Introduction
Cholangiocarcinoma (CCA) is a highly invasive and
metastatic cancer with diagnostic difficulties and high mortality
(1). In recent years, the
incidence and mortality of CCA in the world are increasing rapidly,
according to epidemiological studies (1,2).
However, the pathogenesis of CCA still remains unclear (3). The male/female ratio in incidence of
CCA is 1.4:1, and it often occurs in men aged between 50 and 70
years old (4,5). Hepatitis B and hepatitis C infection
is potentially associated with the occurrence of CCA (6). Surgery is the main effective
treatment for CCA; however, the postoperative recurrence rate is
high, and the 5-year survival rate is only 5% (7,8). For
the majority of patients with terminal CCA, surgical treatment and
liver transplantation are not the ideal treatment. Conventional
radiotherapy and chemotherapy exhibit limited therapeutic effect on
patients with CCA. Therefore, it is imperative to seek effective
treatments and identify sensitive biomarkers for early diagnosis of
CCA. Molecular targeted therapy possesses wide application
potentially with highly targeted and fewer adverse reactions, which
may control cancer cell proliferation, and prevent or delay
recurrence metastasis (9).
Therefore, there is an urgent need to understand of the molecular
mechanisms underlying carcinogenesis and progression of CCA, which
contribute to finding novel diagnosis markers and novel therapeutic
targets.
In recent years, noncoding RNAs, including long
noncoding RNAs (lncRNAs) and microRNAs (miRNAs/miRs), have been
demonstrated to have important functions in various biological
processes, particularly in cancers (10–15).
The research of oncogenes and noncoding RNAs has been useful for
further investigation of the pathogenesis of CCA and novel
molecular targeted therapy (16,17).
Recent evidences demonstrated that lncRNAs can regulate tumor
development and progression by acting as competing endogenous RNAs
in various cancers. For instance, the long noncoding RNA taurine
upregulated 1 acts as a competing endogenous RNA to sponge miR-132
in hepatocellular carcinoma (18).
Long non-coding RNA metastasis associated lung adenocarcinoma
transcript 1 (MALAT1) sponges miR-144-3p to promote metastasis and
proliferation in osteosarcoma cells (19). miRNAs are a group of small (19–25
nucleotides) endogenous non-coding RNA molecules, which act as gene
regulators by binding to partially complementary target sites in
mRNA 3′ untranslated regions (3′UTRs) that results in degradation
of the target mRNAs or translational repression of the encoded
proteins (20). Wang et al
(21) reported that downregulation
of miR-138 enhanced the proliferation, migration and invasion of
CCA cells through the upregulation of RhoC/phospho-ERK/matrix
metalloproteinase-2/9. However, the roles of noncoding RNAs in CCA
still remain largely unknown.
In this study, the expression patterns of long
intergenic non-protein coding RNA 1296 (LINC01296) and its
biological functions in CCA were investigated. The further
investigations were conducted to determine the molecular mechanism
by which LINC01296 regulates CCA development. Finally, it was
demonstrated that LINC0129 interacted with miR-5095 to enhance the
expression of MYCN proto-oncogene bHLH transcription factor (MYCN)
and promoted CCA development and progression.
Materials and methods
Patient and tissue samples
Paired CCA tissues (n=57) and matched peritumor
samples were obtained from a tissue bank of samples collected from
patients that underwent surgical treatment between January 2010 and
December 2016 at the Second Affiliated Hospital of Guangzhou
Medical University (Guangzhou, China) (Table I). Tissue samples were snap frozen
in liquid nitrogen immediately following surgical resection and
stored at −80°C. Written informed consent was obtained from all
patients. This study did not include patients that had received
radiotherapy and/or immunotherapy prior to or following surgical
treatment. The study protocol conformed to the ethical guidelines
of the 1975 Declaration of Helsinki, and was approved by The Second
Affiliated Hospital of Guangzhou Medical University.
 | Table IAssociation between
clinicopathological features and LINC01296 expression in 57
patients with cholangiocarcinoma. |
Table I
Association between
clinicopathological features and LINC01296 expression in 57
patients with cholangiocarcinoma.
| Feature | LINC01296
expression
| P-value |
|---|
| Low | High |
|---|
| All cases | 35 | 22 | |
| Age (year) | | | 0.553 |
| <60 | 34 | 20 | |
| ≥60 | 1 | 2 | |
| Sex | | | 0.789 |
| Male | 19 | 13 | |
| Female | 16 | 9 | |
| Size (cm) | | | 0.003 |
| <3 | 21 | 4 | |
| ≥3 | 14 | 18 | |
| Lymph node
metastasis | | | 0.031 |
| No | 22 | 7 | |
| Yes | 13 | 15 | |
| TNM | | | 0.024 |
| I/II | 26 | 9 | |
| III/IV | 9 | 13 | |
Fluorescence in situ hybridization
(FISH)
A FISH kit was purchased from Guangzhou Boye
Biological Technology Co., Ltd. (Guangzhou, China) and fluorescein
isothiocyanate (FITC)-conjugated LINC01296 DNA probe (5′-TATGGG
AAGGGGACTGTCTG-3′) was used for RNA-FISH as previously described
(22). In brief, CCA tissues were
fixed in 4% paraformaldehyde for 48 h at 37°C. Paraffin sections (7
µm in thickness) were processed by dewaxing, protein removal
and pre-hybridization, and then hybridized with probe. DAPI
staining was performed and observed with a FV1000 confocal laser
microscope (Olympus Corporation, Tokyo, Japan).
In situ hybridization (ISH)
ISH was performed as previously described (23). In brief, samples were fixed and
embedded in paraffin. Then, sample sections were incubated in
graded alcohols and incubated in 3% hydrogen peroxide
(H2O2) for 30 min. Biotin-conjugated probes
and streptavidin-horseradish peroxidase conjugate were used for
ISH. The samples were finally stained with hematoxylin and observed
with a light microscope (Nikon Corporation, Tokyo, Japan). The
LINC01296 DNA probe sequence was 5′-CTCCCTCAAATCAGGATGGG-3′
Cell culture and transfection
CCA cell lines, including HIBEC noncancerous
cholangiocyte cell line, and RBE, CCLP1, HuCCT1 and HCCC-9810
cholangiocarcinoma cell lines, were purchased from the Chinese
Academy of Sciences Cell Bank (Shanghai, China). All cells were
cultured in RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS;
Invitrogen; Thermo Fisher Scientific, Inc.) in humidified 5%
CO2 at 37°C. miR-5095 mimics
(5′-UUACAGGCGUGAACCACCGCG-3′), inhibitor
(5′-CGCGGTGGTTCACGCCTGTAA-3′), short hairpin RNA (shRNA) which
targeted LINC0129 (shLINC0129, 5′-GGUUCAUCUGUGUUGCUCU-3′) and
relative controls (5′-AATTCTCCGAACGTGTCACGT-3′) were obtained from
GenePharma Co., Ltd. (Shanghai, China). The transfection was
conducted by using Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) in 5×106 cells at a
final concentration of 50 nM. Subsequent assays were performed at
48 h after transfection. To overexpress LINC01296, CCA cells were
transfected with pCDNA3-LINC01296 using Lipofectamine®
2000. After transfection for 24 h, expression of LINC01296 was
validated by reverse transcription-quantitative polymerase chain
reaction (RT-qPCR).
