Introduction
Genome-wide association studies have identified
numerous single nucleotide polymorphisms (SNPs) that are associated
with human diseases, including type 2 diabetes mellitus (T2DM)
(1). However, the majority of
these SNPs are located in the non-coding regions of the genome and
the mechanisms by which they affect disease risk are poorly
understood. Potassium voltage-gated channel subfamily Q member 1
(KCNQ1) is a gene that is significantly associated with T2DM
in different ethnic groups, particularly in patients of East Asian
origin (1–3). Numerous T2DM-associated SNPs,
including rs2237892, rs2237895, rs151290, rs2283228 and rs2074196,
have been located in a KCNQ1 intron (2–4). A
recent study has also indicated that increased KCNQ1 protein
expression can limit insulin secretion from pancreatic β cells by
regulating potassium channel currents (5). However, to the best of our
knowledge, there are currently no data regarding the association
between disease-associated variants located in a KCNQ1
intron and the expression of KCNQ1.
Studies from the Encyclopedia of DNA Elements
(ENCODE) project have confirmed that disease-associated variants
are generally enriched in regulatory DNA, and that promoters and
distal elements engage in multiple long-range interactions to form
complex networks (6–9). For example, obesity-associated
variants within the α-ketoglutarate dependent dioxygenase gene form
long-range functional connections with the iroquois homeobox 3 gene
as a distal enhancer (8).
Therefore, SNPs in the KCNQ1 gene may affect the expression
of nearby or remote genes. Among the diabetes-associated candidate
genes located close to KCNQ1, cyclin-dependent kinase
inhibitor 1C (CDKN1C) is notable. Loss of this gene in focal
hyperinsulinism-associated lesions is correlated with an increased
proliferation of pancreatic β cells (10) and targeting CDKN1C promotes
adult human β cell replication in vitro (11). Furthermore, the long non-coding
RNA (lncRNA), KCNQ1 opposite strand/antisense transcript 1
(KCNQ1OT1), regulates the expression of CDKN1C via
imprinting (12–14). A mouse model has also demonstrated
that a paternal allelic mutation of the KCNQ1 locus reduces
pancreatic β cell mass by epigenetic modification of CDKN1C
through KCNQ1OT1 (15).
In order to analyze the functional impact of SNPs
located in non-coding regions of the genome, our group recently
developed a comparative method using a novel nanobead system. It
was successfully demonstrated that this method was effective for
the identification of allele-specific DNA-binding proteins and may
provide important information to understand the functional impact
of SNPs (16). Our group
demonstrated that nuclear transcription factor Y (NF-Y)
specifically bound to a genomic region containing the non-risk
allele for T2DM of the KCNQ1 SNP rs2074196. NF-Y also
stimulated transcriptional activity in an allele-specific manner in
an artificial promoter containing SNP rs2074196 (16). These results suggest that SNP
rs2074196 modulates the binding activity of the locus for NF-Y and
may induce subsequent changes in gene expression. However, in this
previous study, candidate target genes that may be regulated by the
SNP rs2074196 locus and NF-Y were not identified. Furthermore, our
group detected allele-specific binding proteins for other intronic
SNP (rs2237892, rs151290 and rs2283228) regions of the KCNQ1
gene, which revealed that it is possible that other regions may
also be functional and contribute to T2DM susceptibility (16).
The ENCODE project has also characterized
genome-wide histone modification patterns, including the
acetylation of lysine 27 of histone H3 (H3K27Ac), which is often
found in histones that flank active promoters and enhancers
(17). Among the T2DM-associated
SNPs in the KCNQ1 locus, only KCNQ1 SNP rs163184
exists in a genomic region that co-localizes with H3K27Ac in human
skeletal muscle myoblasts, normal human lung fibroblasts, human
adipose tissue and human pancreatic islets, as demonstrated in the
University of California, Santa Cruz Genome Browser (genome.ucsc.edu/). Covalent modifications of histones
are known to maintain an appropriate balance in stability and
reversibility, and to influence biological processes in the context
of development and cellular responses (18). Therefore, the SNP rs163184 locus
can be considered to be a functional genomic region; although, it
is unclear whether the locus positively or negatively regulates
gene expression. Furthermore, the SNP rs163184, and widely studied
KCNQ1 SNPs such as rs2237892 and rs2237895, are in moderate
linkage disequilibrium with each other (2). This information prompted the
speculation that certain proteins or protein complexes may bind to
the SNP rs163184 locus in an allele-specific manner and may change
the transcriptional regulation of target genes. Therefore, the
present study used our group's previously developed comparative
nanobead method to investigate SNP rs163184 and identify
allele-specific binding proteins for this region. In addition,
potential target genes that are regulated by SNP rs163184 and its
specific binding proteins were investigated.
Materials and methods
Cells
The rat pancreatic β cell line, INS-1 (19), was kindly provided by Dr C.B.
Wollheim and Dr N. Sekine (University of Geneva, Geneva,
Switzerland). The INS-1 cells were cultured at 37°C with 5%
CO2 atmosphere in RPMI-1640 medium (Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany), supplemented with 10% fetal bovine serum
(FBS; Biowest, Nuaille, France), 10 mM HEPES, 1 mM sodium pyruvate
and 50 mM 2-mercaptoethanol. KP-3, KP-4, QGP-1 and HuH-7 cells were
obtained from the Japan Health Science Research Resources Bank
(Osaka, Japan). HeLa, 293T, MCF-7 and Caco-2 cells were sourced
from the American Type Culture Collection (Manassas, VA, USA). KP-3
and QGP-1 cells were cultured in RPMI-1640 medium supplemented with
10% FBS. KP-4 cells were cultured in Iscove's modified Dulbecco's
medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 10% FBS. HuH-7, HeLa and 293T cells were cultured
in Dulbecco's modified Eagle's medium (Sigma-Aldrich; Merck KGaA)
supplemented with 10% FBS. MCF-7 and Caco-2 cells were cultured in
Eagle's minimum essential medium (Sigma-Aldrich; Merck KGaA)
supplemented with 10% FBS. These cell lines (KP-3, KP-4, QGP-1,
HuH-7, HeLa, 293T, MCF-7 and Caco-2) were cultured at 37°C in an
atmosphere with 5% CO2.
Preparation of DNA-immobilized
nanobeads
The oligonucleotides used in the present study were
synthesized by Eurofins Genomics (Tokyo, Japan). These
oligonucleotides contained either risk or non-risk alleles of the
KCNQ1 gene via single-nucleotide substitutions.
DNA-immobilized nanobeads were prepared as previously described
(16). Briefly, two complementary
oligonucleotides were phosphorylated, annealed and ligated to
produce oligomers that ranged from 100-3,000 base pairs. These
oligomers, containing tandemly repeated SNP regions, were applied
to a NICK column (cat. no. 17-0855-02; GE Healthcare, Chicago, IL,
USA), according to the manufacturer's protocol. The second fraction
(100 µg prepared DNA) from each NICK column was mixed with 1
mg FG beads (Tamagawa Seiki Co., Ltd., Nagano, Japan), then a
coupling reaction was conducted at 50°C for 24 h.
Affinity purification
Nuclear extracts were prepared from INS-1 cells
according to the method described by Dignam et al (20). The DNA-immobilized nanobeads or
negative control nanobeads without immobilized DNA (200 µg)
were equilibrated using binding buffer [20 mM HEPES-NaOH (pH 7.9),
10% glycerol, 100 mM KCl, 1 mM MgCl2, 0.2 mM
CaCl2, 0.2 mM EDTA, 0.1% NP-40, 1 mM DTT and 0.2 mM
PMSF]. INS-1 nuclear extracts (200 µg) were mixed with 100
µg of single-stranded DNA (cat. no. AM9680; Ambion; Thermo
Fisher Scientific, Inc.) and 10 µg of
poly(dI-dC):poly(dI-dC) (Sigma-Aldrich; Merck KGaA). These
solutions were then mixed with equilibrated DNA-immobilized
nanobeads or negative control nanobeads without immobilized DNA,
and incubated for 4 h at 4°C with gentle rotation using a RT-50
rotator (Taitec Corp., Saitama, Japan). After washing with binding
buffer three times, bound proteins were eluted with binding buffer
supplemented with 1 M KCl.
Mass spectrometry analysis
Affinity purified proteins were separated using
SDS-PAGE (5–20% gradient gel) and subjected to silver staining
using a protocol described by Shevchenko et al (21). Specific protein bands were excised
and mass spectrometry analysis was performed, as previously
described (22). Band slices were
reduced with 10 mM DTT for 1 h at 56°C and then alkylated with 55
mM iodoacetamide for 45 min at room temperature. The bands were
next digested overnight at 37°C with sequencing grade modified
trypsin (12 mg/ml; Promega Corp., Madison, WI, USA) and desalted
with ZipTip C18 (EMD Millipore, Billerica, MA, USA). Nano liquid
chromatography-tandem mass spectrometry analysis was performed as
previously described (23).
Extracted peptides were separated via nanoflow liquid
chromatography (Paradigm MS4; Michrom BioResources, Tokyo, Japan)
using a reverse phase C18 column (L-Column Micro; Chemicals
Evaluation and Research Institute, Tokyo, Japan). The liquid
chromatography eluent was coupled to a nano-ionspray source
attached to a QSTAR Elite Q-TOF mass spectrometer (Sciex,
Framingham, MA, USA). Peptide identification was performed by
matching raw spectra data against rat proteins from the
International Protein Index database (24) (version 3.87) using ProteinPilot
software (version 3.0; Sciex) and the Paragon algorithm with a
confidence threshold of 95% (ProteinPilot unused score
>1.3).
Immunoblotting
Antibodies directed against transcription factor Sp1
(Sp1; cat. no. 07-645), transcription factor Sp3 (Sp3; cat. no.
07-107), myelin transcription factor 1 (Myt1; cat. no. ABN21),
histone deacetylase 1 (Hdac1; cat. no. 06-720) and REST corepressor
1 (Rcor1; cat. no. MABN486) were obtained from EMD Millipore.
