Introduction
Colorectal cancer (CRC) is one of the most common
malignancies in the world and a major cause of mortality (1). At the time of diagnosis, ~50% of
patients presented with liver metastasis, with 20–25% of these
presenting with synchronous liver metastases (2). Combinations of chemotherapy and
biological therapy have improved disease-free survival for patients
with metastatic CRC (3). Lesions
that underwent curative resection have demonstrated 5-year survival
exceeding 50% (4). However, there
are still disappointingly high rates of local recurrence or distant
metastasis after curative surgical resection (5). Thus, the early detection of liver
metastasis becomes increasingly desirable for patient survival. The
cancer stem cell (CSC) theory presumes that solid tumor initiation,
maintenance, and progression are the result of a small population
of cancer cells with self-renewal and pluripotency capabilities
(6). This population of cells is
often considered to be associated with chemoresistance and
radioresistance that lead to the failure of traditional therapy
(7,8). A better understanding of CSCs will
allow us to detect remote metastasis earlier, target CSC
subpopulations and potentially eradicate tumors (5,9).
Identifying the molecular markers of a metastatic phenotype is a
prerequisite for developing new therapeutic approaches in CRC
(10,11).
CD44, a transmembrane glycoprotein that serves as a
recyclable receptor for hyaluronan, plays important roles in
regulating cell adhesion, proliferation, growth, survival,
motility, migration, angiogenesis and differentiation (12,13).
The CD44 marker can be used to isolate CSC populations from
colorectal tumors (14).
Furthermore, CD44 has been demonstrated to perform an unexpected
tumor-progressing function and lead to tumor metastasis, resulting
in worse prognosis (15).
CD133 (also known as Prominin-1 or AC133), a cell
surface transmembrane glycoprotein, was identified in
subpopulations of cells in colon tumors (16). Subpopulations of CD133-positive
colon cancer cells have shown increased tumorigenic potential in
transplantation studies in vitro and in vivo
(16–18). Moreover, high expression of CD133
is associated with poor prognosis, resistance to chemotherapy
(19,20) and radiotherapy (21) and even distant metastasis (22,23).
Recently, some studies have focused on CD44 and CD133
co-expression. Cells from peritoneal washes of metastatic colon
cancer patients or the metastatic colon cancer HCT116 cell line
positively express CD44 and CD133 (24). CD133+CD44+
cells are undifferentiated, with the abilities of extensive
self-renewal and epithelial lineage differentiation in vitro
(13,25,26).
Moreover, long-term cultured CD133+CD44+
cells can enrich a CD133+CD44high
subpopulation of cells that express epithelial to mesenchymal
transition marker, are more invasive in vitro, and are
responsible for liver metastasis in vivo (26). Other studies revealed that CD44 and
CD133 co-expression is significantly higher in colon cancers with
early liver metastases than in those without early liver metastases
(4,5), and overall survival of patients who
are positive for both CD44 and CD133 is significantly shorter than
that of all other patients (27).
A recent study directly verified the momentous role of
CD133+CD44+ tumor cells in hematogenous
metastasis of liver cancers, suggesting that CD133 is important for
tumor growth and CD44 is responsible for invasion. CD133 and CD44
can be regarded as cooperative markers of liver
metastasis-facilitating pathways (28). In another study, Bellizzi et
al (29) generated stem cell
enriched human colonosphere cultures from fresh samples and
compared the sphere-forming potential of these clones.
CD133+ colon cancer cells were confirmed to play a
crucial role both in primary tumors and in metastatic lesions, but
liver metastasis seemed to be strictly related to
CD133+CD44+.
CD44 and CD133 appear to be useful markers for
isolation and further characterization of colorectal CSCs. However,
the relationships among expression of colon CSC markers, CRC
metastasis, and clinicopathological features are still unknown.
Moreover, there have been few studies focusing on mRNA expression
using simple and practical methods such as reverse transcription
polymerase chain reaction (RT-PCR). The aim of the present study
was to examine CD44 and CD133 mRNA expression in CRC and analyze
the potential correlation with hepatic metastasis and
clinicopathological factors.
Materials and methods
Tissue samples
Tissue samples were obtained from 36 patients
diagnosed with primary CRC with synchronous hepatic metastasis from
2004 to 2011 in the Department of Colorectal Surgery, Chonnam
National University Hwasun Hospital, South Korea.
