Leukemia is a complex hematological malignancy,
characterised by clonal proliferation of malignant hematopoietic
stem cells in the blood and bone marrow (1), with a total of 350,000 new cases and
25,700 deaths annually (2). Genetic
as well as environmental factors have been suggested to be
associated with leukemia, including trisomy 21, gender,
cytotoxicity of anticancer drugs, exposure to benzene and ionising
radiation (3–6). Leukemia is a heterogeneous disease that
comprises acute lymphocytic leukemia (ALL), chronic lymphocytic
leukemia (CLL), acute myelogenous leukemia (AML) and chronic
myelogenous leukemia (CML). ALL accounts for 81% of childhood
leukemia cases, while CLL and AML frequently occur in adults
(7).
Racial and ethnic disparities have been identified
in the expression of leukemia-related genes, the clinical outcome
and the mortality rate of leukemia (8–12). These
disparities are likely due to a combination of genetic,
environmental and socioeconomic factors (13), which may affect epigenetic changes.
Epigenetics, such as DNA methylation, have been shown to play an
important role in cancer susceptibility (14,15).
Therefore, DNA methylation studies may help elucidate these racial
and ethnic disparities in leukemia patients.
Aberrant DNA methylation of genes has been shown to
be associated with a large number of human malignancies (16,17).
Although a recent meta-analysis by our group identified significant
associations between a number of aberrantly methylated genes and
leukemia (18), the majority of the
studies published in the Chinese language are overlooked. Thus, the
aim of the present study was to focus on the association of
aberrant DNA methylation and leukemia susceptibility in the Chinese
population and to investigate ethnic differences in DNA methylation
using subgroup meta-analyses.
A systematic literature search was performed through
the National Center for Biotechnology Information (NCBI) PubMed and
Wanfang literature databases, updated until July 10, 2014. The
search was performed using the keywords ‘leukemia’ and
‘methylation’. Potentially relevant articles were identified by
their titles and abstracts, followed by selection of eligible
studies based on full-text analysis. Case-control studies on gene
methylation in Chinese leukemia patients containing sufficient
information on methylation to calculate the odds ratios (ORs) and
95% confidence intervals (CIs) were considered to be eligible. A
flow chart of the study selection process is shown in Fig. 1.
The following characteristics were extracted from
each eligible study: First author's name, year of publication,
disease category and methylation status of cases and controls. All
the studies included were reviewed by three authors (D.J., Y.S. and
C.X.). For genes with methylation data in other populations, the
corresponding data were retrieved and subjected to meta-analyses
for comparison with the Chinese population.
Review Manager 5.0 software (The Nordic Cochrane
Centre, The Cochrane Collaboration, Copenhagen, Denmark) was used
for the meta-analysis. The ORs and 95% CIs were calculated to
evaluate the association between gene methylation and leukemia.
Heterogeneity of the included studies was assessed using
I2 statistics (19). When
there was significant heterogeneity (I2>50%), the
random-effects model was used to calculate the overall OR and 95%
CI; otherwise, the fixed-effects model was applied (20).
In the present study, eligible studies were
retrieved from the NCBI PubMed and Wanfang literature databases and
a systematic meta-analysis was performed to investigate the
association between the methylation status of 53 genes and
leukemia, with the aim of providing evidence regarding the role of
gene methylation in the pathogenesis of leukemia, particularly in
different leukemia subgroups and ethnic groups.
Aberrant gene promoter methylation, occurring in
almost every tumor type, is one of several mechanisms of gene
inactivation (21). Promoter
hypermethylation of tumor suppressor genes often contributes to
loss of function and cancer development (22,23). One
potential mechanism for hypermethylation-induced silencing is
changing the structure of specific binding sites for certain
transcriptional regulators (24).