Antibodies directed against Myt1-like protein (Myt1l; cat. no.
ab139732), REST corepressor 2 (cat. no. ab113826) and NF-YA (cat.
no. ab6558) were supplied by Abcam (Cambridge, MA, USA). Antibodies
directed against lysine-specific histone demethylase 1A
(Lsd1/Kdm1a; cat. no. 2184), Hdac2 (cat. no. 2540) and Myc
proto-oncogene protein (Myc)-tagged proteins (Myc-tag; cat. no.
2276) were sourced from Cell Signaling Technology, Inc. (Danvers,
MA, USA). The antibody directed against nucleolar protein 4 (Nol4;
cat. no. 14802-1-AP) was purchased from ProteinTech Group, Inc.
(Chicago, IL, USA). The antibody directed against the FLAG-tag
(cat. no. F1804) was purchased from Sigma-Aldrich.
Nuclear extracts (10 µg) from INS-1 cells and
affinity-purified fractions were resolved by SDS-PAGE (5–20%
gradient gel) and electro-transferred to a polyvinylidene
difluoride membrane (EMD Millipore). The membrane was then blocked
with 5% non-fat milk for 1 h at room temperature and then incubated
overnight at 4°C with the primary antibodies as previously
described (16). This was
followed by incubation with a horseradish peroxidase-conjugated
secondary antibody [anti-mouse-immunoglobulin G (IgG), cat. no.
7076; or anti-rabbit-IgG, cat. no. 7074; Cell Signaling Technology,
Inc.] for 1 h at room temperature. Visualization was accomplished
using Immobilon Western Chemiluminescent HRP Substrate (cat. no.
WBKLS0500; EMD Millipore) and a ChemiDoc XRS Plus system (Bio-Rad
Laboratories, Inc., Hercules, CA, USA).
Preparation of radiolabeled recombinant
proteins
Total RNA was isolated from HeLa cells using an
RNeasy mini kit (cat. no. 74106; Qiagen GmbH, Hilden, Germany)
according to the manufacturer's protocol. Complementary DNA (cDNA)
was synthesized from the total RNA using the SuperScript III
First-Strand Synthesis system (cat. no. 18080-051; Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. cDNA for MYT1 and MYT1L were amplified
using PrimeSTAR HS DNA Polymerase (cat. no. R010A; Takara Bio,
Inc., Otsu, Japan) according to the manufacturer's protocol. The
primers used for amplification were as follows: MYT1
forward, 5′-GCCGCCGCGATCGCCATGAGCTTAGAAAATGAAGACAAGC-3′ and
reverse, 5′-GCTCGAGCGGCCGCGTGACCTGGATGCCCCTCACAGC-3′; and
MYT1L forward, 5′′-GCCGCCGCGATCGCCATGGAGGTGGACACCGAGGAG-3′
and reverse, 5′-GCTCGAGCGGCCGCGTGACCTGAATTCCTCTCACAGCC-3′. The
thermocycling conditions were as follows: 98°C for 1 min; followed
by 30 cycles of 98°C for 10 sec, 60°C for 5 sec and 72°C for 4 min;
and 72°C for 5 min.
The cDNA were inserted into a pCMV6-Entry vector
(OriGene Technologies, Inc., Rockville, MD, USA) and sequenced with
an ABI 3700 sequence analyzer (Applied Biosystems; Thermo Fisher
Scientific, Inc.). Expression plasmids for SP1 (cat. no.
RC209154), SP3 (cat. no. RC222027), LSD1/KDM1A (cat.
no. RC230555) and PHD finger protein 21A (PHF21A; cat. no.
RC213929) were obtained from OriGene Technologies, Inc. These are
mammalian expression vectors that express the cloned cDNA
(SP1, SP3, LSD1/KDM1A and PHF21A,
respectively) with an Myc-FLAG-tag. The subsequent preparation of
radiolabeled recombinant protein was performed as previously
described (22). T7
promoter-tagged DNA fragments of these cDNA (MYT1,
MYT1L, SP1, SP3, LSD1/KDM1A and
PHF21A, respectively) were amplified using PrimeSTAR HS DNA
Polymerase (Takara Bio, Inc.) according to the manufacturer's
protocol. The primers used for amplification were as follows:
Forward,
5′-CACCGGTACCTAATACGACTCACTATAGGGAATACAAGCTACTTGTTCTTTTTGCAAGATCTGCCGCCGCGATCGCC-3′
and reverse,
5′-TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTAAACCTTATCGTCGTCATCC-3′. The
aforementioned thermocycling conditions were used. These amplified
DNA fragments were used to synthesize 35S-radiolabeled
recombinant proteins in a coupled transcription/ translation system
(TNT T7 Quick for PCR DNA; cat. no. L5540; Promega Corp.),
following the manufacturer's protocol.
Binding assay
DNA-immobilized nanobeads or negative control
nanobeads without immobilized DNA (200 µg) were equilibrated
with binding buffer, as previously described, and incubated with
200 µl of each of the radiolabeled proteins (Sp1, Sp3,
Lsd1/Kdm1a, Phf21a, Myt1 and Myt1l) at 4°C for 4 h using a RT-50
rotator. After washing with binding buffer, bound proteins were
eluted by boiling for 5 min at 98°C in SDS-PAGE sample buffer [125
mM Tris-HCl (pH 6.8), 250 mM 2-mercaptoethanol, 4% SDS, 0.2%
bromophenol blue and 20% glycerol]. Eluates and inputs were
subjected to SDS-PAGE (5–20% gradient gel). Gels were dried and
auto-radiography was used to visualize the radiolabeled
proteins.
Luciferase assay
To construct reporter plasmids that contained a
luciferase gene under the control of the SNP rs163184 region,
complementary oligonucleotides containing three copies of the SNP
region were annealed and cloned into the pGL4.23 vector (Promega
Corp.). The cloned sequences were verified as correct using an ABI
sequence analyzer 3700 (Thermo Fisher Scientific, Inc.).
The constructed luciferase reporter plasmids (100
ng) and the control Renilla luciferase reporter vector
pGL4.74 (10 ng; Promega Corp.), were transfected into 293T and
HuH-7 cells (2×105 cells/well) with the mammalian
expression vectors (SP1, SP3, LSD1/KDM1A,
PHF21A, MYT1 or MYT1L) using Lipofectamine
2000 (Invitrogen; Thermo Fisher Scientific, Inc.). Cells were
seeded into 24-well plates and 12 h after transfection, the culture
medium was changed. Cell extracts were prepared using Reporter
Lysis Buffer (cat. no. E3971; Promega Corp.), according to the
manufacturer's protocol, 48 h after transfection, and the Firefly
and Renilla luciferase activities were determined using a
Dual-Glo luciferase assay system (Promega Corp.) with a CentroPro
LB 962 luminometer (Berthold Technologies GmbH & Co. KG, Bad
Wildbad, Germany). Luciferase activities were calculated in
relative light units as a ratio of Firefly luciferase activities to
Renilla luciferase activities.
The Lsd1/Kdm1a inhibitor 2-PCPA and the monoamine
oxidase A (Mao) inhibitor pargyline were obtained from
Sigma-Aldrich (Merck KGaA). These reagents (5–500 µM) were
added to the culture when the medium was changed.
Chromatin immunoprecipitation (ChIP)
assay
Several cell lines (HeLa, MCF-7, KP-3, KP-4, QGP-1,
293T and Caco-2; 6×106 cells/dish) were transfected with
16 µg of the expression vector for Sp3-Myc-FLAG (cat. no.
RC222027; OriGene Technologies, Inc.) using Lipofectamine 2000. A
total of 48 h after transfection, ChIP sample preparation was
performed using a Magna ChIP G kit (EMD Millipore) according to the
manufacturer's protocol. Samples were sonicated on ice for 10
rounds of 30 pulses each (0.5 sec on and 0.5 sec off) with
intervals of 10 min using a Branson 450D Sonifier (Emerson,
Danbury, CT, USA) at a maximum amplitude of 15%. The sonicated
samples were immunoprecipitated overnight at 4°C with 2.5 µg
anti-Myc-tag antibody. After immunoprecipitation, DNA fragments
were separated by 2% agarose gel electrophoresis and fragments
between 200 and 1,000 base pairs were isolated using a NucleoSpin
Gel and PCR clean-up kit (cat. no. 740609; Macherey-Nagel GmbH,
Düren, Germany). The amount of DNA in the input and
immunoprecipitated fractions was quantified by quantitative PCR
(qPCR) using a Fast SYBR-Green Master mix and a StepOne Real-Time
PCR system (both from Applied Biosystems; Thermo Fisher Scientific,
Inc.). The thermocycling conditions were as follows: 95°C for 20
sec; followed by 50 cycles of 95°C for 3 sec and 60°C for 30 sec.
The primers used for qPCR were as follows:
5′-TTGCTCAGTAACGGACTGGAC-3′ and 5′-GTCAATGTGGGGAGGTGGTA-3′ for the
SNP rs163184 region; 5′-ATGGAGAGGGCTGAACACG-3′ and
5′-CCCTTGCCTCACATCCTTT-3′ for the 3,000-base pair region upstream
of SNP rs163184; and 5′-AGCCACCAGGAGCAGTTGT-3′ and
5′-TCTGGGAGTTGATGAGGGAAG-3′ for the 3,000-base pair region
downstream from SNP rs163184. The primers for upstream and
downstream regions of SNP rs163184 were designed as reference
regions. Bound to input ratios were determined for each amplicon
[cycle threshold (Ct) values of immunoprecipitated samples were
normalized to Ct values obtained from input samples] (25).
RNA interference and gene expression
analysis
Several cell lines (HeLa, MCF-7, KP-3, KP-4, 293T
and Caco-2) were seeded (2×105 cells/well) into 24-well
plates and trans-fected with either a control small interfering RNA
(siRNA) (10 nM; cat. no. 4390844; Thermo Fisher Scientific, Inc.)
or siRNA (10 nM) targeting human SP3 (cat. no. s13324;
Silencer Select siRNA; Thermo Fisher Scientific, Inc.) using
Lipofectamine 2000.