RNA extraction
One of the successive tissue slides was stained with
hematoxylin and eosin, and two pathologists measured and circled
the areas of normal mucosa, CRC, and hepatic metastasis to indicate
the associated normal or tumor tissue area. Other slides were used
for RNA isolation after removing irrelevant tissue. The slides were
dewaxed in xylene, rinsed in a graded ethanol series, and finally
rehydrated in double-distilled water. The tissue was scraped into a
1.5-ml microcentrifuge tube. The total RNA was isolated using
TRIzol® reagent (Invitrogen, Carlsbad, CA, USA)
according to the manufacturer’s protocol. After 10-min incubation
at room temperature, RNA was extracted by the addition of 200 μl
chloroform (Sigma, St. Louis, MO, USA). The tube was lightly shaken
for 15 sec and then incubated for 10 min. The phases were separated
automatically, followed by a 15-min centrifugation at 13,000 rpm in
a table-top centrifuge at 4°C. The aqueous phase was transferred to
a fresh tube, and an equal volume of isopropanol (Fisher
Scientific, Seoul, Korea) was added. The mixture was incubated at
room temperature for 10 min to precipitate RNA. After another
15-min centrifugation at 13,000 rpm and 4°C, the supernatant was
carefully discarded. One milliliter of 80% ethanol was added to the
tube, which was then centrifuged at 4°C for 5 min at 7,500 rpm.
After the liquid had been discarded, the content of the tube was
dried briefly for ~15 min at room temperature without
centrifugation. The RNA pellet was dissolved in 15 μl of
diethylpyrocarbonate-treated water (Biosesang, Seongnam, Korea),
and the RNA was quantified using a NanoDrop™ ND-1000
spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA) at
260 nm.
cDNA synthesis
Reverse transcription was performed using the
Moloney murine leukemia virus reverse transcriptase (M-MLV RT;
Beams Biotech, Seongnam, Korea). Each total RNA template (2 μg) was
reverse transcribed with mixture of 0.25 μg oligo(dT) primer and
0.5 μg random primers (Promega, Seoul, Korea). The sample tube was
heated to 72°C for 10 min, chilled rapidly on ice, and then spun
briefly to collect the solution at the bottom of the tube. Then 0.5
μl ribonuclease inhibitor (40 U/μl; Beams Biotech), 1X M-MLV
reaction buffer, 10 mM dNTPs, and 0.5 μl M-MLV RT (200 U/μl; Beams
Biotech) were added, and the mixture was incubated for 70 min at
42°C and 10 min at 72°C.
RT-PCR
RT-PCR was performed using nTaq-HOT DNA polymerase
(Enzynomics, Seoul, Korea). Oligonucleotide primers specific for
CD44, CD133 and GAPDH were designed from their GenBank sequences
(AJ251595.1, NM_006017.2 and NM_002046.3, respectively). The GAPDH
gene was used as an internal control. The PCR primer sequences are
listed in Table I. The RT-PCR
products were electrophoresed through a 4% agarose gel at 100 V for
25 min in 1X TBE buffer. After being stained with ethidium bromide
(5 mg/ml), the gels were photographed under ultraviolet light. The
RT-PCR products were quantified using an Image Reader (Mulit
Gauge2.0 Analytical Software; FujiFilm, Tokyo, Japan).
 | Table IPrimer sequences used for RT-PCR. |
Table I
Primer sequences used for RT-PCR.
| Primer (5′-3′,
forward/reverse) | PCR
conditionsa | Product size
(bp) |
|---|
| CD44 |
CTGCAGGTATGGGTTCATAG/ATATGTGTCATACTGGGAGGTG | 60 (35) | 124 |
| CD133 |
GATTAAGTCCATGGCAACAGCG/GCTGGTCAGACTGCTGCTAAGC | 60 (35) | 115 |
| GAPDH |
ACGGGAAGCTTGTCATCAATGG/ATGGTGGTGAAGACGCCAGTGG | 60 (35) | 124 |
Immunohistochemistry
Immunohistochemical staining was performed on
specimens that showed a typical mRNA expression pattern. Sections
of formalin-fixed, paraffin-embedded tissues of CRC, synchronous
hepatic metastasis tissue, and the adjacent normal mucosa were
sliced into 3-μm-thick sections and mounted on silane-coated glass
slides. The slides were dewaxed in xylene, and rehydrated in a
graded ethanol series followed by double-distilled water. For
antigen retrieval, slides for CD44 and CD133 were treated by
pressure cooker for 15 min in sodium citrate buffer (10 mM, pH 6.0;
Sigma-Aldrich, Bangalore, India) and retrieval buffer (pH 9.0;
Dako, Carpinteria, CA, USA), respectively. Specimens were rinsed
and blocked with 3% H2O2 for 10 min to
abolish endogenous peroxidase activity. To reduce non-specific
background staining, 5% normal goat serum was placed on the
sections for 1 h at room temperature, and then drained off.