RT-qPCR
Total RNA was extracted from 1×107 CCA
and noncancerous cholangiocyte cells using the TRIzol reagent
(Thermo Fisher Scientific, Inc.) according to the manufacturer's
instructions. For miRNA analysis, RT-qPCR was performed using the
TaqMan MicroRNA Reverse Transcription kit, TaqMan Universal PCR
Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions with corresponding
primers: Universal miRNA RT-qPCR primer, 5′-AACGAGACGACGACAGAC-3′;
5′-GCAAATTCGTGAAG CGTTCCATA-3′ for U6; and 5′-TACAGG CGTGAACCACC-3′
for miR-5095. For mRNA analysis, RT-qPCR was performed using the
TaqMan High-Capacity cDNA Reverse Transcription Kit, TaqMan Fast
PCR Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions with corresponding
primers. GAPDH was used as an internal control to normalize MYCN.
The RT-qPCR protocol included an initial denaturation step (95°C
for 5 min) and 40 cycles of denaturation (95°C for 10 sec),
annealing (60°C for 20 sec) and extension (72°C for 10 sec). The
relative expression levels were calculated using 2−ΔΔCq
method as previously described (24). The primer sequences were as
follows: LINC01296 (forward, 5′-GAGAAGCAGTGGTGGGTTCC-3′ and
reverse, 5′-GAGCAACACAGATGAACCGC-3′), MYCN (forward,
5′-ACTGTAGCCATCCGAGGACA-3′ and reverse, 5′-CAAGCCCTGCTCCTTACCTC-3′)
and GAPDH (forward, 5′-TCCTCTGACTTCAACAGCGACAC-3′ and reverse,
5′-CAC CCTGTTGCTGTAGCCAAATTC-3′).
Cell proliferation assay
Cells were seeded at 5,000 cells per well in 96-well
plates at 24 h after transfection. Cell proliferation was measured
using the Cell Counting Kit-8 (CCK8; Dojindo Molecular
Technologies, Inc., Kumamoto, Japan) at 24, 48 and 72 h after the
cells were seeded. Absorbance was determined at 450 nm using a
microplate spectrophotometer (Thermo Fisher Scientific, Inc.).
Cell cycle analysis
Cells were harvested at 48 h after transfection. The
cells were washed with PBS and fixed in ethanol at −20°C. The cells
were then washed with PBS, rehydrated and resuspended in propidium
iodide (PI; 10 µl)-RNase A solution (Sigma-Aldrich; Merck
KGaA) at 37°C for 30 min. The stained cells (1×105) were
then analyzed for DNA content with a flow cytometer (BD
Biosciences, Franklin Lakes, NJ, USA). The results were analyzed
with FlowJo 7.6.1 software (FlowJo LLC, Ashland, OR, USA).
Cell apoptosis analysis
Cells were harvested, washed with ice-cold PBS, and
stained with Annexin V-FITC apoptosis detection kits (Nanjing
KeyGen Biotech Co., Ltd., Nanjing, China). Cell apoptosis was
analyzed using a flow cytometer (BD Biosciences). The results were
analyzed with FlowJo 7.6.1 software.
Cell migration and invasion assay
Cell migration was evaluated using a wound-healing
assay. In brief, at 48 h after transfection, cells were cultured in
6-well plates (5×104 cells per well). At 90–95%
confluence, the monolayer of cells was scratched with using a
sterile plastic micropipette tip, and then cell were cultured under
standard conditions for 24 h. Following several washes, recovery of
the wound was observed and imaged using an X71 inverted microscope
(Olympus Corporation).
A Transwell invasion assay was performed to
determine cell invasion. Transfected cells (1×105) were
seeded into the upper chamber of Matrigel-coated inserts with
free-serum medium. Medium with 10% FBS was added to the lower
chamber as chemoattractant. The cells were allowed to invade for 48
h at 37°C with 5% CO2. Then cells invaded to the lower
surface of filter were fixed in 70% ethanol for 30 min and stained
with 0.1% crystal violet for 10 min at 25°C. The number of cells
that migrated to the lower side was counted in five randomly
selected fields under an X71 inverted microscope (Olympus
Corporation).
Luciferase activity assay
Wild-type LINC01296 or MYCN 3′UTR sequences were
amplified from a human cDNA library. Mutations of the miR-5095
binding site were introduced by site-directed mutagenesis using a
fast mutation kit (New England BioLabs, Inc., Ipswich, MA, USA).
The PCR fragment was cloned into psiCHECK-2 vector (Addgene, Inc.,
Cambridge, MA, USA) downstream of the firefly luciferase coding
region within XhoI and NotI (Takara Bio, Inc., Otsu,
Japan). psiCHECK-2-control was used as internal control. The
psiCHECK-2 reporter plasmids (1 µg per well) were tranfected
into 1×106 CCA cells using Lipofectamine®
2000 (Invitrogen; Thermo Fisher Scientific, Inc.). At 48 h after
transfection, the luciferase activity was measured as previously
described (25). The primer
sequences for LINC01296 or MYCN cloning were as follows: LINC01296
(forward, 5′-CCCTGCAGCAGCAGCGGCGTG-3′ and reverse,
5′-ACACTCACACACACTCCCACC-3′) and MYCN (forward,
5′-ACGCTTCTCAAAACTGGACAG-3′ and reverse,
5′-AGCTATTTATTTTCATAAACATG-3′).
Western blot analysis
Total protein lysates were resolved by 10% SDS-PAGE
and transferred to polyvinyl difluoride membranes (EMD Millipore,
Billerica, MA, USA). Following blocking in Tris-buffered saline
containing 0.1% Tween-20 (TBS-T) with 5% nonfat dry milk for 30 min
at 37°C, membranes were washed four times in TBS-T and incubated
with primary antibodies overnight at 4°C. Primary antibodies were
all obtained from Abcam (Cambridge, MA, USA) and used at the
following dilutions: Anti-caspase-3 (1:300; cat. no. 3550914; BD
Biosciences), Twist1 (1:500; cat. no. 46702), anti-GAPDH (1:1,000;
cat. no. 5174), anti-Cyclin D1 (1:1,000; cat. no. 2978),
anti-matrix metalloproteinase-2 (1:1,000; cat. no. 87809) and
anti-MYCN (1:2,000; cat. no. 84406) (all from CST Biological
Reagents Co., Ltd.). Following extensive washing, membranes were
incubated with horseradish peroxidase-linked goat polyclonal
anti-rabbit IgG secondary antibody (cat. no. 7074; CST Biological
Reagents Co., Ltd.) at a dilution of 1:2,000 for 1 h at room
temperature. Immunoreactivity was detected by enhanced
chemiluminescence (Pierce; Thermo Fisher Scientific, Inc.) and
visualized using a ChemiDoc XRS imaging system and analysis
software (Bio-Rad Laboratories, Inc., Hercules, CA, USA). GAPDH
served as the loading control.
In vivo xenograft experiments
Male BALB/c nude mice (6-week-old; n=6) were
purchased from Beijing HFK Bioscience Co. Ltd. (Beijing, China) and
maintained under pathogen-free conditions with approval by the
Committee of the Second Affiliated Hospital of Guangzhou Medical
University. For tumor propagation analysis, 1×107 RBE
tumor cells were subcutaneously injected into BALB/c nude mice.
Tumor volume was calculated using the formula: volume =
πab2/6 (a, tumor length; b, tumor width) at indicative
time points. Tumor weight was measured at week 4 post-injection.
Animal experiments were performed in accordance with relevant
guidelines and regulations of the Animal Care and Use Committees at
the Second Affiliated Hospital of Guangzhou Medical University, and
a signed document issued by the Animal Care and Use Committees that
granted approval was obtained.
Cohort analysis
Online-available data set (GSE61850) was downloaded
from NCBI (ncbi.nlm.nih.gov/gds/?term¼). R language (r-project.org) and Bioconductor (biocon-ductor.org/) were used for background
correction, normalization, expression calculation and annotation.