A total of 48 h after transfection, total RNA was
isolated from cells using an RNeasy mini kit (Qiagen) according to
the manufacturer's protocol. For RT-qPCR analysis, cDNA was
synthesized from the total RNA using the SuperScript III
First-Strand Synthesis system (Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. qPCR was then performed
using the TaqMan Gene Expression Master mix (Applied Biosystems;
Thermo Fisher Scientific, Inc.) for each TaqMan probe according to
the manufacturer's protocol. The thermocycling conditions were as
follows: 95°C for 20 sec; followed by 40 cycles of 95°C for 1 sec
and 60°C for 20 sec. Relative mRNA abundance was calculated using
the 2−ΔΔCq method (25) and normalized to the corresponding
values for β-actin (ACTB).
TaqMan Gene Expression assays for SP3
(UniGene ID, Hs01595811_m1), CD81 molecule (CD81; UniGene
ID, Hs01002167_m1), tumor suppressing subtransferable candidate 4
(TSSC4; UniGene ID, Hs00185082_m1), transient receptor
potential cation channel subfamily M member 5 (TRPM5;
UniGene ID, Hs00175822_m1), KCNQ1 (UniGene ID,
Hs00165003_m1), KCNQ1OT1 (UniGene ID, Hs03665990_s1),
CDKN1C (UniGene ID, Hs00175938_m1), solute carrier family 22
member 18 (SLC22A18; UniGene ID, Hs00180039_m1),
SLC22A18 anti-sense (SLC22A18AS; UniGene ID,
Hs00757934_m1), pleckstrin homology like domain family A member 2
(PHLDA2; UniGene ID, Hs00169368_m1), nucleosome assembly
protein 1-like 4 (NAP1L4; UniGene ID, Hs00924275_m1),
cysteinyl-tRNA synthetase (CARS; UniGene ID, Hs01000965_m1),
oxysterol binding protein like 5 (OSBPL5; UniGene ID,
Hs00957760_m1), MAS related GPR family member G (MRGPRG;
UniGene ID, Hs01010229_s1) and ACTB (UniGene ID, 4352935)
were obtained from Applied Biosystems (Thermo Fisher Scientific,
Inc.).
Immunoprecipitation and immunoblotting
analysis
The expression vectors (4 µg each) for
Sp3-Myc-FLAG (cat. no. RC222027; OriGene Technologies, Inc.) and
NF-Ys (NF-YA, NF-YB-HA and NF-YC-Myc) were transfected into 293T
cells (6×106 cells/ dish) using Lipofectamine 2000. The
constructions of the expression vectors for NF-Ys (NF-YA, NF-YB-HA
and NF-YC-Myc) were previously described (16). A total of 48 h after transfection,
cells were lysed in lysis buffer [50 mM Tris-HCl (pH 8.0),
containing 150 mM NaCl and 1.0% NP-40] with protease inhibitor
cocktail (cat. no. 25955-11; Nacalai Tesque, Inc., Kyoto, Japan).
Immunoprecipitation from transfected cell lysates was performed
using anti-FLAG M2 Magnetic beads (cat. no. M8823; Sigma-Aldrich;
Merck KGaA), and the beads were washed three times with lysis
buffer. Immunoprecipitates or total cell lysates were analyzed by
immunoblotting using anti-FLAG-tag antibody (1:1,000; cat. no.
F1804; Sigma-Aldrich; Merck KGaA) and anti-NF-YA antibody (1:1,000;
cat. no. ab6558; Abcam), using the aforementioned protocol.
Bioinformatics
Image data involving the information on the genomic
positions of H3K27Ac mark and SNPs in the KCNQ1 locus were
downloaded from the University of California, Santa Cruz Genome
Browser (genome.ucsc.edu/).
Statistical analysis
All data are expressed as the mean ± standard
deviation from at least three independent experiments. Statistical
analysis was performed using an unpaired two-tailed Student's
t-test for comparisons of two groups or by two-way non-repeated
measures analysis of variance, followed by the Bonferroni post hoc
test, for the comparisons of multiple groups. These statistical
analyses were performed using Microsoft Excel 2011 for Mac
(Microsoft Corporation, Redmond, WA, USA) with the add-in software,
Excel Statistical Program File ystat2008 (Igakutosho-shuppan, Ltd.,
Tokyo, Japan). P<0.01 was considered to indicate a statistically
significant difference.
Results
Purification of allele-specific
DNA-binding proteins using magnetic nanobeads
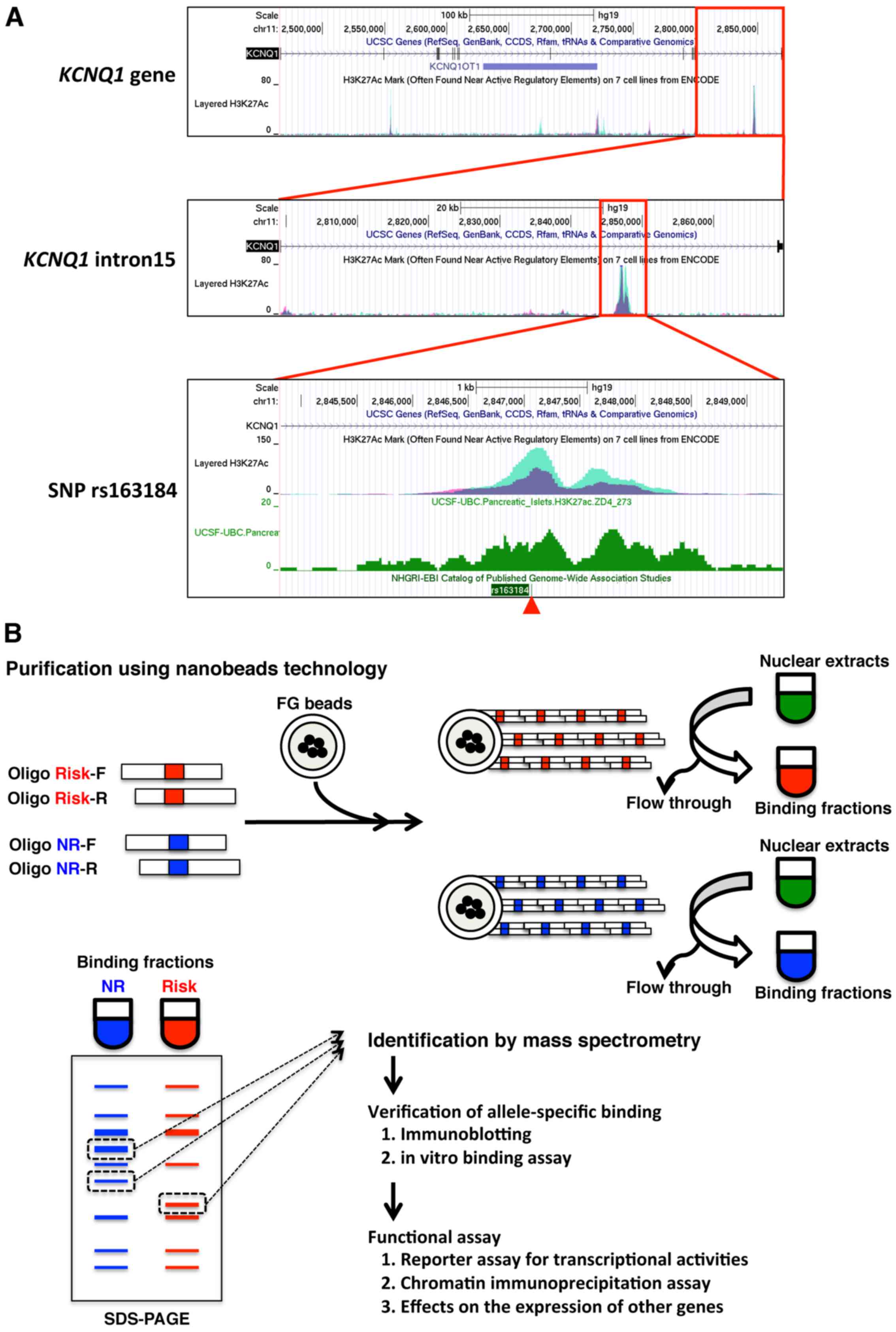
One of the SNPs that is most highly associated with
T2DM is the KCNQ1 SNP rs163184 (2). The position of this SNP overlaps
with a H3K27Ac, which has been shown by the ENCODE project to mark
active regulatory elements (Fig.
1A). The present study aimed to identify proteins that bind the
genomic region that encompasses the SNP rs163184 region (Fig. 1B). To identify allele-specific
binding proteins, DNA-immobilized beads were prepared by binding
each SNP rs163184-containing allele onto magnetic nanobeads
(Fig. 2A). Using these nanobeads,
DNA-binding proteins were purified from the nuclear extract of
INS-1 pancreatic β cells and bound proteins were analyzed using
SDS-PAGE (Fig. 2B). This revealed
that the binding activities of several proteins differed between
the alleles for the region containing SNP rs163184. Notably, in the
sample using oligo 3, many extra bands were detected, which were
not observed, or less observed, in the sample using oligos 4 or 5
(Fig. 2B and C).