Finally, the sections were incubated overnight at 4°C with the CD44
monoclonal antibody (Cell Signaling Technology, Danvers, MA, USA)
and AC133 monoclonal antibody (Miltenyi Biotec GmbH, Bergisch
Gladbach, Germany), respectively, both at a dilution of 1:100. The
slides were washed three times with 1X TBS-T, followed by
incubations for 1 h with HRP (Cell Signaling Technologies) and
polymer HRP anti-rabbit and mouse IgG (Life Technologies, Grand
Island, NY, USA), respectively. Immunostaining was visualized using
3,3′-diami-nobenzidine (DAB) (Dako). All the slides were
counterstained with hematoxylin, dehydrated and mounted. The
negative control was performed by incubating samples with 1X TBS-T
(data not shown). All the sections were independently examined for
their protein expressions and assessed by comparing the staining
among the normal mucosa, CRC, and hepatic metastasis regions under
microscopic examination. All the histological slides were examined
by two experienced gastrointestinal pathologists who were unaware
of the clinical data or disease outcomes.
Statistical analysis
All statistical analyses were performed with SPSS
version 17.0 (SPSS Inc., Chicago, IL, USA). The associations
between the mRNA expression of CD44 and CD133 and the
clinicopathological parameters were examined using the Chi-squared
test. To evaluate the associations with disease-free survival, the
median normalized CD44 and CD133 value (metastasis/normal, 1.25 and
1.22, respectively) was chosen as the cut-off point for
discriminating the 36 patients into two subgroups (low vs. high).
The Kaplan-Meier method was used to estimate survival as a function
of time, and survival differences were assessed with the log-rank
test. A value of P<0.05 was considered statistically
significant.
Results
Typical results of gene expression by reverse
transcriptase polymerase chain reaction are shown in Fig. 1. The mean ± SD intensities of CD44
gene expression with ratio of GAPDH in enrolled 36 samples were
0.5443±0.1955 in colorectal tumor, 0.6226±0.2037 in hepatic
metastasis tissue, and 0.5081±0.1882 in adjacent normal tissue. For
CD133 gene expression, mean intensities were 0.5889±0.1564 in