The gene expression profiles were evaluated for further analyses
using Excel (Microsoft Corporation, Redmond, WA, USA), SPSS 19.0
statistical software (version 19.0; IBM Corp., Armonk, NY, USA) or
GraphPad Prism 5 software (GraphPad Software, Inc., La Jolla, CA,
USA).
Colony formation assays
Colony formation assays were used to assess
clonogenic ability of transfected cells. CCA cells
(2×103/well) were trypsinized in a single-cell
suspension and seeded in 6-well dishes. Subsequently, the cells
were maintained in RPMI-1640 supplemented with 10% fetal bovine
serum for about 2 weeks. The visible colonies were fixed in 4%
paraformaldehyde for 4 h at 37°C and stained with 0.5% crystal
violet for 2 h at 37°C (Beyotime Institute of Biotechnology,
Haimen, China). Colony numbers were counted under a light
microscope (Olympus Corporation).
Statistical analysis
Data from at least three independent experiments are
expressed as the mean ± standard deviation. Statistical analysis
was performed using IBM SPSS 19.0 statistical software. Statistical
analysis was performed using Student's t-test or ANOVA. Pearson's
χ2 tests were used for analysis of the correlation
between clinicopathological features and LINC01296 expression in
CCA patients. Kaplan-Meier survival analysis was used for analysis
of survival rate and P-value was calculated by the log-rank test.
Spearman's correlation analysis was used to determine the
correlations between the levels of LINC01296 and miR-5095 in CCA
tissues. P<0.05 was considered to indicate a statistically
significant difference.
Results
LINC01296 is upregulated in human
CCA
To explore important lncRNAs in human CCA, an online
dataset (GSE61850) was analyzed and a functionally undefined lncRNA
LINC01296 that was greatly upregulated in human CCA tissues was
identified (Fig. 1A). To validate
this analysis, the expression levels of LINC01296 were measured in
57 pairs of human CCA samples by RT-qPCR, and LINC01296 expression
was significantly upregulated in CCA tissues compared with nontumor
tissues (Fig. 1B). Additionally,
FISH and ISH assays were performed with pairs of CCA samples, which
demonstrated that the expression of LINC01296 was higher in tumor
tissues than nontumor tissues (Fig. 1C
and D). Furthermore, RT-qPCR also showed that the expression of
LINC01296 was higher in CCA cell lines (RBE, CCLP1, HuCCT1 and
HCCC-9810) compared with HIBEC noncancerous cholangiocyte cell line
(Fig. 1E). To further investigate
the clinicopathological significance of LINC01296 level in patients
with CCA, The 57 patients were divided into 2 subgroups based on
the mean value: Low LINC01296 group (35 cases) and a high LINC01296
group (22 cases). As presented in Table I, LINC01296 levels in CCA tissues
were positively associated with advanced clinical stages and lymph
node metastasis. Furthermore, patients with higher expression of
LINC01296 had a lower survival rate (Fig. 1F).
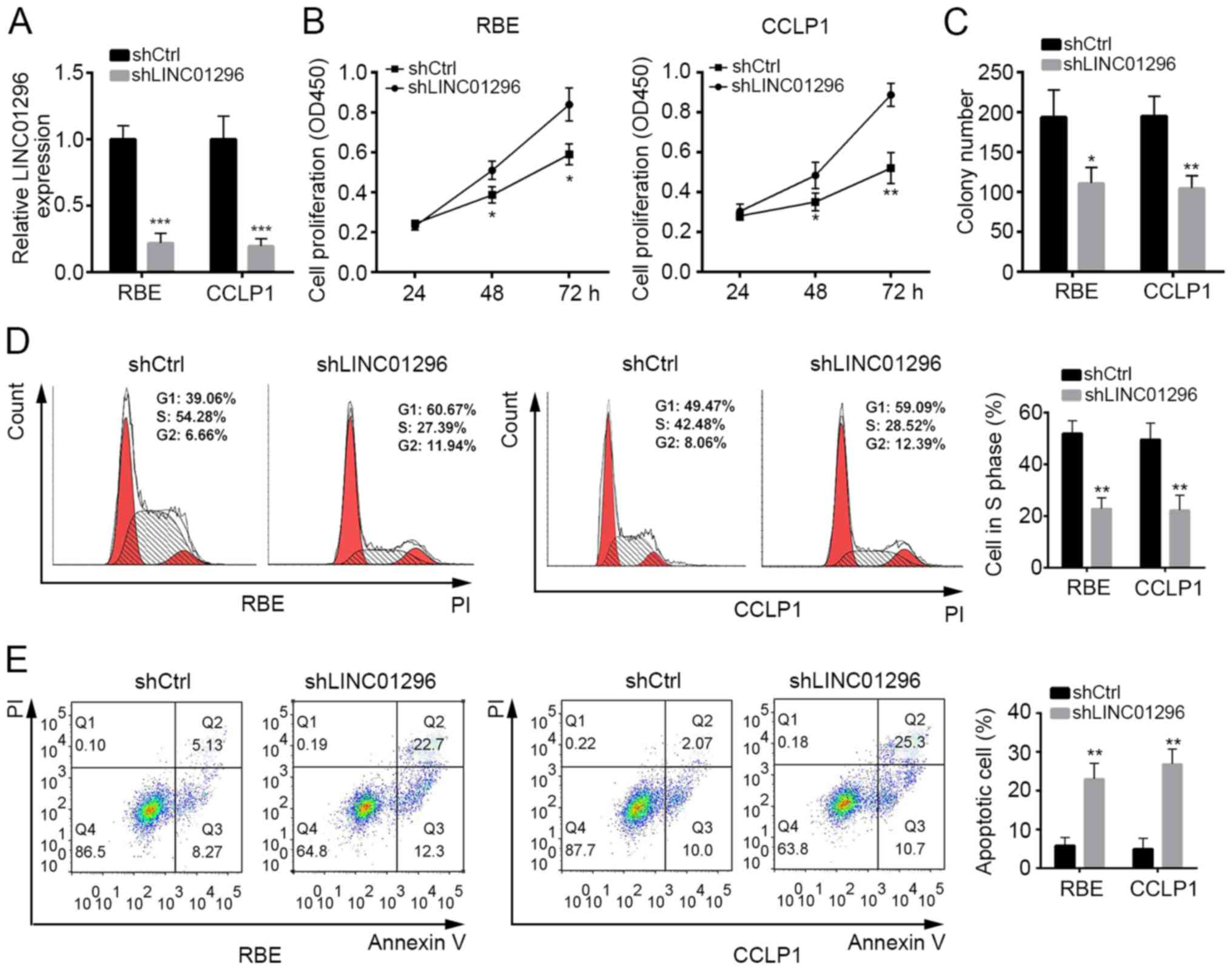
LINC01296 knockdown suppresses cell
proliferation and promotes cell apoptosis
LINC01296 expression was highest in RBE and CCLP1
cells among the CCA cell lines (Fig.
1E). Thus, these two cell lines were selected for use in
subsequent experiments. To explore whether LINC01296 influences the
proliferation of RBE and CCLP1 cells, LINC01296 was knocked down by
transfection with shLINC01296 plasmid (Fig. 2A). CCK8 and colony formation assays
demonstrated that LINC01296 knockdown significantly suppressed the
viability and proliferation of RBE and CCLP1 cells (Fig. 2B and C). To further investigate the
mechanism through which LINC01296 inhibited cell proliferation, the
effects of LINC01296 on cell cycle distribution were determined.
Flow cytometry analysis demonstrated that the percentage of cells
in S phase was decreased in RBE and CCLP1 cells transfected with
shLINC01296 compared with control shRNA (Fig. 2D), which suggested that LINC01296
promoted cell proliferation by regulating the cell cycle.