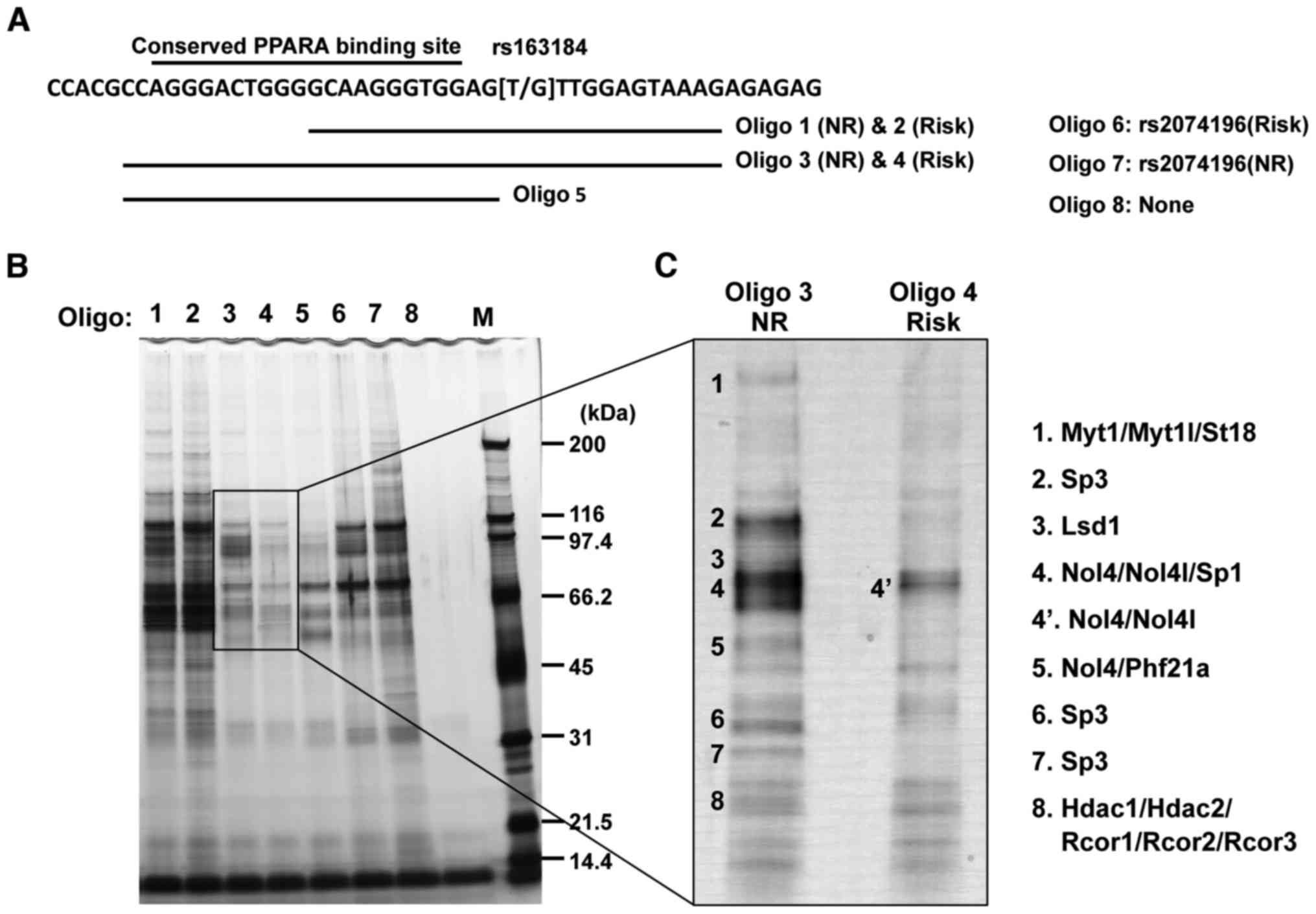 | Figure 2Purification and identification of
allele-specific proteins bound to the region containing SNP
rs163184. (A) Oligos immobilized on the nanobeads. Oligos 6 and 7
were designed for the risk and the NR alleles of SNP rs2074196,
respectively, as described in our previous study (16), and were included to demonstrate
the reproducibility of the purification system using nuclear
extracts from INS-1 cells. Oligo 5 was designed against the
predicted PPARA binding site, which was also included in oligos 3
and 4. However, further analysis identified no binding between
PPARA and this region. (B) Aliquots of purified fractions were
separated using SDS-PAGE (5–20% gradient gel) and visualized using
silver staining. (C) The same samples (panel B, lanes 3 and 4) were
separated using longer SDS-PAGE (5–20% gradient gel) and visualized
using silver staining to allow for the excision of the specific
protein bands. The names and locations on the gel of polypeptides
that were analyzed using ion-spray mass spectrometry are indicated
by corresponding numbers. Polypeptide band 4 was broad and part of
the band was also present in the oligo 4 (risk) lane, designated as
polypeptide band 4′ and used in further analysis. SNP, single
nucleotide polymorphism; oligo, oligonucleotides; NR, non-risk. |
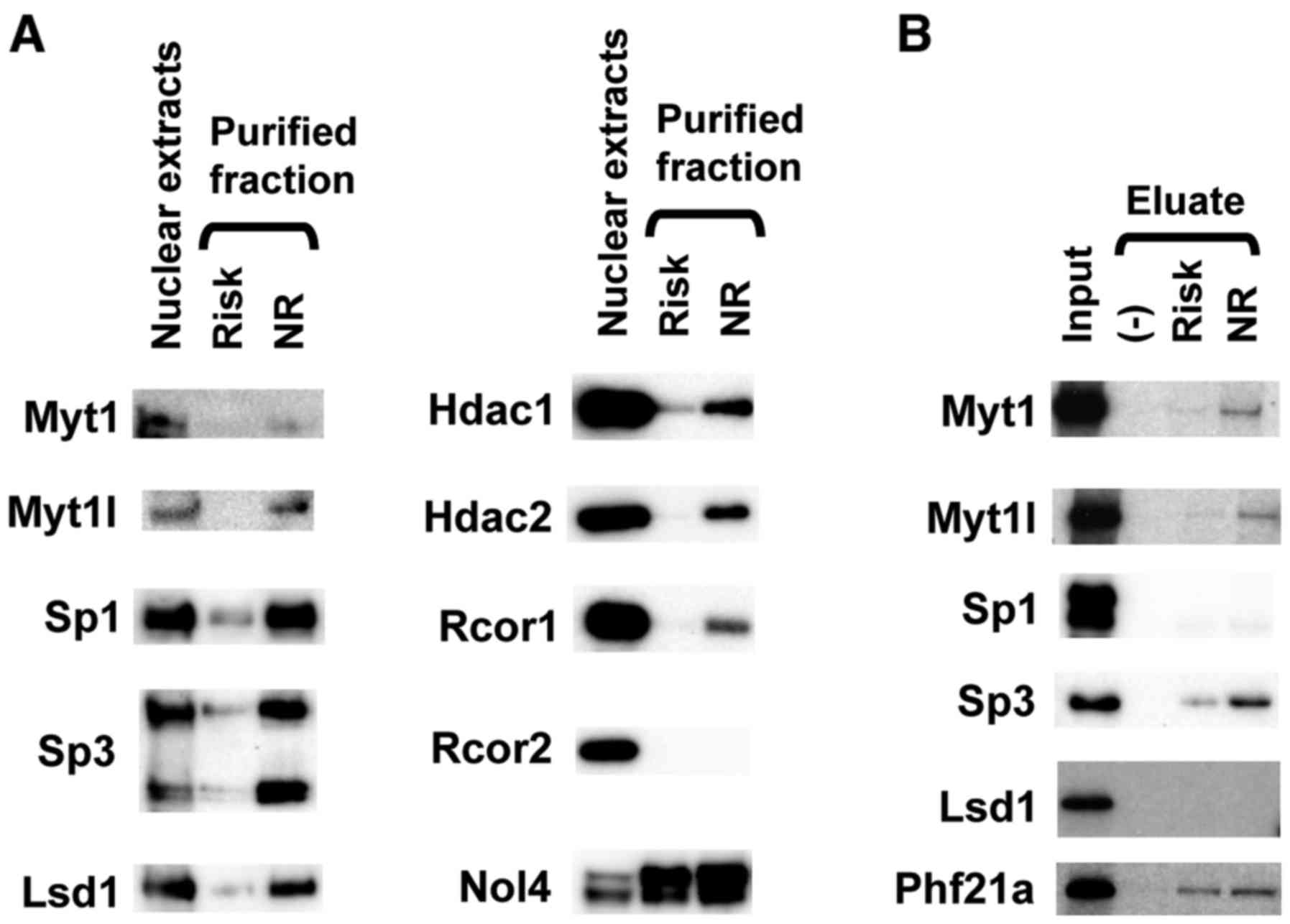
Following mass spectrometry analysis, the proteins
described in Fig. 2C were
identified, including Myt1/Myt1l/St18, Sp3, Lsd1, Nol4/Nol4l/Sp1,
Nol4/Phf21a, Hdac1/Hdac2/Rcor1/Rcor2/ Rcor3 and Nol4/Nol4l.
Immunoblotting with specific antibodies directed against these
proteins confirmed that Myt1, Myt1l, Sp1, Sp3, Lsd1/Kdm1a, Hdac1,
Hdac2 and Rcor1 were purified in an allele-specific manner
(Fig. 3A). It was not possible to
obtain a specific antibody directed against rat Phf21a that worked
effectively. Nol4 only exhibited non-specific binding activity
against the SNP rs163184 genomic region. Sp3 exhibited the highest
recovery efficiency among these proteins.
Next, the in vitro DNA-binding activities of
Myt1, Myt1l, Sp1, Sp3, Lsd1/Kdm1a and Phf21a were examined using
recombinant proteins. Myt1, Myt1l, Sp3 and Phf21a each exhibited
higher specific binding activity against the non-risk allele of the
genomic region that includes SNP rs163184 (Fig. 3B). The DNA-binding specificities
of recombinant Myt1, Myt1l, and Sp3 against the non-risk allele
were consistent with those observed in the immunoblotting of
purified proteins from INS-1 nuclear extracts (Fig. 3). Lsd1/Kdm1a exhibited no DNA
binding activity (Fig. 3B). This
indicates that Lsd1/Kdm1a is recruited to the rs163184 region by
other proteins, such as Sp3, which can bind to the region in an
allele-specific manner. Unexpectedly, the transcription factor Sp1
also exhibited no observable binding activity for this SNP region
(Fig. 3B). This suggests that Sp1
is also recruited to the SNP 163184 region by other proteins, or
due to post-translational modifications that are not present in the
recombinant Sp1 synthesized in the cell-free system and are
required for binding to this SNP region. Sp3 exhibited the highest
DNA-binding activity among the proteins examined for this SNP
region.