colorectal tumor, 0.6354±0.1696 in hepatic metastasis tissue and
0.5396±0.1356 in adjacent normal tissue.
mRNA expression
The CD44 expression in the groups was statistically
different between normal tissue and colorectal tumor (P=0.008),
between normal tissue and hepatic metastasis tissue (P<0.001)
and between colorectal tumor and hepatic metastasis tissue
(P<0.001). CD133 expression was also significantly different
between normal tissue and colorectal tumor (P=0.024), between
normal tissue and hepatic metastasis tissue (P<0.001), and
between colorectal tumor and hepatic metastasis tissue (P=0.040)
(Fig. 2).
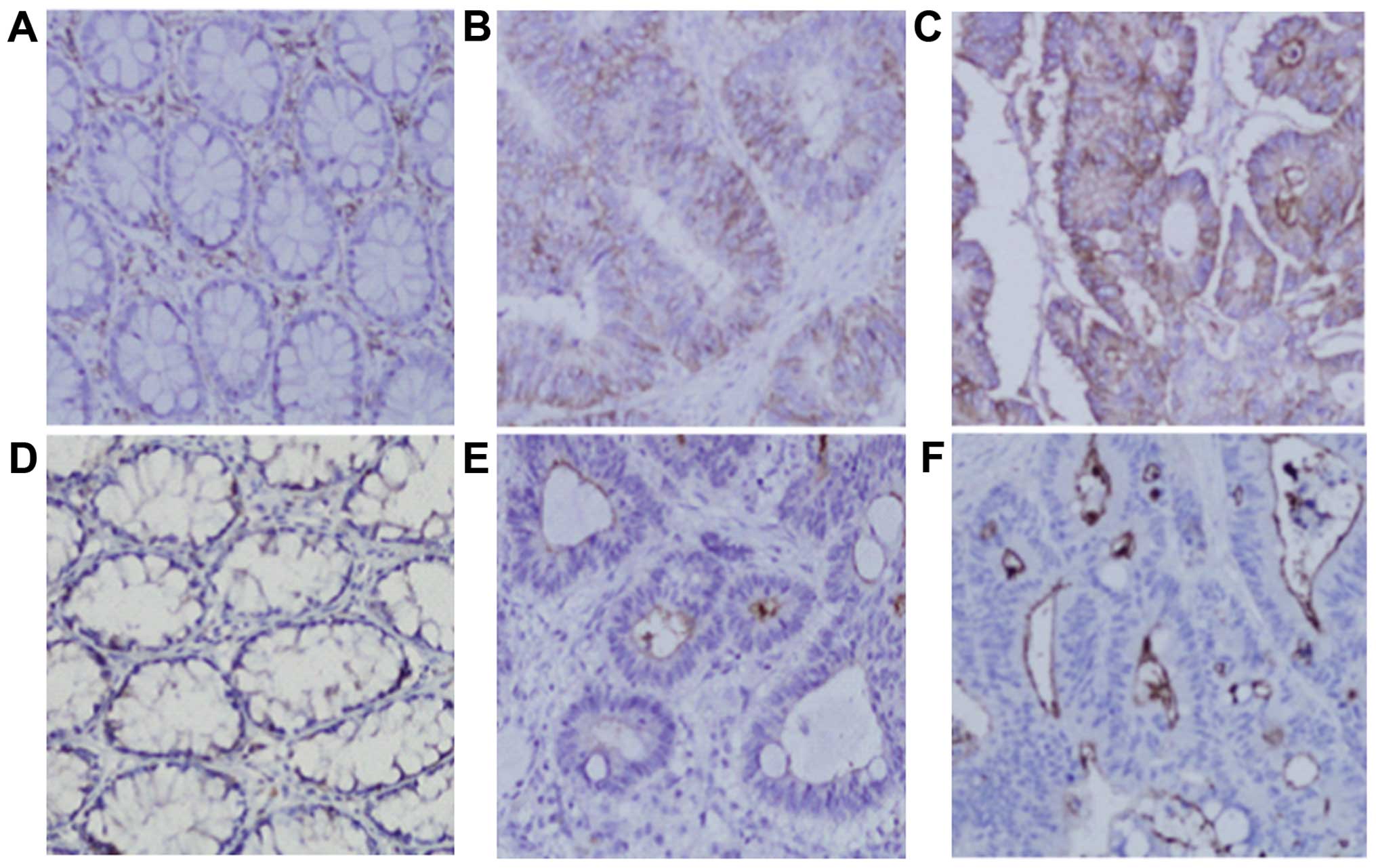
Immunohistochemistry
To investigate expression of CD44 and CD133
proteins, immunohistochemical analysis was performed on samples
with typical mRNA expression patterns. Consistent with previous
reports (22,30–32),
the CD44 and CD133 immunoreactivities were visualized as brown
membrane and lumina surface staining, respectively (Fig. 3). Hepatic metastasis tissue
exhibited the highest expression of CD44 and CD133 proteins,
followed by CRC and normal mucosa, consistent with previous
reports.
Clincopathological parameters
This analysis included tissue samples from 21 (58%)
male patients and 15 (42%) female patients with a median age of 66
years (range, 42–91 years). Twenty-one (58%) tumors were located in
the colon, and 15 (42%) were in the rectum. The median tumor size
was 4.6 cm (range, 1.5–8.5 cm). The relations between CD44 and
CD133 mRNA expression and the clinicopathological factors are shown
in Table II. Tumors in colon and
poorly differentiated tumors showed high CD44 mRNA expression with
statistical significance. However, there were no significant
differences in CD44 and CD133 mRNA expression according to age,
gender, tumor size, depth of invasion, lymphovascular invasion,
perineural invasion or preoperative CEA level.
 | Table IICorrelation between mRNA expression
of CD44 and CD133 and the clinicopathological parameters of
colorectal cancer metastasis. |
Table II
Correlation between mRNA expression
of CD44 and CD133 and the clinicopathological parameters of
colorectal cancer metastasis.