Furthermore, Annexin V/PI staining demonstrated that LINC01296
knockdown significantly promoted the apoptosis of RBE and CCLP1
cells (Fig. 2E).
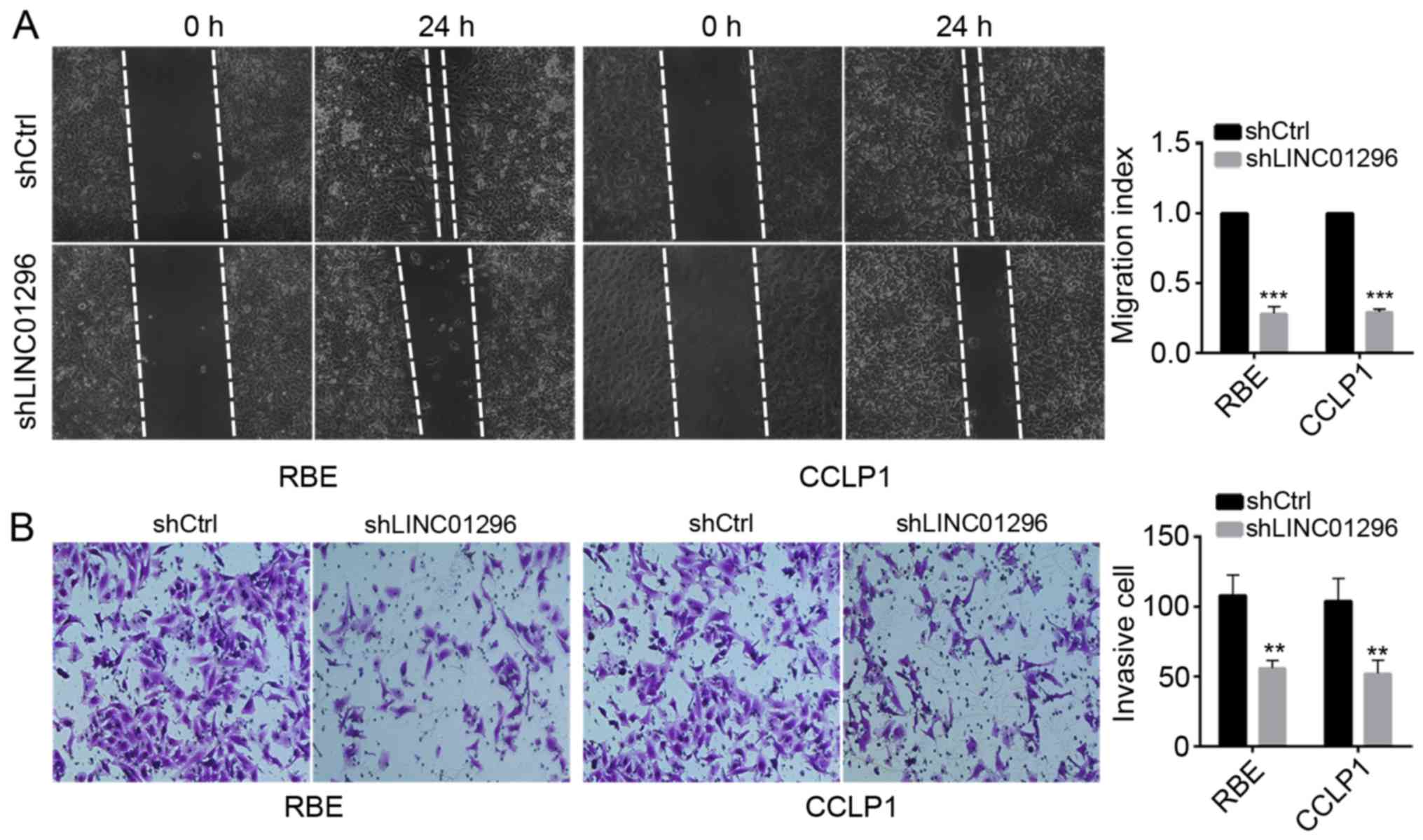
LINC01296 knockdown inhibits the
migration and invasion of RBE and CCLP1 cells
To further define the effects of LINC01296 on tumor
metastasis, wound healing assay and Transwell invasion assay were
performed. LINC01296 knockdown significantly inhibited cell
motility (Fig. 3A) and suppressed
cell invasion (Fig. 3B) compared
with control shRNA. These data suggested that LINC01296 inhibited
metastatic capacity of RBE and CCLP1 cells.
LINC01296 interacts with miR-5095 in RBE
and CCLP1 cells
To explore the molecular mechanism by which
LINC01296 inhibit CCA progression, the target miRNAs of LINC01296
were predicted with computational algorithms, including TargetScan
(targetscan.org/vert_71/) and miRanda
(34.236.212.39/microrna/getGeneForm.do). Among all
candidates, miR-5095 ranked top. The predicted miR-5095 interaction
site in LINC01296 is illustrated in Fig. 4A. RT-qPCR demonstrated that
overexpression of LINC01296 reduced the level of miR-5095, while
the miR-5095 mimic reduced LINC01296 levels in RBE and CCLP1 cells
(Fig. 4B and C). To further
validate the interaction between LINC01296 and miR-5095, a
dual-luciferase reporter assay was performed. Overexpression of
miR-5095 significantly decreased the luciferase activity of the
reporter containing wild-type LINC01296 in RBE cells, while it
failed to repress the reported with mutated miR-5095 binding site
(Fig. 4D). miR-5095 inhibitor also
produced similar results (Fig.
4D). Furthermore, RT-qPCR demonstrated that miR-5095 expression
was lower in CCA cell lines than in HIBEC cells (Fig. 4E), and the expression of miR-5095
was lower in CCA tissues compared with nontumor tissues (Fig. 4F). Finally, in CCA tissues,
miR-5095 expression was also inversely correlated with LINC01296
expression (P<0.001, Spearman's rank test) (Fig. 4G).