SNP rs163184 modulates Sp3-dependent
transcriptional activity
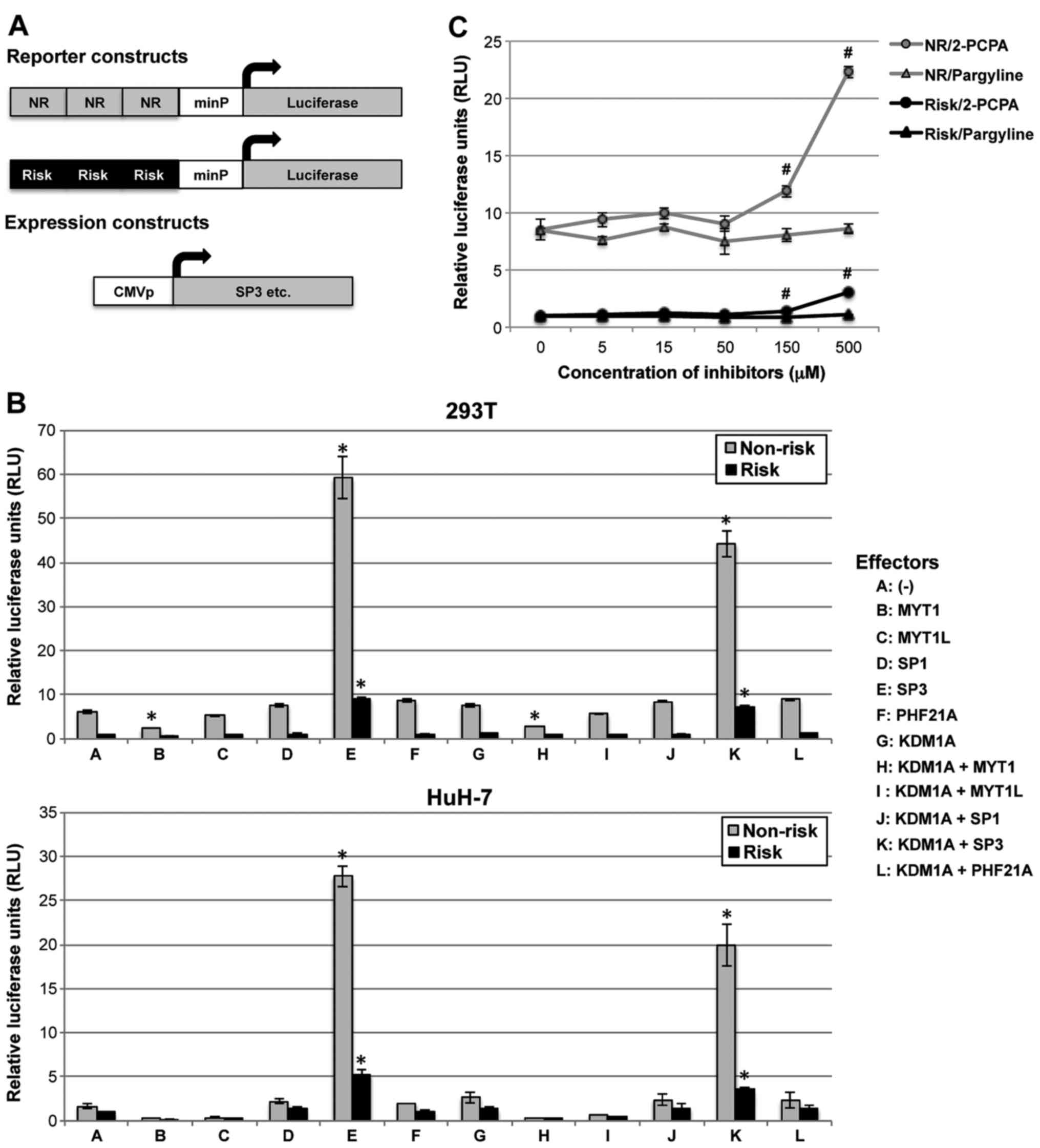
The transcriptional activities of Myt1, Myt1l, Sp1,
Sp3, Phf21a and Lsd1/Kdm1a were investigated using a luciferase
reporter gene assay. As illustrated in Fig. 4A, two types of reporter constructs
that were either risk or non-risk alleles were prepared. The basal
luciferase activities, mediated by endogenous transcription factors
(that may include Sp3), were markedly different between the two
types of constructs (Fig. 4A).
The relative luciferase activities (RLUs) of the non-risk allele
reporter construct were higher than those of the risk allele
reporter construct in both 293T and HuH-7 cells.
When the expression construct containing SP3
was cotransfected with each of the luciferase reporter plasmids
under the control of the SNP rs163184 region, RLUs of the non-risk
allele reporter construct were increased (RLU, 6.16 vs. 59.17 in
293T cells; RLU, 1.68 vs. 27.79 in HuH-7 cells) (Fig. 4B). The RLUs of the risk allele
reporter construct were also increased; however, these were still
much lower than the RLUs of the non-risk allele reporter
constructs. In HuH-7 cells, Sp3-induced a larger fold increase in
luciferase activities in non-risk allele constructs (RLU, 1.68 vs.
27.79; fold change, 16.57) compared to those in risk allele
constructs (RLU, 1 vs. 5.32; fold change, 5.32). These results
suggest that Sp3 may modulate the transcription of some specific
genes using the genomic region encompassing KCNQ1 SNP
rs163184 as a cis-regulatory element, particularly in the non-risk
allele.
When the expression construct containing MYT1
was cotransfected, the RLUs of each reporter construct were
decreased (Fig. 4B). This
suggests that Myt1 downregulates transcription controlled by the
genomic region flanking SNP rs163184.
When the expression construct containing
KDM1A was cotransfected with that of SP3,
Sp3-dependent luciferase activity was decreased compared to that in
cells transfected with SP3 alone (Fig. 4B). This indicates that Lsd1/Kdm1a
negatively regulates Sp3-dependent transcription under the control
of the SNP rs163184 region.
In order to confirm the suppressive effect
demonstrated by Lsd1/Kdm1a, the Lsd1/Kdm1a inhibitor 2-PCPA, which
reduces Lsd1/Kdm1a, Mao-A and Mao-B activity, was used. The Mao
inhibitor pargyline, which inhibits Mao-A and Mao-B, but is less
potent against Lsd1/Kdm1a, was also used. The Lsd1/Kdm1a inhibitor
2-PCPA enhanced the luciferase activities of the non-risk allele
reporter construct (RLU, 8.53 vs. 22.32) (Fig. 4C). However, the Mao inhibitor
pargyline had little effect on the RLUs of the non-risk allele
reporter construct (RLU, 8.53 vs. 8.58) (Fig. 4C). In addition, 2-PCPA enhanced
the RLUs of the risk allele reporter construct (RLU, 1 vs. 2.98),
although it was still much lower than that of the non-risk allele
reporter construct (Fig. 4C).
SNP rs163184 modulates the binding
activity of the locus for Sp3
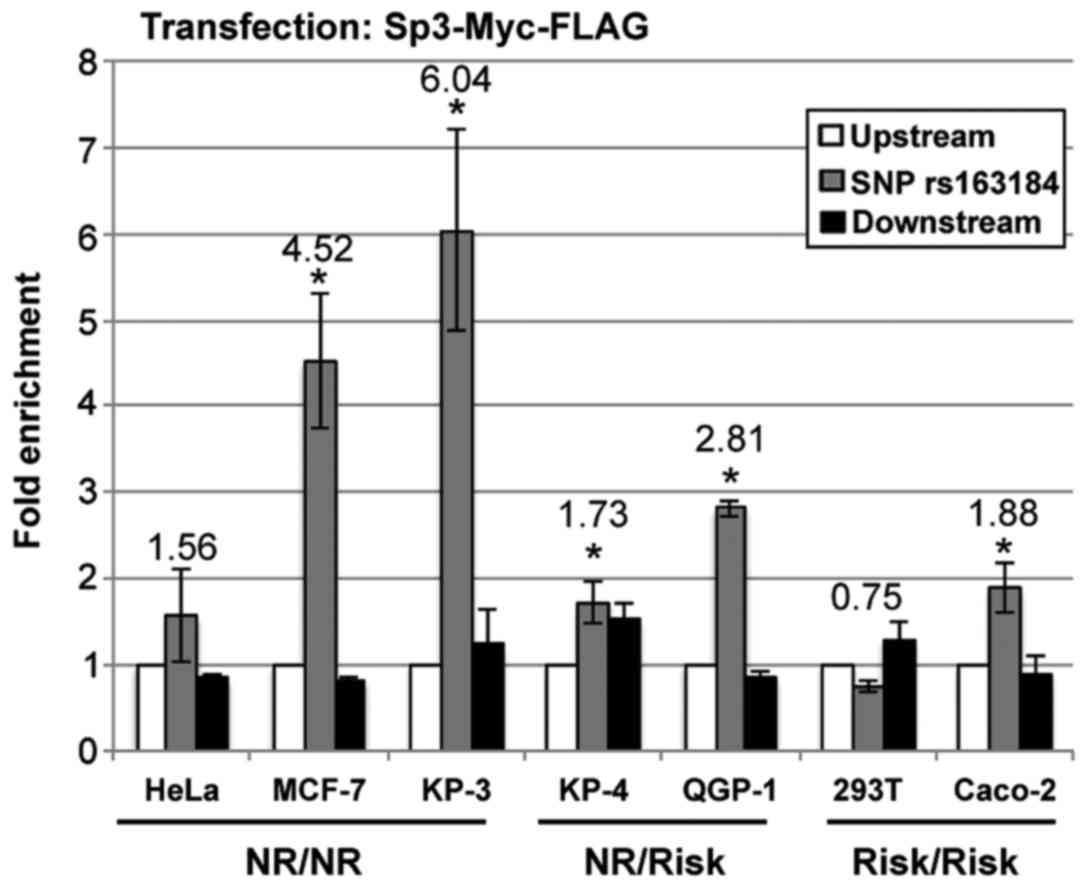
The DNA-binding activities of Sp3 were further
examined using a ChIP assay in HeLa, MCF-7, KP-3, KP-4, QGP-1, 293T
and Caco-2 cells. HeLa, MCF-7 and KP-3 cells were identified to be
homozygous for the non-risk allele, while KP-4 and QGP-1 cells were
heterozygous, and 293T and Caco-2 cells were homozygous for the
risk-associated allele. These cell lines were transfected with
expression vectors containing Sp3-Myc-FLAG. ChIP analysis revealed
that the genomic region containing the KCNQ1 SNP rs163184
was highly enriched for Sp3 in MCF-7 and KP-3 cells, moderately
enriched in QGP-1 cells, slightly enriched in HeLa, KP-4 and Caco-2
cells, and not significantly enriched in 293T cells (Fig. 5). Taken together with the data
from the in vitro binding assay, these results suggest that
Sp3 preferentially binds the non-risk allele of the endogenous
KCNQ1 intronic region that contains SNP rs163184. However,
the possibility that these cell lines may have additional genetic
variants that impact on the recruitment of Sp3 to the SNP rs163184
region cannot be excluded.
