
and Diez O: Ionizing radiation or mitomycin-induced micronuclei in
lymphocytes of BRCA1 or BRCA2 mutation carriers. Breast Cancer Res
Treat. 127:611–622. 2011.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Trenz K, Rothfuss A, Schütz P and Speit G:
Mutagen sensitivity of peripheral blood from women carrying a BRCA1
or BRCA2 mutation. Mutat Res. 500:89–96. 2002.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Ernestos B, Nikolaos P, Koulis G, Eleni R,
Konstantinos B, Alexandra G and Michael K: Increased chromosomal
radiosensitivity in women carrying BRCA1/BRCA2 mutations assessed
with the G2 assay. Int J Radiat Oncol Biol Phys. 76:1199–1205.
2010.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Becker AA, Graeser MK, Landwehr C, Hilger
T, Baus W, Wappenschmidt B, Meindl A, Weber RG and Schmutzler RK: A
24-color metaphase-based radiation assay discriminates heterozygous
BRCA2 mutation carriers from controls by chromosomal
radiosensitivity. Breast Cancer Res Treat. 135:167–175.
2012.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Kote-Jarai Z, Salmon A, Mengitsu T,
Copeland M, Ardern-Jones A, Locke I, Shanley S, Summersgill B, Lu
YJ, Shipley J and Eeles R: Increased level of chromosomal damage
after irradiation of lymphocytes from BRCA1 mutation carriers. Br J
Cancer. 94:308–310. 2006.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Frankenberg-Schwager M and Gregus A:
Chromosomal instability induced by mammography X-rays in primary
human fibroblasts from BRCA1 and BRCA2 mutation carriers. Int J
Radiat Biol. 88:846–857. 2012.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Barwell J, Pangon L, Georgiou A, Kesterton
I, Langman C, Arden-Jones A, Bancroft E, Salmon A, Locke I,
Kote-Jarai Z, et al: Lymphocyte radiosensitivity in BRCA1 and BRCA2
mutation carriers and implications for breast cancer
susceptibility. Int J Cancer. 121:1631–1636. 2007.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Baert A, Depuydt J, Van Maerken T, Poppe
B, Malfait F, Storm K, van den Ende J, Van Damme T, De Nobele S,
Perletti G, et al: Increased chromosomal radiosensitivity in
asymptomatic carriers of a heterozygous BRCA1 mutation. Breast
Cancer Res. 18(52)2016.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Baert A, Depuydt J, Van Maerken T, Poppe
B, Malfait F, Van Damme T, De Nobele S, Perletti G, De Leeneer K,
Claes KB and Vral A: Analysis of chromosomal radiosensitivity of
healthy BRCA2 mutation carriers and non-carriers in BRCA families
with the G2 micronucleus assay. Oncol Rep. 37:1379–1386.
2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
O'Keefe EP: siRNAs and shRNAS: Tools for
protein knockdown by Gene Silencing. Mater Methods. 3(197)2013.
View Article : Google Scholar
|
|
21
|
Navaraj A, Finnberg N, Dicker DT, Yang W,
Matthew EM and El-Deiry WS: Reduced cell death, invasive and
angiogenic features conferred by BRCA1-deficiency in mammary
epithelial cells transformed with H-Ras. Cancer Biol Ther.
8:2417–2444. 2009.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Hellemans J, Mortier G, De Paepe A,
Speleman F and Vandesompele J: qBase relative quantification
framework and software for management and automated analysis of
real-time quantitative PCR data. Genome Biol. 8(R19)2007.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Li J and Xu X: DNA double-strand break
repair: A tale of pathway choices. Acta Biochim Biophys Sin
(Shanghai). 48:641–646. 2016.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Mladenov E, Magin S, Soni A and Iliakis G:
DNA double-strand-break repair in higher eukaryotes and its role in
genomic instability and cancer: Cell cycle and
proliferation-dependent regulation. Semin Cancer Biol. 37-38:51–64.
2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Ceccaldi R, Rondinelli B and D'Andrea AD:
Repair pathway choices and consequences at the double-strand break.
Trends Cell Biol. 26:52–64. 2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Rothkamm K, Barnard S, Moquet J, Ellender
M, Rana Z and Burdak-Rothkamm S: DNA damage foci: Meaning and
significance. Environ Mol Mutagen. 56:491–504. 2015.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Willers H, Taghian AG, Luo CM,
Treszezamsky A, Sgroi DC and Powell SN: Utility of DNA repair
protein foci for the detection of putative BRCA1 pathway defects in
breast cancer biopsies. Mol Cancer Res. 7:1304–1309.
2009.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Vaclová T, Gómez-López G, Setién F, Bueno
JM, Macías JA, Barroso A, Urioste M, Esteller M, Benítez J and
Osorio A: DNA repair capacity is impaired in healthy BRCA1
heterozygous mutation carriers. Breast Cancer Res Treat.
152:271–282. 2015.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Sioftanos G, Ismail A, Föhse L, Shanley S,
Worku M and Short SC: BRCA1 and BRCA2 heterozygosity in embryonic
stem cells reduces radiation-induced Rad51 focus formation but is
not associated with radiosensitivity. Int J Radiat Biol.