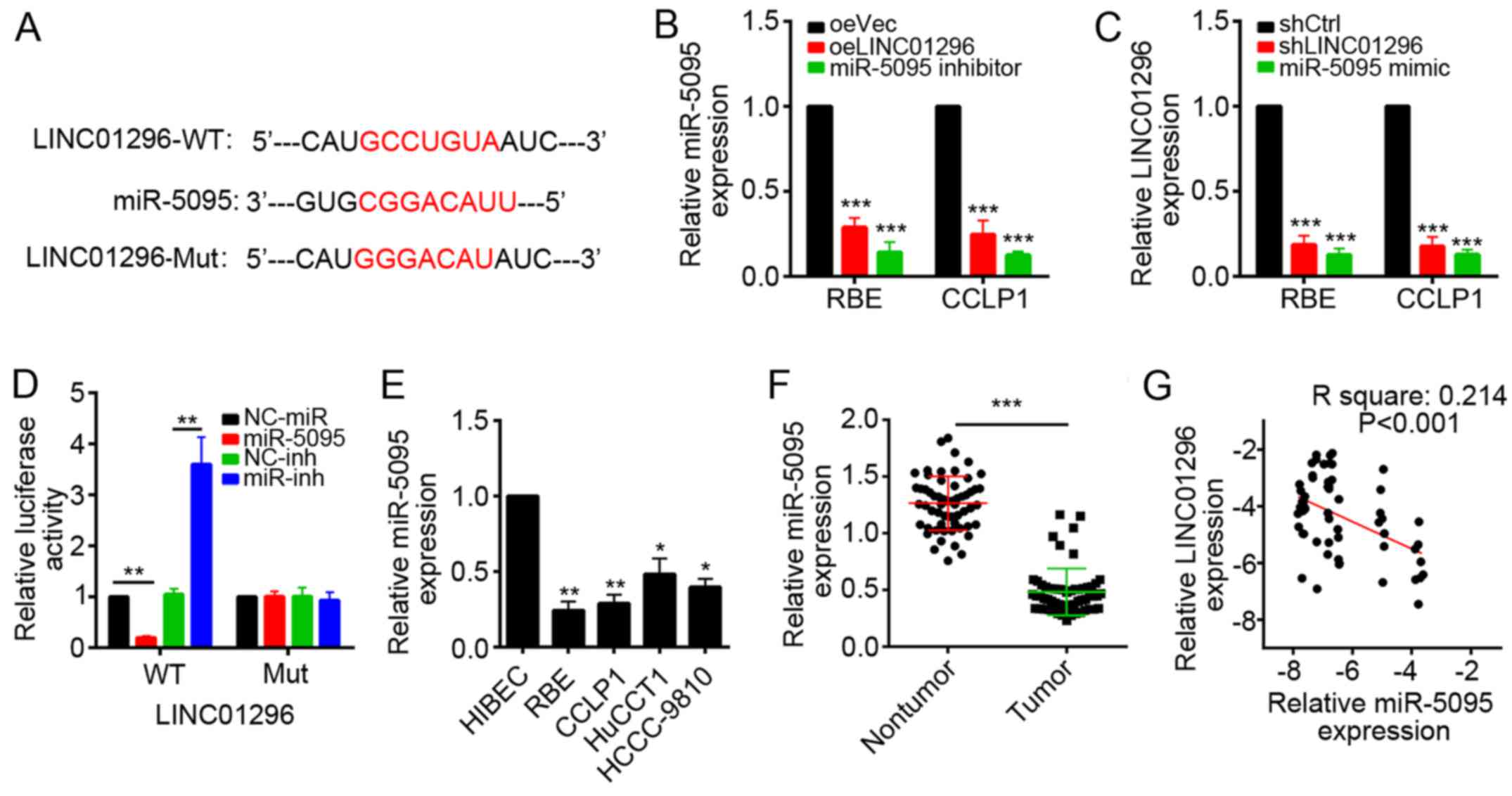 | Figure 4LINC01296 interacts with miR-5095 in
RBE and CCLP1 cells. (A) miR-5095 binding sites in LINC01296
predicted by bioinformatics analysis. (B) Analysis of miR-5095
expression levels in RBE and CCLP1 cells transfected with LINC01296
overexpressing plasmid or miR-5095 inhibitor by RT-qPCR. (C)
Analysis of LINC01296 expression levels in RBE and CCLP1 cells
transfected with shLINC01296 or miR-5095 mimic by RT-qPCR.
***P<0.001 vs. oeVec. (D) Luciferase reporter assays
were performed using RBE cells co-transfected with the miR-5095
mimic or inhibitor and LINC01296-wt or LINC01296-mut reporter
plasmid. **P<0.01. (E) The expression levels of
miR-5095 were determined in CCA cell lines (RBE, CCLP1, HuCCT1 and
HCCC-9810) and noncancerous cholangiocyte cell line (HIBEC) by
RT-qPCR. *P<0.05 and **P<0.01 vs.
HIBEC. (F) The expression of miR-5095 in CCA samples and nontumor
tissues was determined by RT-qPCR. ***P<0.001. (G)
Spearman's correlation analysis was used to determine the
correlations between the levels of LINC01296 and miR-5095 in human
CCA (n=57). Data are presented as the mean ± standard deviation.
RT-qPCR, reverse transcription-quantitative polymerase chain
reaction; CCA, cholangiocarcinoma; LINC01296, long intergenic
non-protein coding RNA 1296; WT, wild-type; Mut, mutated; miR,
microRNA; oeVec, overexpression empty vector; oeLINC01296,
LINC01296 overexpression vector; sh, short hairpin RNA; Ctrl,
control; NC, negative control; inh, miR inhibitor. |
MYCN is a target of miR-5095
To further explore the downstream target genes of
miR-5095, prediction of miR-5095 targets was performed with
computational algorithms, including TargetScan and miRanda. MYCN
was selected. MYCN has been proven to promote tumor initiation and
metastasis in various cancer types (26–28).
The predicted miR-5095 interaction site in the MYCN 3′UTR is
illustrated in Fig. 5A. To further
validate the interaction between miR-5095 and MYCN, dual-luciferase
reporter assay was performed. miR-5095 mimic significantly
decreased the luciferase activity of the wild-type MYCN 3′UTR in
RBE cells, while it failed to repress luciferase activity of the
reported with mutated MYCN 3′UTR, and the miR-5095 inhibitor had
the opposing effect (Fig. 5B).
RT-qPCR and western blot analysis demonstrated that miR-5095 mimic
suppressed the expression of MYCN in RBE and CCLP1 cells, and the
miR-5095 inhibitor had the opposing effect (Fig. 5C and D). Furthermore, the
expression of MYCN was upregulated in CCA samples compared with
nontumor tissues (Fig. 5E).
Additionally, in CCA tissues, miR-5095 expression was inversely
correlated with MYCN expression (P<0.001, Spearman's rank test)
(Fig. 5F).
Restoration of MYCN reverses LINC01296
knockdown-induced inhibition of cell viability, migration, and
invasion of RBE and CCLP1 cells
To investigate whether the regulation of cell
proliferation, migration and invasion of RBE and CCLP1 cells by
LINC01296 is MYCN-dependent, knockdown of LINC01296, miR-5095, MYCN
and/or overexpression of MYCN were performed (Fig. 6A). Cell viability, apoptosis,
migration and invasion were analyzed using CCK8, flow cytometry and
Transwell assays, respectively. Knockdown of LINC01296 or MYCN and
overexpression of miR-5095 suppressed cell viability, migration and
invasion in RBE and CCLP1 cells, while miR-5095 inhibition or
overexpression of MYCN reversed LINC01296 knockdown-induced
inhibition of cell viability, migration and invasion (Fig. 6B–E).