| | CD44
expression | | CD133
expression | |
|---|
| |
| |
| |
|---|
| Parameters | No. | Low (n=20) | High (n=16) | P-value | Low (n=27) | High (n=9) | P-value |
|---|
| Age (years) |
| ≤60 | 15 | 7 | 8 | 0.500 | 11 | 4 | 1.000 |
| >60 | 21 | 13 | 8 | | 16 | 5 | |
| Gender |
| Male | 21 | 11 | 10 | 0.741 | 16 | 5 | 1.000 |
| Female | 15 | 9 | 6 | | 11 | 4 | |
| Tumor location |
| Colon | 21 | 8 | 13 | 0.019 | 16 | 5 | 1.000 |
| Rectum | 15 | 13 | 3 | | 11 | 4 | |
| Histology |
| Well +
moderate | 20 | 15 | 5 | 0.026 | 17 | 3 | 0.202 |
| Poor | 15 | 5 | 10 | | 9 | 6 | |
| Tumor size |
| ≤5 | 21 | 13 | 8 | 0.500 | 14 | 7 | 0.252 |
| >5 | 15 | 7 | 8 | | 13 | 2 | |
| Depth of invasion
(T) |
| T1+T2 | 32 | 17 | 15 | 0.613 | 24 | 8 | 1.000 |
| T3+T4 | 4 | 3 | 1 | | 3 | 1 | |
| Lymphovascular
invasion |
| Negative | 28 | 17 | 11 | 0.422 | 20 | 8 | 0.648 |
| Positive | 8 | 3 | 5 | | 7 | 1 | |
| Perineural
invasion |
| Negative | 22 | 13 | 9 | 0.734 | 17 | 5 | 0.712 |
| Positive | 14 | 7 | 7 | | 10 | 4 | |
| Preoperative CEA
(ng/ml) |
| ≤5 | 17 | 10 | 7 | 0.749 | 11 | 6 | 0.255 |
| >5 | 19 | 10 | 9 | | 16 | 3 | |
Prognosis
With a median follow-up period of 38 months, the
5-year disease-free survival of the patients with high CD44 mRNA
expression in CD44 hepatic metastasis tissue was significantly
lower than that of the patients with low expression (P=0.002;
Fig. 4). The mRNA expressions of
CD44 in colorectal tumor, CD133 in colorectal tumor, and CD133 in
hepatic metastasis tissue were not significantly associated with
survival.
Discussion
The CSC theory has significant implications in
cancer therapy. CSCs, which are characterized by a slow cell cycle,
have been reported to be resistant to anticancer therapies by
intrinsic defense mechanisms, such as quiescence, efflux pumps, and
detoxifying enzymes, and by induction of anti-apoptotic proteins
(5,33,34).
The result is that conventional cytotoxic therapies initially
shrink the bulk of a tumor but fail to eradicate it, resulting in
tumor recurrence, because current chemotherapeutics interfere with
the ability of rapidly growing cells and, therefore, might spare
CSCs (35,36). Treatment approaches that target
CSCs may increase the efficacy of current treatment regimens and
reduce the risk of tumor relapse and metastasis (37).
One of the major findings of the present study is
that CD44 and CD133 mRNA were more highly co-expressed in hepatic
metastases tissue than in CRC and normal tissue, which is
consistent with other evidence, such as protein expression
(11), suggesting that CD44 and
CD133 might be potential biomarkers for predicting liver metastases
of CRC. Even though the underlying mechanism of CD44 and CD133
co-expression is unknown, studies of their co-expression in protein
and mRNA levels are reliable (4,26,29).
Moreover, one study demonstrated the critical role of
CD133+CD44+ tumor cells in hematogenous
metastasis of liver cancers, suggesting that CD133 is responsible
for tumor growth and CD44 is important for invasion, two important
factors in tumor metastasis (28).
CD44 mRNA expression in the liver metastasis tissue
group showed statistically significant association with colon
tumors (P=0.019) and poorly differentiated tumors (P=0.026), but
not with rectal tumors and well and moderately differentiated
tumors. The difference in results from colon and rectal tumors
might be due to embryologic, morphologic, physiologic,
histochemical and biological differences between colon cancer and
rectal cancer (38). The
difference in results between poorly differentiated and well and
moderately differentiated tumors is inconsistent with reports that
poorly differentiated CRC is responsible for more extensive
invasiveness (39) and poorer
prognosis (40), and might be
associated with more frequent microsatellite instability, a cause
of genomic instability (41).
The evidence regarding the use of currently known
colon CSC markers as prognostic indicators is contradictory. In the
present study, disease-free survival of the patients with high CD44
mRNA expression in the liver metastasis tissue group was
significantly lower than that of patients with low expression, but
the CRC groups did not show a similar pattern. Expression of CD44
is associated with worse outcome for CRC patients. Mulder et
al (42) reported that
increased CD44 expression is associated with increased
tumor-related death. However, some studies failed to detect a
relationship between CD44 expression and patient outcome or tumor
progression (22,43). Notably, less membranous CD44 has
been linked to worse survival and remote metastasis in other
studies (11,44).
For CD133, Horst et al (45) found that increased CD133 expression
assessed by immunohistochemistry is associated with worse outcome.
They also compared the prognostic value of colon CSC markers CD133,
CD44 and CD166 in CRC. CD133 turned out to be the best prognostic
marker and indicated significantly worse prognosis (22,27).
In another study, CD133 expression was associated with worse
outcome when comparing well and moderately differentiated cancer
(46). In addition, CD133
expression increased in stage IIIB colon cancer samples, indicative
of worse outcome (47). In
contrast, Choi et al (48)
discovered that none of the colon CSC markers CD133, CD44 or CD24
were significant prognostic predictors of survival. Furthermore,
Lugli et al (44), using
tissue microarrays, failed to demonstrate an association between
CD133 expression and tumor progression or patient survival.