86:1095–1105. 2010.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Naipal KA, Verkaik NS, Ameziane N, van
Deurzen CH, Ter Brugge P, Meijers M, Sieuwerts AM, Martens JW,
O'Connor MJ, Vrieling H, et al: Functional ex vivo assay to select
homologous recombination-deficient breast tumors for PARP inhibitor
treatment. Clin Cancer Res. 20:4816–4826. 2014.PubMed/NCBI View Article : Google Scholar
|
|
32
|
AlHilli MM, Becker MA, Weroha SJ, Flatten
KS, Hurley RM, Harrell MI, Oberg AL, Maurer MJ, Hawthorne KM, Hou
X, et al: In vivo anti-tumor activity of the PARP inhibitor
niraparib in homologous recombination deficient and proficient
ovarian carcinoma. Gynecol Oncol. 143:379–388. 2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Mukhopadhyay A, Elattar A, Cerbinskaite A,
Wilkinson SJ, Drew Y, Kyle S, Los G, Hostomsky Z, Edmondson RJ and
Curtin NJ: Development of a functional assay for homologous
recombination status in primary cultures of epithelial ovarian
tumor and correlation with sensitivity to poly(ADP-ribose)
polymerase inhibitors. Clin Cancer Res. 16:2344–2351.
2010.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Depuydt J, Baert A, Vandersickel V,
Thierens H and Vral A: Relative biological effectiveness of
mammography X-rays at the level of DNA and chromosomes in
lymphocytes. Int J Radiat Biol. 89:532–538. 2013.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Barazzuol L, Jena R, Burnet NG, Meira LB,
Jeynes JC, Kirkby KJ and Kirkby NF: Evaluation of poly (ADP-ribose)
polymerase inhibitor ABT-888 combined with radiotherapy and
temozolomide in glioblastoma. Radiat Oncol. 8(65)2013.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Vandersickel V, Mancini M, Slabbert J,
Marras E, Thierens H, Perletti G and Vral A: The radiosensitizing
effect of Ku70/80 knockdown in MCF10A cells irradiated with X-rays
and p(66)+Be(40) neutrons. Radiat Oncol. 5(30)2010.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Vandersickel V, Slabbert J, Thierens H and
Vral A: Comparison of the colony formation and crystal violet cell
proliferation assays to determine cellular radiosensitivity in a
repair-deficient MCF10A cell line. Radiat Measurements. 46:72–75.
2011. View Article : Google Scholar
|
|
38
|
Slabbert JP, Theron T, Serafin A, Jones
DT, Böhm L and Schmitt G: Radiosensitivity variations in human
tumor cell lines exposed in vitro to p(66)/Be neutrons or 60Co
gamma-rays. Strahlenther Onkol. 172:567–572. 1996.PubMed/NCBI
|
|
39
|
Imbalzano KM, Tatarkova I, Imbalzano AN
and Nickerson JA: Increasingly transformed MCF-10A cells have a
progressively tumor-like phenotype in three-dimensional basement
membrane culture. Cancer Cell Int. 9(7)2009.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Sedic M and Kuperwasser C:
BRCA1-hapoinsufficiency: Unraveling the molecular and cellular
basis for tissue-specific cancer. Cell Cycle. 15:621–627.
2016.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Warren M, Lord CJ, Masabanda J, Griffin D
and Ashworth A: Phenotypic effects of heterozygosity for a BRCA2
mutation. Hum Mol Genet. 12:2645–2656. 2003.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Kraakman-van der Zwet M, Overkamp WJ, van
Lange RE, Essers J, van Duijn-Goedhart A, Wiggers I, Swaminathan S,
van Buul PP, Errami A, Tan RT, et al: Brca2 (XRCC11) deficiency
results in radioresistant DNA synthesis and a higher frequency of
spontaneous deletions. Mol Cell Biol. 22:669–679. 2002.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Yuan SS, Lee SY, Chen G, Song M, Tomlinson
GE and Lee EY: BRCA2 is required for ionizing radiation-induced
assembly of Rad51 complex in vivo. Cancer Res. 59:3547–3551.
1999.PubMed/NCBI
|
|
44
|
Keimling M, Volcic M, Csernok A, Wieland
B, Dörk T and Wiesmüller L: Functional characterization connects
individual patient mutations in ataxia telangiectasia mutated (ATM)
with dysfunction of specific DNA double-strand break-repair
signaling pathways. FASEB J. 25:3849–3860. 2011.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Pathania S, Bade S, Le Guillou M, Burke K,
Reed R, Bowman-Colin C, Su Y, Ting DT, Polyak K, Richardson AL, et
al: BRCA1 haploinsufficiency for replication stress suppression in
primary cells. Nat Commun. 5(5496)2014.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Karanam K, Kafri R, Loewer A and Lahav G:
Quantitative live cell imaging reveals a gradual shift between DNA
repair mechanisms and a maximal use of HR in mid S phase. Mol Cell.
47:320–329. 2012.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Erratum for the Perspective: ‘Laying a
trap to kill cancer cells: PARP inhibitors and their mechanisms of
action’ by Y. Pommier, M. J. O'Connor, J. de Bono. Sci Transl Med
8: 368er7, 2016.
|
|
48
|
Khodyreva SN and Lavrik OI:
Poly(ADP-Ribose) polymerase 1 as a key regulator of DNA repair. Mol
Biol (Mosk). 50:655–673. 2016.(In Russian). PubMed/NCBI View Article : Google Scholar
|
|
49
|
Murai J, Huang SY, Das BB, Renaud A, Zhang
Y, Doroshow JH, Ji J, Takeda S and Pommier Y: Trapping of PARP1 and
PARP2 by clinical PARP inhibitors. Cancer Res. 72:5588–5599.
2012.PubMed/NCBI View Article : Google Scholar
|