In addition, in order to examine the interaction
between Sp3 and NF-Y, which binds the SNP rs2074196 region in the
KCNQ1 locus and possibly induces subsequent change in gene
expression, immunoprecipitation and immunoblotting experiments were
performed. These results demonstrated possible interaction between
Sp3 and NF-Y (data not shown).
CDKN1C is a candidate target gene
regulated by the SNP rs163184 locus
In order to identify target genes that are regulated
by Sp3 and the locus encompassing the KCNQ1 SNP rs163184,
the expression of genes flanking KCNQ1 after siRNA-mediated
knockdown of endogenous SP3 was examined in HeLa, MCF-7,
KP-3, KP-4, 293T and Caco-2 cells. QGP-1 cells were excluded from
this analysis as endogenous SP3 could not be sufficiently
reduced (data not shown). In the luciferase reporter gene assay, it
was demonstrated that Sp3 upregulated the expression of the
reporter gene under the control of the KCNQ1 SNP rs163184
region and that Lsd1/Kdm1a downregulated Sp3-dependent
transcription under the control of the SNP rs163184 region. The
present study indicated that Lsd1/Kdm1a itself does not exhibit
DNA-binding activity, at least to the SNP rs163184 region, and
seems to be recruited to this genomic region by other transcription
factors such as Sp3. Therefore, certain genes are upregulated by
Sp3 alone and others are suppressed by Sp3 and Lsd1/Kdm1a, which
are possible targets of Sp3.
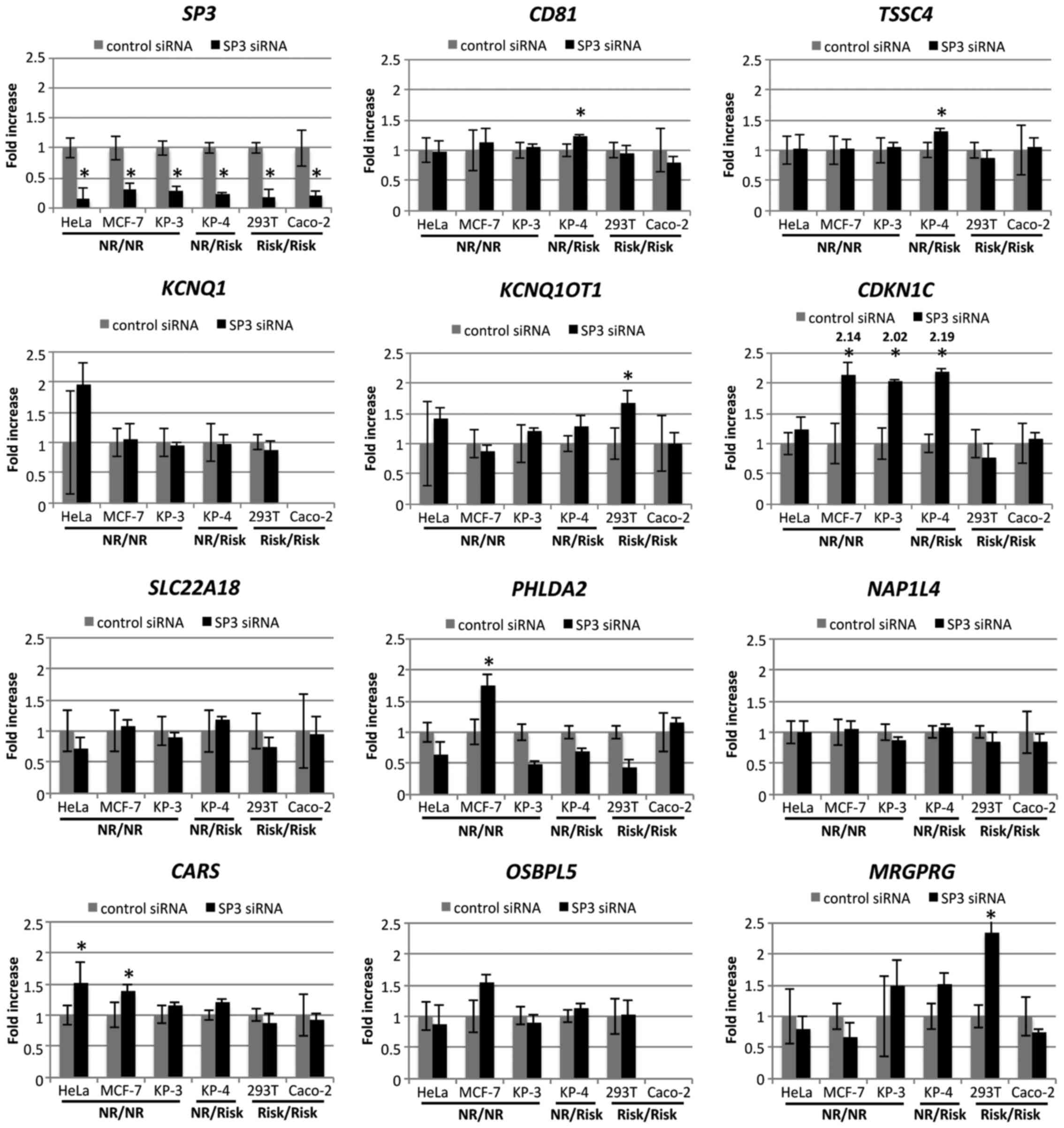
The expression levels of genes following SP3
knockdown, relative to cells transfected with control siRNA, are
presented in Fig. 6. The
downstream target gene was chosen depending on the following
criteria: i) Gene expression was upregulated or downregulated after
SP3 knockdown in the cell lines that possessed one or two
non-risk alleles (HeLa, MCF-7, KP-3 and KP-4); or ii) gene
expression was hardly changed after SP3 knockdown in the
cell lines that only possessed the risk allele (293T and Caco-2).
The expression of CDKN1C after SP3 knockdown was
significantly upregulated in MCF-7 (fold change, 2.14), KP-3 (fold
change, 2.02) and KP-4 (fold change, 2.19) cells. By contrast, the
expression of CDKN1C revealed little observable change in
293T and Caco-2 cells. None of the other genes in close proximity
that were examined fit these criteria, despite certain genes being
upregulated or downregulated depending on the particular cell
line.
These results indicate that the SNP rs163184 locus
exerts a negative impact on CDKN1C expression through
regulation by Sp3 and Lsd1/Kdm1a, suggesting that CDKN1C is
a downstream target gene of the SNP rs163184 locus.
Discussion
Previous genome-wide association studies have
indicated that SNPs in the KCNQ1 locus are associated with
development of T2DM (2–4). As the majority of these SNPs are
located in the intronic regions of KCNQ1 and are associated
with impaired insulin secretion, it was hypothesized that several
of these SNP-containing regions participate in regulating gene
expression in pancreatic β cells. These may include cis-regulatory
elements, such as enhancers; SNPs may also affect associated
activity at the KCNQ1 locus. In a previous comparative
analysis between alleles using DNA-immobilized nanobeads, our group
systematically identified allele-specific DNA-binding proteins in
several SNP regions (rs2237892, rs151290, rs2283228 and rs2074196)
(16). In addition, it was
demonstrated that the KCNQ1 SNP rs2074196 modulates the
binding activity of the locus for NF-Y, which may in turn induce
changes in gene expression (16).