In the present study, consistent with previous
results, we found that CD133 mRNA expression in colorectal tumor
and liver metastasis tissue was not significantly associated with
disease-free survival. There are large disparities in the reported
prognostic potential of colon CSC markers. The different results
among the groups could be explained by the varied methods and
cut-offs used in tissue staining and scoring (49,50).
Moreover, this study had some limitations. First, the number of
samples in this study was not large, allowing the possibility of a
type II error. Second, CD44 and CD133 mRNA expression level was
analyzed by RT-PCR with 35 cycles, which may be slightly too many
cycles, since formalin-fixed, paraffin-embedded tumor specimens
were used (49). Galizia et
al (5) demonstrated the
critical role of CD44 and CD133 co-expression by
fluorescence-activated cell sorting, which is a complex and
time-consuming method. In the present study, we used much simpler
techniques (RT-PCR and immunohistochemistry), which might be
clinically promising to perform. Although our findings were
insufficient to draw definite conclusions, further investigations
with more samples may demonstrate associations between expression
of the CSC markers CD44 and CD133, liver metastasis and
clinicopathological parameters.
In conclusion, CD44 and CD133 mRNA were highly
co-expressed in CRC with hepatic metastases. CD44 expression was an
independent factor associated with patient survival, while CD133
did not show this pattern. Thus, CD44 is a more reliable marker for
predicting hepatic metastases and survival. Further prospective
controlled studies with larger numbers are warranted to support
these findings.
Acknowledgements
The present study was supported by Chonnam National
University, 2012–2684 and Research Grant 0720570 from the National
Cancer Center and by a Grant from the Chonnam National Univeristy
Biomedical Research Institute (CRI 13042-21, 22), South Korea.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar
|
|
2
|
Sakai Y: Current topics in colorectal
liver metastasis. Int J Clin Oncol. 16:451. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Van Cutsem E, Lambrechts D, Prenen H, Jain
RK and Carmeliet P: Lessons from the adjuvant bevacizumab trial on
colon cancer: what next? J Clin Oncol. 29:1–4. 2011. View Article : Google Scholar
|
|
4
|
Huang X, Sheng Y and Guan M: Co-expression
of stem cell genes CD133 and CD44 in colorectal cancers with early
liver metastasis. Surg Oncol. 21:103–107. 2012. View Article : Google Scholar
|
|
5
|
Galizia G, Gemei M, Del Vecchio L, et al:
Combined CD133/CD44 expression as a prognostic indicator of
disease-free survival in patients with colorectal cancer. Arch
Surg. 147:18–24. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Anderson EC, Hessman C, Levin TG, Monroe
MM and Wong MH: The role of colorectal cancer stem cells in
metastatic disease and therapeutic response. Cancers (Basel).
3:319–339. 2011. View Article : Google Scholar :
|
|
7
|
Hu Y and Fu L: Targeting cancer stem
cells: a new therapy to cure cancer patients. Am J Cancer Res.
2:340–356. 2012.PubMed/NCBI
|
|
8
|
Yu ZR, Pestell TG, Lisanti MP and Pestell
RG: Cancer stem cells. Int J Biochem Cell Biol. 44:2144–2151. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Soltanian S and Matin MM: Cancer stem
cells and cancer therapy. Tumour Biol. 32:425–440. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wakamatsu Y, Sakamoto N, Oo HZ, Naito Y,
Uraoka N, Anami K, Sentani K, Oue N and Yasui W: Expression of
cancer stem cell markers ALDH1, CD44 and CD133 in primary tumor and
lymph node metastasis of gastric cancer. Pathol Int. 62:112–119.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bendardaf R, Algars A, Elzagheid A,
Korkeila E, Ristamaki R, Lamlum H, Collan Y, Syrjanen K and
Pyrhonen S: Comparison of CD44 expression in primary tumours and
metastases of colorectal cancer. Oncol Rep. 16:741–746.
2006.PubMed/NCBI
|
|
12
|
Bird NC, Mangnall D and Majeed AW: Biology
of colorectal liver metastases: a review. J Surg Oncol. 94:68–80.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jaggupilli A and Elkord E: Significance of
CD44 and CD24 as cancer stem cell markers: an enduring ambiguity.
Clin Dev Immunol. May 30–2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dalerba P, Dylla SJ, Park IK, et al:
Phenotypic characterization of human colorectal cancer stem cells.