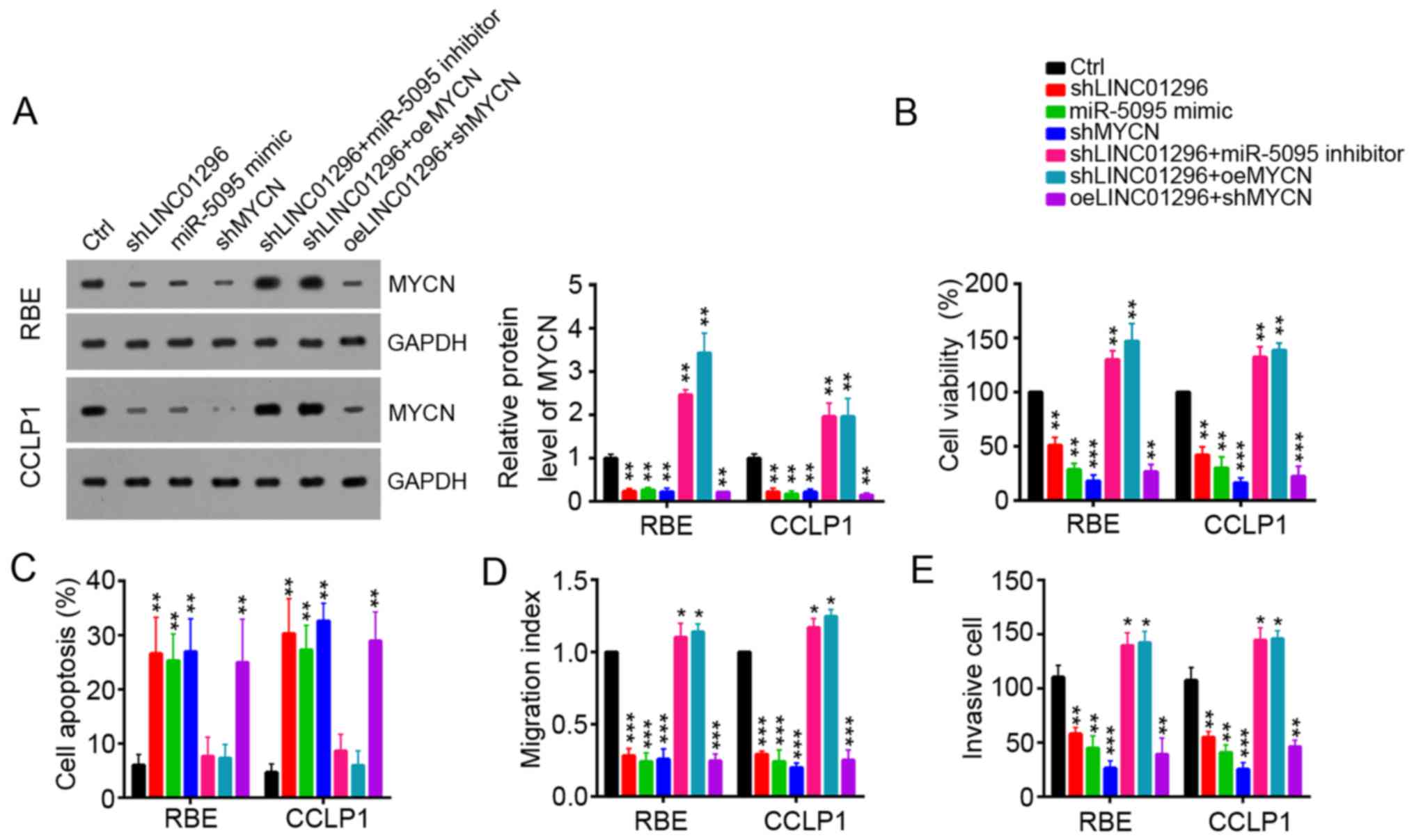 | Figure 6Restoration of MYCN reverses
LINC01296 knockdown-induced inhibition of cell proliferation,
migration, and invasion of RBE and CCLP1 cells. (A) Knockdown of
LINC01296 or miR-5095 inhibited the level of MYCN protein in RBE
and CCLP1 cells as demonstrated by western blot analysis. (B) Cell
viability, (C) apoptosis, (D) migration and (E) invasion were
determined in RBE and CCLP1 cells transfected with shLINC01296,
miR-5095 mimic, shMYCN, miR-5095 inhibitor or MYCN-overexpressing
plasmid by CCK8, flow cytometry, wound-healing, and invasion
assays, respectively. Data are presented as the mean ± standard
deviation. *P<0.05, **P<0.01 and
***P<0.001 vs. Ctrl. Ctrl, control; sh, short hairpin
RNA; LINC01296, long intergenic non-protein coding RNA 1296; miR,
microRNA; MYCN, MYCN proto-oncogene bHLH transcription factor; oe,
overexpression vector. |
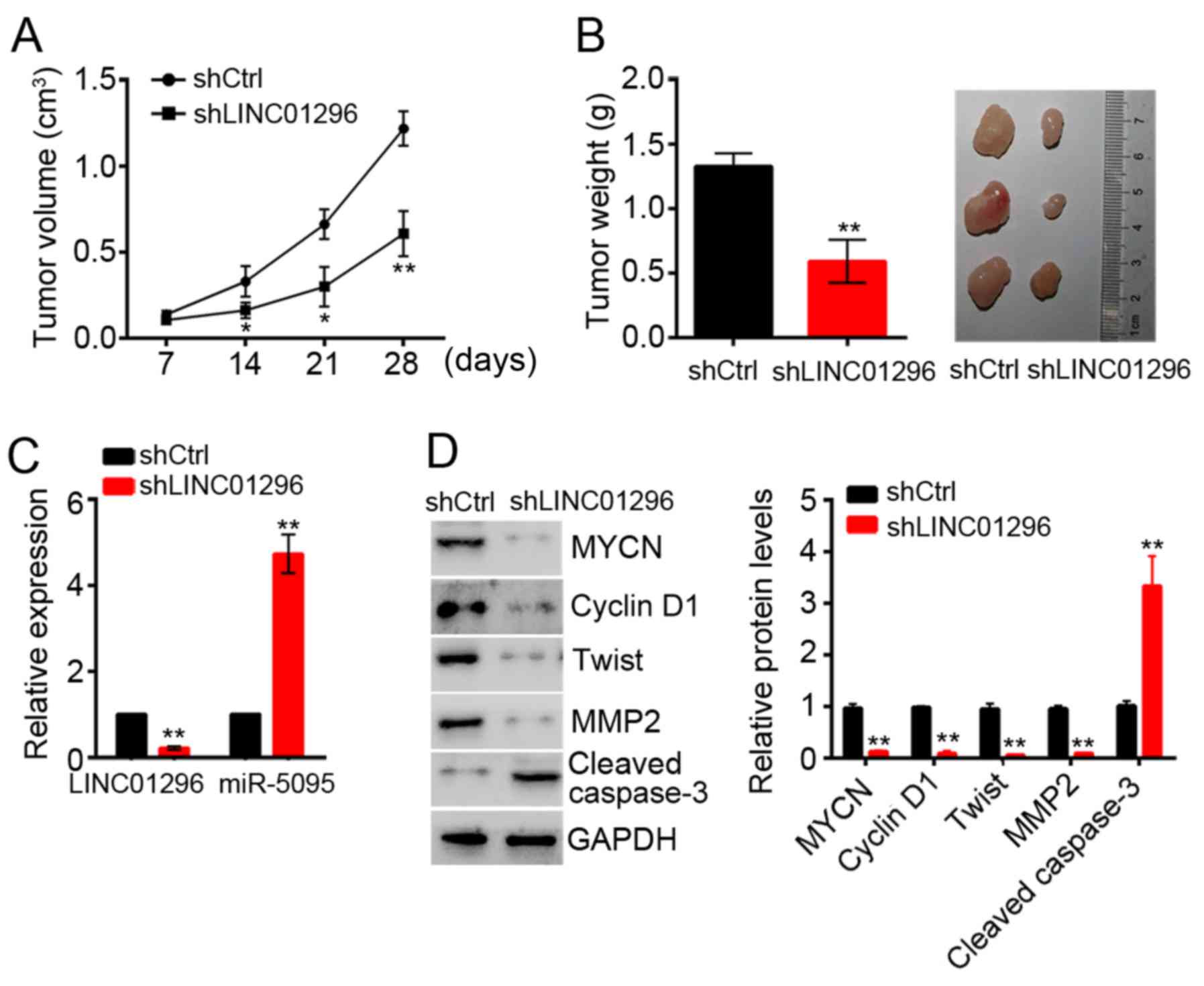
LINC01296 promoted tumor growth in vivo
by activation of MYCN
The biological effects of LINC01296 on CCA
development were evaluated in a xenograft mouse model. RBE cells
transfected with shCtrl or shLINC01296 were implanted
subcutaneously into nude mice. Then tumor growth was evaluated
every 7 days. LINC01296 knockdown significantly delayed tumor
growth in vivo (Fig. 7A).
At 5 weeks post-implantation, the nude mice were sacrificed, and
tumors were harvested and weighed. LINC01296 knockdown
significantly decreased the tumor size and weight (Fig. 7B). Subsequently, the expression
levels of LINC01296, miR-5095 and MYCN were determined in tumor
tissues by RT-qPCR and western blot analysis, respectively. Tumors
derived from shLINC01296 cells exhibited a significant decrease in
LINC01296 and MYCN expression, and an increase in miR-5095
expression (Fig. 7C and D).