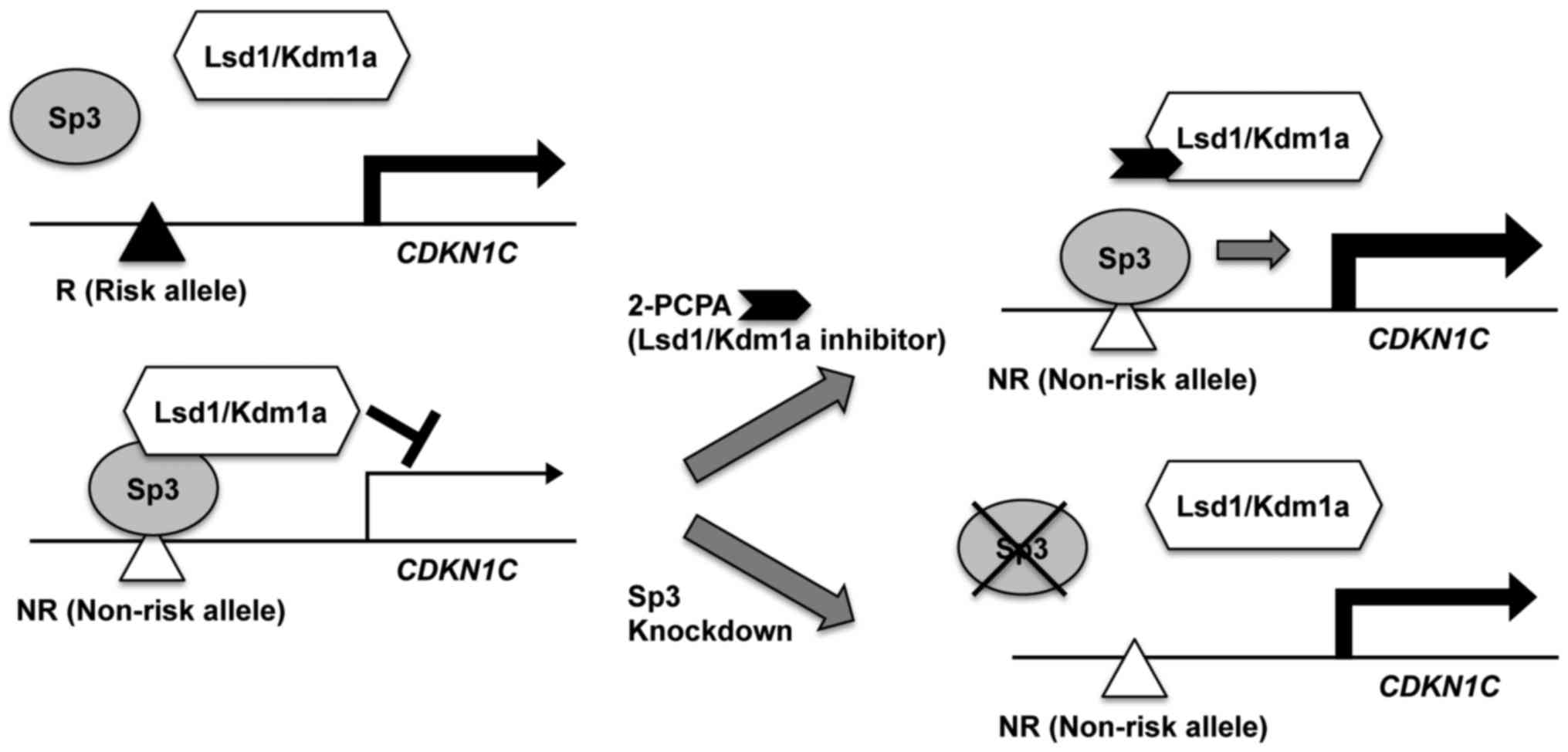
In the present study, it was indicated that the
function of the SNP rs163184 region was allele-specific and
context-sensitive (Fig. 7). It
was demonstrated that, for several proteins, binding activities for
the SNP rs163184 region in the KCNQ1 gene locus differed
between risk and non-risk alleles. Sp3 preferentially bound to the
non-risk allele of the KCNQ1 SNP rs163184 region and
stimulated transcriptional activity of an artificial promoter
containing the SNP rs163184 region. Lsd1/Kdm1a also preferentially
bound to the non-risk allele of the SNP rs163184 region indirectly,
possibly via the formation of complexes with other proteins, such
as Sp3. This suppressed the Sp3-dependent transcriptional activity
of the artificial promoter containing the SNP rs163184 region.
Furthermore, CDKN1C was indicated to be a functional target
gene regulated by Sp3, Lsd1/Kdm1a and the locus containing SNP
rs163184.
CDKN1C is a cyclin-dependent kinase inhibitor
and a negative regulator of cellular proliferation, which is
located in close proximity to KCNQ1 and SNP rs163184.
CDKN1C has been demonstrated to regulate pancreatic β cell
proliferation, and the loss of CDKN1C in focal
hyperinsulinism-associated lesions is associated with an increased
proliferation of pancreatic β cells (10). Additionally, targeting
CDKN1C promotes adult human β cell replication in
vitro (11). A recent study
has also indicated that a novel variant of CDKN1C is
associated with early-adulthood-onset diabetes, as well as
intrauterine growth restriction and a short stature (26). Therefore, overexpression of
CDKN1C may lead to reduced insulin production and the onset
of diabetes due to decreased proliferation of β cells during
development. In the present study, the risk-associated allele of
SNP rs163184 may have induced overexpression of CDKN1C
because of decreased binding activity for Sp3 and Lsd1/Kdm1a at
that locus. Therefore, it was hypothesized that SNP rs163184 in the
KCNQ1 locus increases T2DM susceptibility, at least in part,
through the modulation of CDKN1C expression.
Several mechanisms may contribute to the regulation
of CDKN1C gene expression. Firstly, the activity of the
CDKN1C promoter is regulated by the transcription factors
Sp1 and early growth response protein 1 (27,28). Secondly, the lncRNA
KCNQ1OT1 regulates the expression of CDKN1C via
imprinting (12–14). Finally, the present study revealed
that the KCNQ1 intron 15 region is also involved in
regulation of CDKN1C expression via the recruitment of Sp3
and Lsd1/Kdm1a. Lsd1/ Kdm1a is a component of the B-Raf
proto-oncogene serine/threonine kinase-HDAC repression complex,
which includes Hdac1, Hdac2, Rcor1, Phf21a and high mobility group
20B (29). This multimeric
complex regulates histone acetylation and methylation (29). In human breast cancer cells,
depletion of Lsd1/Kdm1a by siRNA enhances CDKN1C gene
expression (30). Therefore,
Lsd1/Kdm1a is a negative regulator of CDKN1C expression.
Although the factor that recruits Lsd1/Kdm1a to genomic regions
remains unknown, Sp3 and Phf21a are considered potential
candidates. Phf21a was reported to form a protein complex with
Lsd1/Kdm1a (29), although the
binding activity of Phf21a for this SNP region appears weak. Sp3
has a high binding activity for this SNP region and it was reported
that SUMOylation of Sp3 switched its function from an activator to
a repressor via an association with Lsd1/Kdm1a (31,32). Although the function of Sp3 in
pancreatic β cells is not well understood, several studies have
identified Sp3-regulating genes in pancreatic β cells, including
pancreatic and duodenal homeobox 1, glucagon receptor, potassium
voltage-gated channel subfamily J member 11 and gastric inhibitory
polypeptide receptor (33–36).
In addition to these observations, the present study revealed that
Sp3 and Lsd1/ Kdm1a regulated the expression of CDKN1C,
suggesting that Sp3 participates in the regulation of β cell
development and β cell function.
It is also necessary to consider several additional
points. Firstly, it is possible that other genomic regions in
KCNQ1 harboring intronic SNPs, such as rs2237892, rs2237895
and rs2074196, are also functional and contribute to T2DM
susceptibility. As our groups has previously revealed, SNP
rs2074196 modulates the binding activity of the locus for NF-Y and
possibly induces subsequent changes in gene expression (16). In the present study, our group has
also demonstrated a possible interaction between Sp3 and NF-Y. One
explanation for this is that the KCNQ1 SNP rs163184 and
KCNQ1 SNP rs2074196 regions recruit and interact with Sp3
and NF-Y, respectively, and that each have functional target genes.
Another possible interpretation is that CDKN1C expression
may be cooperatively regulated by the rs163184 and rs2074196 SNP
regions by recruiting and interacting with Sp3 and NF-Y together.
Furthermore, our group has already detected allele-specific binding
proteins for other KCNQ1 SNP regions, including rs2237892,
rs151290 and rs2283228, that are also highly associated with T2DM
(16). Secondly, one of the major
limitations of the present study is that the data examining
CDKN1C expression were obtained from human cell lines, which
were not derived from pancreatic β cells. For the purification of
proteins binding to the SNP rs163184 region, nuclear extracts
prepared from rat INS-1 cells were used, which have been widely
used for the study of islet biology. However, this cell line could
not be used for the analysis of the functional impact of the
intrinsic SNP region (ChIP assay and endogenous gene expression
analysis), as the sequence homology for the genomic region
including SNP rs163184 is very low between humans and rodents. In
addition, there are very few human islet cell lines established for
research. Thirdly, we have examined only genes close to
KCNQ1 as potential candidate targets. The ENCODE project has
revealed that there can be long-range interactions between gene
promoters and disease-associated variants (7). We cannot, therefore, exclude the
possibility that remote genes may also be functional targets.
Further experiments employing each allele-specific binding protein
to each T2DM-associated KCNQ1 SNPs and examining the
interactions between these SNPs using chromosome conformation
capture assays should clarify any molecular relationship between
KCNQ1 SNPs and T2DM susceptibility.
Although recent genome-wide association studies
have been extremely successful, it remains a challenge to
functionally annotate SNPs that localize to non-coding regions in
susceptibility genes. In order to understand the functional impact
of individual disease-associated SNPs, it is essential to identify
DNA-binding proteins that recognize the nucleotide sequence
surrounding these disease-associated variants. The comparative
analysis conducted in the present study between alleles using
DNA-immobilized nanobeads has proven to be a useful and powerful
tool that can be used to investigate the molecular mechanisms that
link non-coding genetic variants to the risks of developing common
diseases.
Acknowledgments
The authors would like to thank Dr Wataru
Nishimura, Dr Miho Kawaguchi, Mr. Dai Suzuki, Ms. Kazuko Nagase and
Ms. Keiko Kano (National Center for Global Health and Medicine,
Tokyo, Japan) for their technical assistance. The present study was
supported by the Japan Society for the Promotion of Science
Grants-in-Aid for Scientific Research (grant no. 22510216; to
M.H.), the Japan Diabetes Foundation (to M.H.), Suzuken Memorial
Foundation (to M.H.), the Leading Project of Ministry of Education,
Culture, Sports, Science and Technology, Japan (to K.Y.), National
Institute of Biomedical Innovation (to K.Y.) and National Center
for Global Health and Medicine (to K.Y.).
Abbreviations:
|
SNP
|
single nucleotide polymorphism
|
|
T2DM
|
type 2 diabetes mellitus
|
|
KCNQ1
|
potassium voltage-gated channel
subfamily Q member 1
|
|
Lsd1/Kdm1a
|
lysine-specific histone demethylase
1A
|
|
Sp3
|
transcription factor Sp3
|
|
CDKN1C
|
cyclin-dependent kinase inhibitor
1C
|
References
|
1
|
Imamura M and Maeda S: Genetics of type 2
diabetes: The GWAS era and future perspectives (Review). Endocr J.