Proc Natl Acad Sci USA. 104:10158–10163. 2012. View Article : Google Scholar
|
|
15
|
Su YJ, Lai HM, Chang YW, Chen GY and Lee
JL: Direct reprogramming of stem cell properties in colon cancer
cells by CD44. EMBO J. 30:3186–3199. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ricci-Vitiani L, Lombardi DG, Pilozzi E,
Biffoni M, Todaro M, Peschle C and De Maria R: Identification and
expansion of human colon cancer-initiating cells. Nature.
445:111–115. 2007. View Article : Google Scholar
|
|
17
|
Haraguchi N, Ohkuma M, Sakashita H,
Matsuzaki S, Tanaka F, Mimori K, Kamohara Y, Inoue H and Mori M:
CD133+CD44+ population efficiently enriches
colon cancer initiating cells. Ann Surg Oncol. 15:2927–2933. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
O’Brien CA, Pollett A, Gallinger S and
Dick JE: A human colon cancer cell capable of initiating tumour
growth in immunodeficient mice. Nature. 445:106–110. 2007.
View Article : Google Scholar
|
|
19
|
Bao SD, Wu QL, McLendon RE, Hao YL, Shi Q,
Hjelmeland AB, Dewhirst MW, Bigner DD and Rich JN: Glioma stem
cells promote radioresistance by preferential activation of the DNA
damage response. Nature. 444:756–760. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Frank NY, Pendse SS, Lapchak PH, Margaryan
A, Shlain D, Doeing C, Sayegh MH and Frank MH: Regulation of
progenitor cellfusionbyABCB5P-glycoprotein, a novel human
ATP-binding cassette transporter. J Biol Chem. 278:47156–47165.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Frank NY, Margaryan A, Huang Y, Schatton
T, Waaga-Gasser AM, Gasser M, Sayegh MH, Sadee W and Frank MH:
ABCB5-mediated doxorubicin transport and chemoresistance in human
malignant melanoma. Cancer Res. 65:4320–4333. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Horst D, Kriegl L, Engel J, Kirchner T and
Jung A: Prognostic significance of the cancer stem cell markers
CD133, CD44, and CD166 in colorectal cancer. Cancer Invest.
27:844–850. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Saigusa S, Tanaka K, Toiyama Y, Yokoe T,
Okugawa Y, Ioue Y, Miki C and Kusunoki M: Correlation of CD133,
OCT4, and SOX2 in rectal cancer and their association with distant
recurrence after chemoradiotherapy. Ann Surg Oncol. 16:3488–3498.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Botchkina IL, Rowehl RA, Rivadeneira DE,
Karpeh MS Jr, Crawford H, Dufour A, Ju J, Wang Y, Leyfman Y and
Botchkina GI: Phenotypic subpopulations of metastatic colon cancer
stem cells: genomic analysis. Cancer Genomics Proteomics. 6:19–29.
2009.PubMed/NCBI
|
|
25
|
Botchkina GI, Zuniga ES, Das M, et al:
New-generation taxoid SB-T-1214 inhibits stem cell-related gene
expression in 3D cancer spheroids induced by purified colon
tumor-initiating cells. Mol Cancer. 9:1922010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen KL, Pan F, Jiang H, Chen JF, Pei L,
Xie FW and Liang HJ: Highly enriched CD133(+)CD44(+) stem-like
cells with CD133(+) CD44(high) metastatic subset in HCT116 colon
cancer cells. Clin Exp Metastasis. 28:751–763. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nagata T, Sakakura C, Komiyama S, et al:
Expression of cancer stem cell markers CD133 and CD44 in
locoregional recurrence of rectal cancer. Anticancer Res.
31:495–500. 2011.PubMed/NCBI
|
|
28
|
Hou Y, Zou QF, Ge RL, Shen F and Wang YZ:
The critical role of CD133+CD44+/high tumor
cells in hematogenous metastasis of liver cancers. Cell Res.
22:259–272. 2012. View Article : Google Scholar :
|
|
29
|
Bellizzi A, Sebastian S, Ceglia P, et al:
Co-expression of CD133+/CD44+ in human colon
cancer and liver metastasis. J Cell Physiol. 228:408–415. 2013.
View Article : Google Scholar
|
|
30
|
Huh JW, Kim HR, Kim YJ, Lee JH, Park YS,
Cho SH and Joo JK: Expression of standard CD44 in human colorectal
carcinoma: association with prognosis. Pathol Int. 59:241–246.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ohji Y, Yao T, Eguchi T, Yamada T,
Hirahashi M, Iida M and Tsuneyoshi M: Evaluation of risk of liver
metastasis in colorectal adenocarcinoma based on the combination of
risk factors including CD10 expression: Multivariate analysis of
clinicopathological and mmunohistochemical factors. Oncol Rep.