Furthermore, LINC01296 knockdown significantly inhibited the
expression of proteins associated with tumor cell proliferation,
migration and invasion, and increased the expression of caspase-3,
which is crucial for cell apoptosis (Fig. 7D).
Discussion
In recent decades, various diagnostic advances and
therapeutic strategies have been used to treat CCA. However, the
overall survival rates of patients with CCA remain low due to tumor
recurrence and metastasis (17).
Thus, in order to develop novel and effective therapeutic
approaches for CCA treatment, there is an urgent need to fully
understand the molecular mechanisms that regulate CCA initiation
and progression.
Recent evidence has indicated that 97% transcripts
produced from the human genome are short or lncRNAs with no, or
limited, protein-coding potential (29). The role of lncRNAs in human cancer
has been a developing field in the recent years. A large number of
studies demonstrated that lncRNAs are closely associated with the
development and progression of various tumor types, including CCA
(16). For instance, the lncRNA
HOX transcript antisense RNA activates the Hippo pathway in renal
cell carcinoma (30). lncRNA HOXA
cluster antisense RNA 3 promotes tumor progression and predicts
poor prognosis in glioma (31).
However, the functions of the majority of lncRNAs in CCA remain
elusive. In the current study, LINC01296 was significantly
upregulated in CCA. A previous study reported that LINC01296 is
associated with poor prognosis in prostate cancer and promotes
cancer cell proliferation and metastasis (32). LINC01296 was also demonstrated to
promote colorectal cancer and bladder cancer progression in
vitro (33,34). However, the functions of LINC01296
in CCA have not been defined. In the current study, LINC01296
knockdown significantly inhibited cell proliferation, migration and
invasion, and led to increased cell apoptosis in CCA. These
findings suggested that LINC01296 may have an oncogenic role in CCA
progression.
A previous study reported that lncRNAs can exert
biological functions via a variety of mechanisms, including
transcriptional and post-transcriptional regulation. For example,
lymphocyte-specific protein 1 pseudogene recruited DNA-binding
protein SATB1 to initiate transcription of hes family bHLH
transcription factor 6 and promote hepatocellular carcinoma
progression (13). Additionally,
lncRNA MALAT1 acted as a competing endogenous RNA of miR-144-3p to
promote metastasis and proliferation in osteosarcoma cells
(19). Zhu et al (22) reported that lnc-β-Catm interacts
with β-catenin to prevent it degradation and then sustains liver
cancer stem cell self-renewal. In the current study, LINC01296 was
predicted to interact with miR-5095 by computational algorithms. A
luciferase activity reporter assay then validated that LINC01296
sponged miR-5095 in CCA cells. Additionally, expression of
LINC01296 was negatively correlated with miR-5095 in CCA. miRNAs
can target the 3′UTR of mRNAs for degradation and then regulate
gene expression. The functions of miR-5095 were unknown prior to
the current study. Computational algorithms predicted that MYCN, an
oncogene in various types of human cancer (28,35,36),
was a potential target of miR-5095. Luciferase activity assay,
RT-qPCR and western blot, demonstrated that miR-5095 bound to the
3′UTR of MYCN and downregulated its expression in CCA. Finally,
restoration of MYCN expression in RBE and CCLP1 cells rescued the
LINC01296 knockdown-induced inhibition of cell viability, migration
and invasion. Therefore, our findings indicated that LINC01296
promoted cell proliferation, migration and invasion by sponging
miR-5095 to upregulate MYCN expression.
In summary, the results of the current study
demonstrated that LINC01296 was significantly upregulated in CCA
tumors and cell lines compared with normal tissue and cells, and
LINC01296 was associated with cell growth, migration and invasion
in vitro and in vivo. The present study revealed a
novel mechanism of a LINC01296/miR-5095/MYCN axis that regulates
CCA development and progression. The study demonstrated that
LINC01296 may act as a novel diagnostic and prognostic indicator,
and a therapeutic target for CCA treatment.
Acknowledgments
Not applicable.
Funding
This study was supported by the National Natural
Science Foundation of Guangdong (grant nos. 2014A030310160,
2014A030310015 and 2015A030313466), the Science and Technology
Planning Project of Guangdong (grant nos. 2016A02021521 and
2016A020215038), the Science and Technology Program of Guangzhou
(grant nos. 201707010469 and 201607010033), and the National
Natural Science Foundation Project (grant nos. 81602109 and
81602331).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
DZ and HL designed and performed the experiments;
JX, DJ, LC and XJ contributed to the data analysis; XY enrolled
patients and measured the RNA levels in the clinical samples; PX
initiated the work and wrote the manuscript; and all authors have
read and approved the final version of the manuscript.
Ethics approval and consent to
participate
The use of human tissues was approved by the Ethics
Committee of the Second Affiliated Hospital of Guangzhou Medical
University and patient consent was obtained.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Razumilava N and Gores GJ:
Cholangiocarcinoma. Lancet. 383:2168–2179. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shaib YH, Davila JA, McGlynn K and
El-Serag HB: Rising incidence of intrahepatic cholangiocarcinoma in
the United States: A true increase? J Hepatol. 40:472–477. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Welzel TM, Graubard BI, El-Serag HB, Shaib
YH, Hsing AW, Davila JA and McGlynn KA: Risk factors for
intrahepatic and extrahepatic cholangiocarcinoma in the United
States: A population-based case-control study. Clin Gastroenterol
Hepatol. 5:1221–1228. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Xiong J, Wang Y, Huang H, Bian J, Wang A,
Long J, Zheng Y, Sang X, Xu Y, Lu X, et al: Systematic review and
meta-analysis: Cholecystectomy and the risk of cholangiocarcinoma.
Oncotarget. 8:59648–59657. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu S, Jiang B, Li H, He Z, Lv P, Peng C,
Wang Y, Cheng W, Xu Z, Chen W, et al: Wip1 is associated with
tumorigenity and metastasis through MMP-2 in human intrahepatic
cholangiocarcinoma. Oncotarget. 8:56672–56683. 2017.PubMed/NCBI
|
|
6
|
Dover LL, Jacob R, Wang TN, Richardson JH,
Redden DT, Li P and DuBay DA: Improved postoperative survival for
intraductal-growth subtype of intrahepatic cholangiocarcinoma. Am
Surg. 82:1133–1139. 2016.
|
|
7
|
Smith-Cohn MA, Gill D, Voorhies BN,
Agarwal N and Garrido-Laguna I: Case report: Pembrolizumab-induced
type 1 diabetes in a patient with metastatic cholangiocarcinoma.
Immunotherapy. 9:797–804. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang J, Liu Y, Wang B, Lan H, Liu Y, Chen
F, Zhang J and Luo J: Sumoylation in p27kip1 via RanBP2 promotes
cancer cell growth in cholangiocarcinoma cell line QBC939. BMC Mol
Biol. 18:232017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Loosen SH, Roderburg C, Kauertz KL,
Pombeiro I, Leyh C, Benz F, Vucur M, Longerich T, Koch A,
Braunschweig T, et al: Elevated levels of circulating osteopontin
are associated with a poor survival after resection of
cholangiocarcinoma. J Hepatol. 67:749–757. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Svoboda P: Long and small noncoding RNAs
during oocyte-to-embryo transition in mammals. Biochem Soc Trans.
45:1117–1124. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu B, Ye B, Yang L, Zhu X, Huang G, Zhu
P, Du Y, Wu J, Qin X, Chen R, et al: Long noncoding RNA lncKdm2b is
required for ILC3 maintenance by initiation of Zfp292 expression.