58:723–739. 2011. View Article : Google Scholar
|
|
2
|
Yasuda K, Miyake K, Horikawa Y, Hara K,
Osawa H, Furuta H, Hirota Y, Mori H, Jonsson A, Sato Y, et al:
Variants in KCNQ1 are associated with susceptibility to type 2
diabetes mellitus. Nat Genet. 40:1092–1097. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Unoki H, Takahashi A, Kawaguchi T, Hara K,
Horikoshi M, Andersen G, Ng DP, Holmkvist J, Borch-Johnsen K,
Jørgensen T, et al: SNPs in KCNQ1 are associated with
susceptibility to type 2 diabetes in East Asian and European
populations. Nat Genet. 40:1098–1102. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Voight BF, Scott LJ, Steinthorsdottir V,
Morris AP, Dina C, Welch RP, Zeggini E, Huth C, Aulchenko YS,
Thorleifsson G, et al: MAGIC investigators; GIANT Consortium:
Twelve type 2 diabetes susceptibility loci identified through
large-scale association analysis. Nat Genet. 42:579–589. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yamagata K, Senokuchi T, Lu M, Takemoto M,
Fazlul Karim M, Go C, Sato Y, Hatta M, Yoshizawa T, Araki E, et al:
Voltage-gated K+ channel KCNQ1 regulates insulin
secretion in MIN6 β-cell line. Biochem Biophys Res Commun.
407:620–625. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sanyal A, Lajoie BR, Jain G and Dekker J:
The long-range interaction landscape of gene promoters. Nature.
489:109–113. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Maurano MT, Humbert R, Rynes E, Thurman
RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, et
al: Systematic localization of common disease-associated variation
in regulatory DNA. Science. 337:1190–1195. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Smemo S, Tena JJ, Kim KH, Gamazon ER,
Sakabe NJ, Gómez-Marín C, Aneas I, Credidio FL, Sobreira DR,
Wasserman NF, et al: Obesity-associated variants within FTO form
long-range functional connections wit IRX3. Nature. 507:371–375.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pasquali L, Gaulton KJ, Rodríguez-Seguí
SA, Mularoni L, Miguel-Escalada I, Akerman İ, Tena JJ, Morá I,
Gómez-Marín C and van de Bunt M: Pancreatic islet enhancer clusters
enriched in type 2 diabetes risk-associated variants. Nat Genet.
46:136–143. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kassem SA, Ariel I, Thornton PS, Hussain
K, Smith V, Lindley KJ, Aynsley-Green A and Glaser B: 57 (KIP2)
expression in normal islet cells and in hyperinsulinism of infancy.
Diabetes. 50:2763–2769. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Avrahami D, Li C, Yu M, Jiao Y, Zhang J,
Naji A, Ziaie S, Glaser B and Kaestner KH: Targeting the cell cycle
inhibitor 57Kip2 promotes adult human β cell replication. J Clin
Invest. 124:670–674. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Horike S, Mitsuya K, Meguro M, Kotobuki N,
Kashiwagi A, Notsu T, Schulz TC, Shirayoshi Y and Oshimura M:
Targeted disruption of the human LIT1 locus defines a putative
imprinting control element playing an essential role in
Beckwith-Wiedemann syndrome. Hum Mol Genet. 9:2075–2083. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fitzpatrick GV, Soloway PD and Higgins MJ:
Regional loss of imprinting and growth deficiency in mice with a
targeted deletion of KvDMR1. Nat Genet. 32:426–431. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Arima T, Kamikihara T, Hayashida T, Kato
K, Inoue T, Shirayoshi Y, Oshimura M, Soejima H, Mukai T and Wake
N: ZAC, LIT1 (KCNQ1OT1) and 57KIP2 (CDKN1C) are in an imprinted
gene network that may play a role in Beckwith-Wiedemann syndrome.
Nucleic Acids Res. 33:2650–2660. 2005. View Article : Google Scholar :
|
|
15
|
Asahara S, Etoh H, Inoue H, Teruyama K,
Shibutani Y, Ihara Y, Kawada Y, Bartolome A, Hashimoto N, Matsuda
T, et al: Paternal allelic mutation at the Kcnq1 locus reduces
pancreatic β-cell mass by epigenetic modification oCdkn1c. Proc
Natl Acad Sci USA. 112:8332–8337. 2015. View Article : Google Scholar
|
|
16
|
Hiramoto M, Udagawa H, Watanabe A,
Miyazawa K, Ishibashi N, Kawaguchi M, Uebanso T, Nishimura W, Nammo
T and Yasuda K: Comparative analysis of type 2 diabetes-associated
SNP alleles identifies allele-specific DNA-binding proteins for the
KCNQ1 locus. Int J Mol Med. 36:222–230. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dunham I, Kundaje A, Aldred SF, Collins
PJ, Davis CA, Doyle F, Epstein CB, Frietze S, Harrow J, Kaul R, et
al: ENCODE Project Consortium: An integrated encyclopedia of DNA
elements in the human genome. Nature. 489:57–74. 2012. View Article : Google Scholar
|
|
18
|
Bhaumik SR, Smith E and Shilatifard A:
Covalent modifications of histones during development and disease
pathogenesis. Nat Struct Mol Biol. 14:1008–1016. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Asfari M, Janjic D, Meda P, Li G, Halban
PA and Wollheim CB: Establishment of 2-mercaptoethanol-dependent
differentiated insulin-secreting cell lines. Endocrinology.
130:167–178. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dignam JD, Lebovitz RM and Roeder RG:
Accurate transcription initiation by RNA polymerase II in a soluble
extract from isolated mammalian nuclei. Nucleic Acids Res.
11:1475–1489. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shevchenko A, Wilm M, Vorm O and Mann M:
Mass spectrometric sequencing of proteins silver-stained
polyacrylamide gels. Anal Chem. 68:850–858. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hiramoto M, Maekawa N, Kuge T, Ayabe F,
Watanabe A, Masaike Y, Hatakeyama M, Handa H and Imai T:
High-performance affinity chromatography method for identification
of L-arginine interacting factors using magnetic nanobeads. Biomed
Chromatogr. 24:606–612. 2010.
|
|
23
|
Takahashi E, Okumura A, Unoki-Kubota H,
Hirano H, Kasuga M and Kaburagi Y: Differential proteome analysis
of serum proteins associated with the development of type 2
diabetes mellitus in the KK-Ay mouse model using the
iTRAQ technique. J Proteomics. 84:40–51. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kersey PJ, Duarte J, Williams A,
Karavidopoulou Y, Birney E and Apweiler R: The International
Protein Index: An integrated database for proteomics experiments.
Proteomics. 4:1985–1988. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
26
|
Kerns SL, Guevara-Aguirre J, Andrew S,
Geng J, Guevara C, Guevara-Aguirre M, Guo M, Oddoux C, Shen Y,
Zurita A, et al: A novel variant in CDKN1C is associated with
intrauterine growth restriction, short stature, and
early-adulthood-onset diabetes. J Clin Endocrinol Metab.
99:E2117–E2122. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cucciolla V, Borriello A, Criscuolo M,
Sinisi AA, Bencivenga D, Tramontano A, Scudieri AC, Oliva A, Zappia
V and Della Ragione F: Histone deacetylase inhibitors upregulate
57Kip2 level by enhancing its expression through Sp1 transcription
factor. Carcinogenesis. 29:560–567. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Figliola R, Busanello A, Vaccarello G and
Maione R: Regulation of 57(KIP2) during muscle differentiation:
Role of Egr1, Sp1 and DNA hypomethylation. J Mol Biol. 380:265–277.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shi YJ, Matson C, Lan F, Iwase S, Baba T
and Shi Y: Regulation of LSD1 histone demethylase activity by its
associated factors. Mol Cell. 19:857–864. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Vasilatos SN, Katz TA, Oesterreich S, Wan
Y, Davidson NE and Huang Y: Crosstalk between lysine-specific
demethylase 1 (LSD1) and histone deacetylases mediates
antineoplastic efficacy of HDAC inhibitors in human breast cancer
cells. Carcinogenesis. 34:1196–1207. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ross S, Best JL, Zon LI and Gill G: SUMO-1
modification represses Sp3 transcriptional activation and modulates
its subnuclear localization. Mol Cell. 10:831–842. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rosendorff A, Sakakibara S, Lu S, Kieff E,
Xuan Y, DiBacco A, Shi Y, Shi Y and Gill GX: P-2 association with
SUMO-2 depends on lysines required for transcriptional repression.
Proc Natl Acad Sci USA. 103:5308–5313. 2006. View Article : Google Scholar
|
|
33
|
Ben-Shushan E, Marshak S, Shoshkes M,
Cerasi E and Melloul D: A pancreatic beta-cell-specific enhancer in
the human PDX-1 gene is regulated by hepatocyte nuclear factor
3beta (HNF-3beta), HNF-1alpha, and SPs transcription factors. J
Biol Chem. 276:17533–17540. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Geiger A, Salazar G and Kervran A: Role of
the Sp family of transcription factors on glucagon receptor gene
expression. Biochem Biophys Res Commun. 285:838–844. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hashimoto T, Nakamura T, Maegawa H, Nishio
Y, Egawa K and Kashiwagi A: Regulation of ATP-sensitive potassium
channel subunit Kir6.2 expression in rat intestinal
insulin-producing progenitor cells. J Biol Chem. 280:1893–1900.
2005. View Article : Google Scholar
|
|
36
|
Baldacchino V, Oble S, Décarie PO,
Bourdeau I, Hamet P, Tremblay J and Lacroix A: The Sp transcription
factors are involved in the cellular expression of the human
glucose-dependent insulinotropic polypeptide receptor gene and
overexpressed in adrenals of patients with Cushing's syndrome. J
Mol Endocrinol. 35:61–71. 2005. View Article : Google Scholar : PubMed/NCBI
|