17:525–530. 2007.PubMed/NCBI
|
|
32
|
Park JJ, Kwon JH, Oh SH, et al:
Differential expression of CD133 based on microsatellite
instability status in human colorectal cancer. Mol Carcinog.
53:E1–E10. 2014. View
Article : Google Scholar
|
|
33
|
Jones RJ: Cancer stem cells - clinical
relevance. J Mol Med (Berl). 87:1105–1110. 2009. View Article : Google Scholar
|
|
34
|
Papailiou J, Bramis KJ, Gazouli M and
Theodoropoulos G: Stem cells in colon cancer. A new era in cancer
theory begins. Int J Colorectal Dis. 26:1–11. 2011. View Article : Google Scholar
|
|
35
|
Baumann M, Krause M and Hill R: Exploring
the role of cancer stem cells in radioresistance. Nat Rev Cancer.
8:545–554. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
36
|
Eyler CE and Rich JN: Survival of the
fittest: cancer stem cells in therapeutic resistance and
angiogenesis. J Clin Oncol. 26:2839–2845. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Frank NY, Schatton T and Frank MH: The
therapeutic promise of the cancer stem cell concept. J Clin Invest.
120:41–50. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li M, Li JY, Zhao AL and Gu J: Colorectal
cancer or colon and rectal cancer? Clinicopathological comparison
between colonic and rectal carcinomas. Oncology. 73:52–57. 2007.
View Article : Google Scholar
|
|
39
|
Chung CK, Zaino RJ and Stryker JA:
Colorectal carcinoma: evaluation of histologic grade and factors
influencing prognosis. J Surg Oncol. 21:143–148. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Umpleby HC, Bristol JB, Rainey JB and
Williamson RC: Survival of 727 patients with single carcinomas of
the large bowel. Dis Colon Rectum. 27:803–810. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kazama Y, Watanabe T, Kanazawa T, Tanaka
J, Tanaka T and Nagawa H: Poorly differentiated colorectal
adenocarcinomas show higher rates of microsatellite instability and
promoter methylation of p16 and hMLH1: a study matched for T
classification and tumor location. J Surg Oncol. 97:278–283. 2008.
View Article : Google Scholar
|
|
42
|
Mulder JW, Kruyt PM, Sewnath M, Oosting J,
Seldenrijk CA, Weidema WF, Offerhaus GJ and Pals ST: Colorectal
cancer prognosis and expression of exon-v6- containing CD44
proteins. Lancet. 344:1470–1472. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Weg-Remers S, Anders M, von Lampe B,
Riecken EO, Schuder G, Feifel G, Zeitz M and Stallmach A: Decreased
expression of CD44 splicing variants in advanced colorectal
carcinomas. Eur J Cancer. 34:1607–1611. 1998. View Article : Google Scholar
|
|
44
|
Lugli A, Iezzi G, Hostettler I, Muraro MG,
Mele V, Tornillo L, Carafa V, Spagnoli G, Terracciano L and Zlobec
I: Prognostic impact of the expression of putative cancer stem cell
markers CD133, CD166, CD44s, EpCAM, and ALDH1 in colorectal cancer.
Br J Cancer. 103:382–390. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Horst D, Kriegl L, Engel J, Kirchner T and
Jung A: CD133 expression is an independent prognostic marker for
low survival in colorectal cancer. Br J Cancer. 99:1285–1289. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kojima M, Ishii G, Atsumi N, Fujii S,
Saito N and Ochiai A: Imunohistochemical detection of CD133
expression in colorectal cancer: a clinicopathological study.
Cancer Sci. 99:1578–1583. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Li CY, Li BX, Liang Y, et al: Higher
percentage of CD133(+) cells is associated with poor prognosis in
colon carcinoma patients with stage IIIB. J Transl Med. 7:562009.
View Article : Google Scholar
|
|
48
|
Choi D, Lee HW, Hur KY, Kim JL, Park GS,
Jang SH, Song YS, Lang KS and Paik SS: Cancer stem cell markers
CD133 and CD24 correlate with invasiveness and differentiation in
colorectal adenocarcinoma. World J Gastroenterol. 15:2258–2264.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Huh JW, Park YS, Lee JH, Kim HR, Shin MG
and Kim YJ: CD133 mRNA expression and microsatellite instability in
colorectal carcinoma. J Surg Oncol. 102:765–770. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zlobec I, Terracciano L, Jass JR and Lugli
A: Value of staining intensity in the interpretation of
immunohistochemistry for tumor markers in colorectal cancer.
Virchows Arch. 451:763–769. 2007. View Article : Google Scholar : PubMed/NCBI
|