Nat Immunol. 18:499–508. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhu P, Wang Y, Wu J, Huang G, Liu B, Ye B,
Du Y, Gao G, Tian Y, He L, et al: LncBRM initiates YAP1 signalling
activation to drive self-renewal of liver cancer stem cells. Nat
Commun. 7:136082016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xu YC, Liang CJ, Zhang DX, Li GQ, Gao X,
Fu JZ, Xia F, Ji JJ, Zhang LJ, Li GM, et al: LncSHRG promotes
hepatocellular carcinoma progression by activatingHES6. Oncotarget.
8:70630–70641. 2017.PubMed/NCBI
|
|
14
|
Monteleone NJ and Lutz CS: miR-708-5p: A
microRNA with emerging roles in cancer. Oncotarget. 8:71292–71316.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Faghihi F and Moustafa AA: Impaired
neurogenesis of the dentate gyrus is associated with pattern
separation deficits: A computational study. J Integr Neurosci.
15:277–293. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xu Y, Leng K, Li Z, Zhang F, Zhong X, Kang
P, Jiang X and Cui Y: The prognostic potential and carcinogenesis
of long non-coding RNA TUG1 in human cholangiocarcinoma.
Oncotarget. 8:65823–65835. 2017.PubMed/NCBI
|
|
17
|
Shi X, Zhang H, Wang M, Xu X, Zhao Y, He
R, Zhang M, Zhou M, Li X, Peng F, et al: LncRNA AFAP1-AS1 promotes
growth and metastasis of cholangiocarcinoma cells. Oncotarget.
8:58394–58404. 2017.PubMed/NCBI
|
|
18
|
Li J, Zhang Q, Fan X, Mo W, Dai W, Feng J,
Wu L, Liu T, Si L, Xu S, et al: The long noncoding RNA TUG1 acts as
a competing endogenous RNA to regulate the Hedgehog pathway by
targeting miR-132 in hepatocellular carcinoma. Oncotarget.
8:65932–65945. 2017.PubMed/NCBI
|
|
19
|
Wang Y, Zhang Y, Yang T, Zhao W, Wang N,
Li P, Zeng X and Zhang W: Long non-coding RNA MALAT1 for promoting
metastasis and proliferation by acting as a ceRNA of miR-144-3p in
osteosarcoma cells. Oncotarget. 8:59417–59434. 2017.PubMed/NCBI
|
|
20
|
Puik JR, Meijer LL, Le Large TYS, Prado
MM, Frampton AE, Kazemier G and Giovannetti E: miRNA profiling for
diagnosis, prognosis and stratification of cancer treatment in
cholangiocarcinoma. Pharmacogenomics. 18:1343–1358. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang Q, Tang H, Yin S and Dong C:
Downregulation of microRNA-138 enhances the proliferation,
migration and invasion of cholangiocarcinoma cells through the
upregulation of RhoC/p-ERK/MMP-2/MMP-9. Oncol Rep. 29:2046–2052.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhu P, Wang Y, Huang G, Ye B, Liu B, Wu J,
Du Y, He L and Fan Z: lnc-β-Catm elicits EZH2-dependent β-catenin
stabilization and sustains liver CSC self-renewal. Nat Struct Mol
Biol. 23:631–639. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen ZZ, Huang L, Wu YH, Zhai WJ, Zhu PP
and Gao YF: LncSox4 promotes the self-renewal of liver
tumour-initiating cells through Stat3-mediated Sox4 expression. Nat
Commun. 7:125982016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang P and Lv L: miR-26a induced the
suppression of tumor growth of cholangiocarcinoma via KRT19
approach. Oncotarget. 7:81367–81376. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhao L and Zhang Y and Zhang Y: Long
noncoding RNA CASC2 regulates hepatocellular carcinoma cell
oncogenesis through miR-362-5p/Nf-κB axis. J Cell Physiol. Jan
10–2018.Epub ahead of print. View Article : Google Scholar
|
|
26
|
Zhu S, Zhang X, Weichert-Leahey N, Dong Z,
Zhang C, Lopez G, Tao T, He S, Wood AC, Oldridge D, et al: LMO1
synergizes with MYCN to promote neuroblastoma initiation and
metastasis. Cancer Cell. 32:310–323.e5. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cao Y, Jin Y, Yu J, Wang J, Qiu Y, Duan X,
Ye Y, Cheng Y, Dong L, Feng X, et al: Clinical evaluation of
integrated panel testing by next-generation sequencing for somatic
mutations in neuroblastomas with MYCN unamplification. Oncotarget.
8:49689–49701. 2017.PubMed/NCBI
|
|
28
|
Ambrosio S, Amente S, Saccà CD, Capasso M,
Calogero RA, Lania L and Majello B: LSD1 mediates MYCN control of
epithelial-mesenchymal transition through silencing of metastatic
suppressor NDRG1 gene. Oncotarget. 8:3854–3869. 2017. View Article : Google Scholar :
|
|
29
|
Zhang JY, Weng MZ, Song FB, Xu YG, Liu Q,
Wu JY, Qin J, Jin T and Xu JM: Long noncoding RNA AFAP1-AS1
indicates a poor prognosis of hepatocellular carcinoma and promotes
cell proliferation and invasion via upregulation of the RhoA/Rac2
signaling. Int J Oncol. 48:1590–1598. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hu G, Dong B, Zhang J, Zhai W, Xie T,
Huang B, Huang C, Yao X, Zheng J, Che J, et al: The long noncoding
RNA HOTAIR activates the Hippo pathway by directly binding to SAV1
in renal cell carcinoma. Oncotarget. 8:58654–58667. 2017.PubMed/NCBI
|
|
31
|
Wu F, Zhang C, Cai J, Yang F, Liang T, Yan
X, Wang H, Wang W, Chen J and Jiang T: Upregulation of long
noncoding RNA HOXA-AS3 promotes tumor progression and predicts poor
prognosis in glioma. Oncotarget. 8:53110–53123. 2017.PubMed/NCBI
|
|
32
|
Wu J, Cheng G, Zhang C, Zheng Y, Xu H,
Yang H and Hua L: Long noncoding RNA LINC01296 is associated with
poor prognosis in prostate cancer and promotes cancer-cell
proliferation and metastasis. Onco Targets Ther. 10:1843–1852.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yuan Z, Yu X, Ni B, Chen D, Yang Z, Huang
J, Wang J, Chen D and Wang L: Overexpression of long non-coding
RNA-CTD903 inhibits colorectal cancer invasion and migration by
repressing Wnt/β-catenin signaling and predicts favorable
prognosis. Int J Oncol. 48:2675–2685. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Seitz AK, Christensen LL, Christensen E,
Faarkrog K, Ostenfeld MS, Hedegaard J, Nordentoft I, Nielsen MM,
Palmfeldt J, Thomson M, et al: Profiling of long non-coding RNAs
identifies LINC00958 and LINC01296 as candidate oncogenes in
bladder cancer. Sci Rep. 7:3952017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tsubota S, Kishida S, Shimamura T, Ohira
M, Yamashita S, Cao D, Kiyonari S, Ushijima T and Kadomatsu K:
PRC2-mediated transcriptomic alterations at the embryonic stage
govern tumorigenesis and clinical outcome in MYCN-Driven
neuroblastoma. Cancer Res. 77:5259–5271. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sun G, Lu J, Zhang C, You R, Shi L, Jiang
N, Nie D, Zhu J, Li M and Guo J: MiR-29b inhibits the growth of
glioma via MYCN dependent way. Oncotarget. 8:45224–45233.
2017.PubMed/NCBI
|