Epigenetic silencing of genes by promoter hypermethylation is
associated with the loss of tumor suppression, increasing tumor
severity and reducing patient survival (25). The present meta-analysis revealed
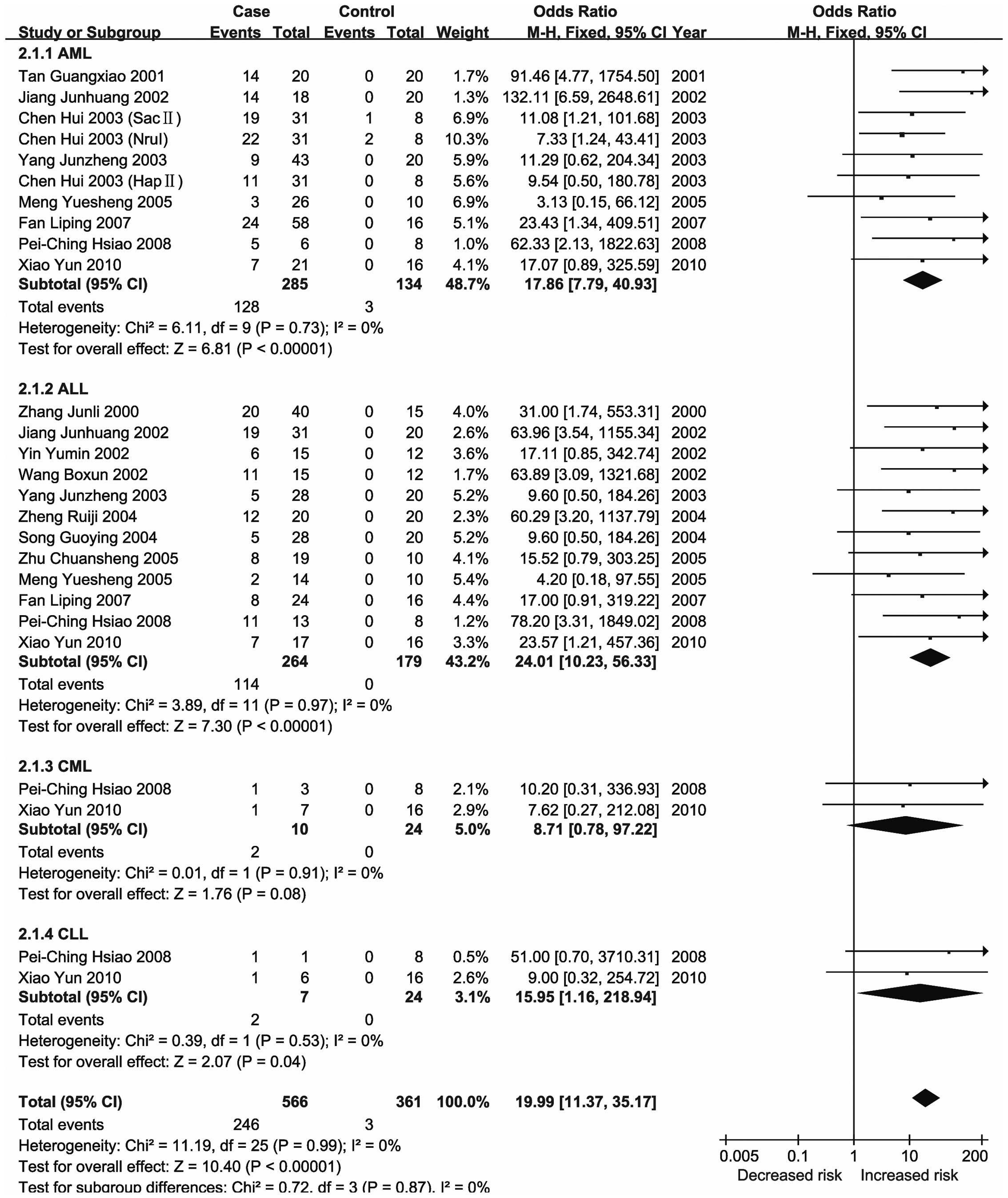
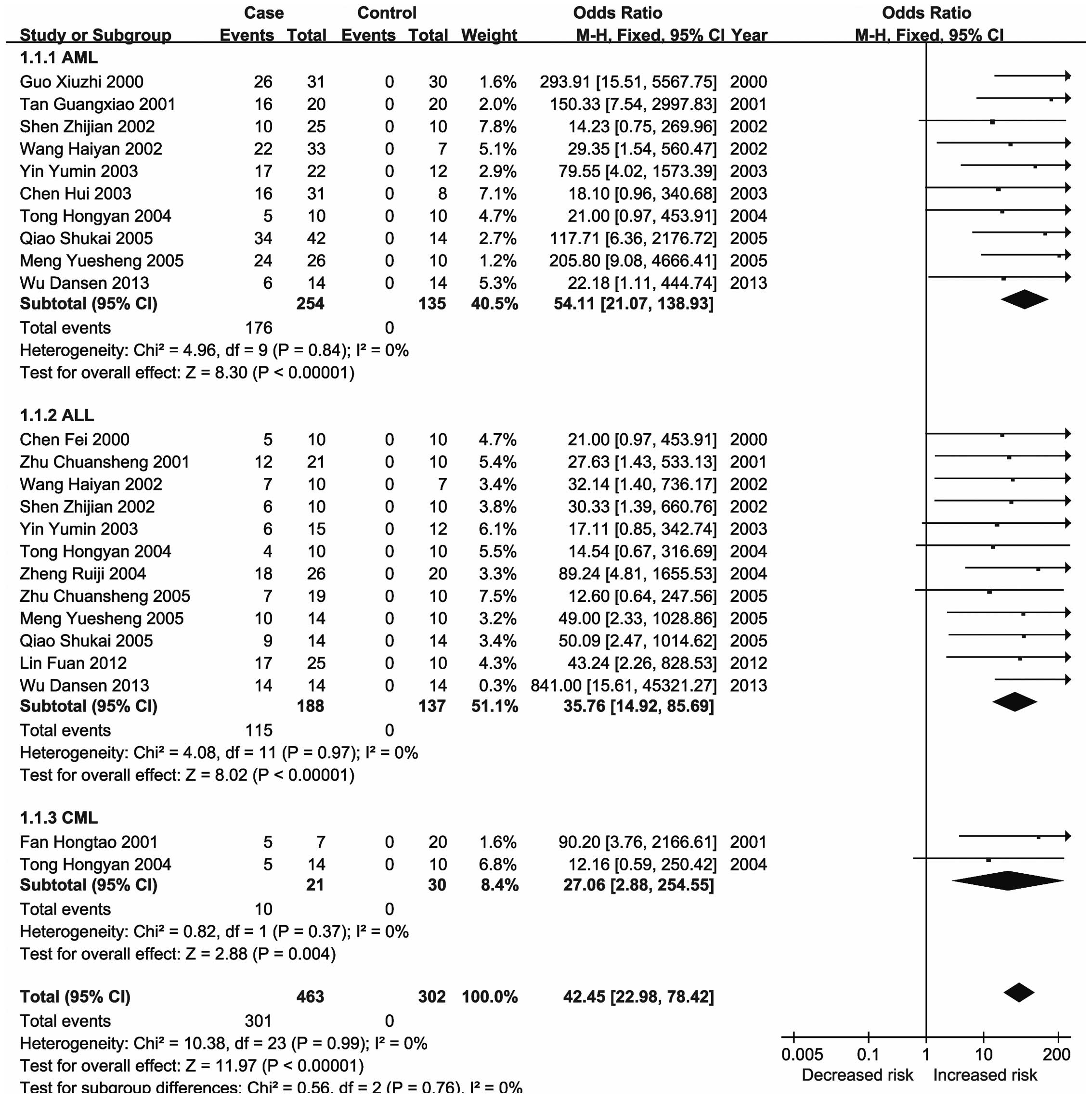
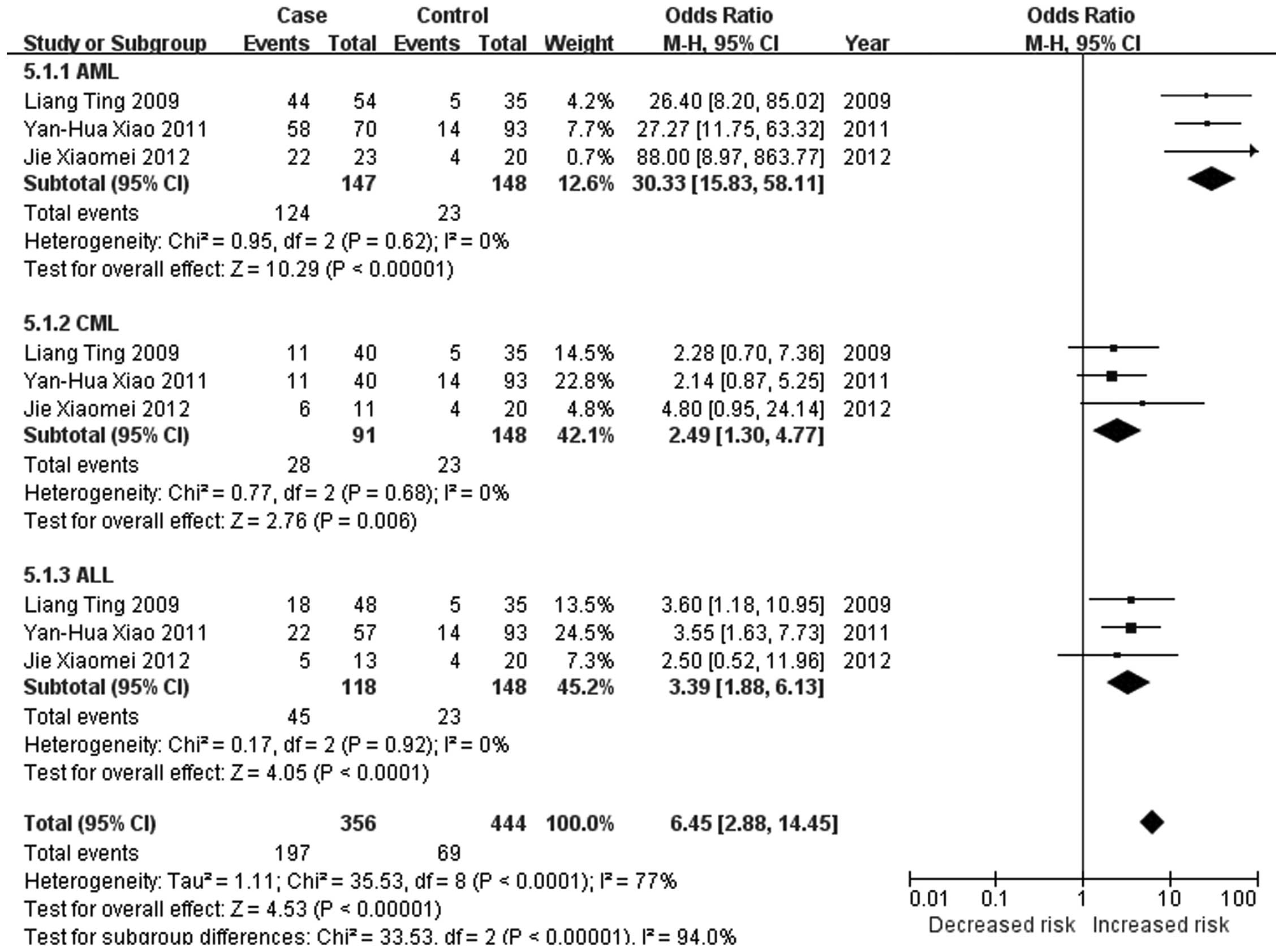
significant changes in the methylation status of the CDKN2A,
CDKN2B, ID4, GliPR1, p73 and Wilms'
tumor 1 (WT1) genes in the major types of leukemia (21,23,26–28).
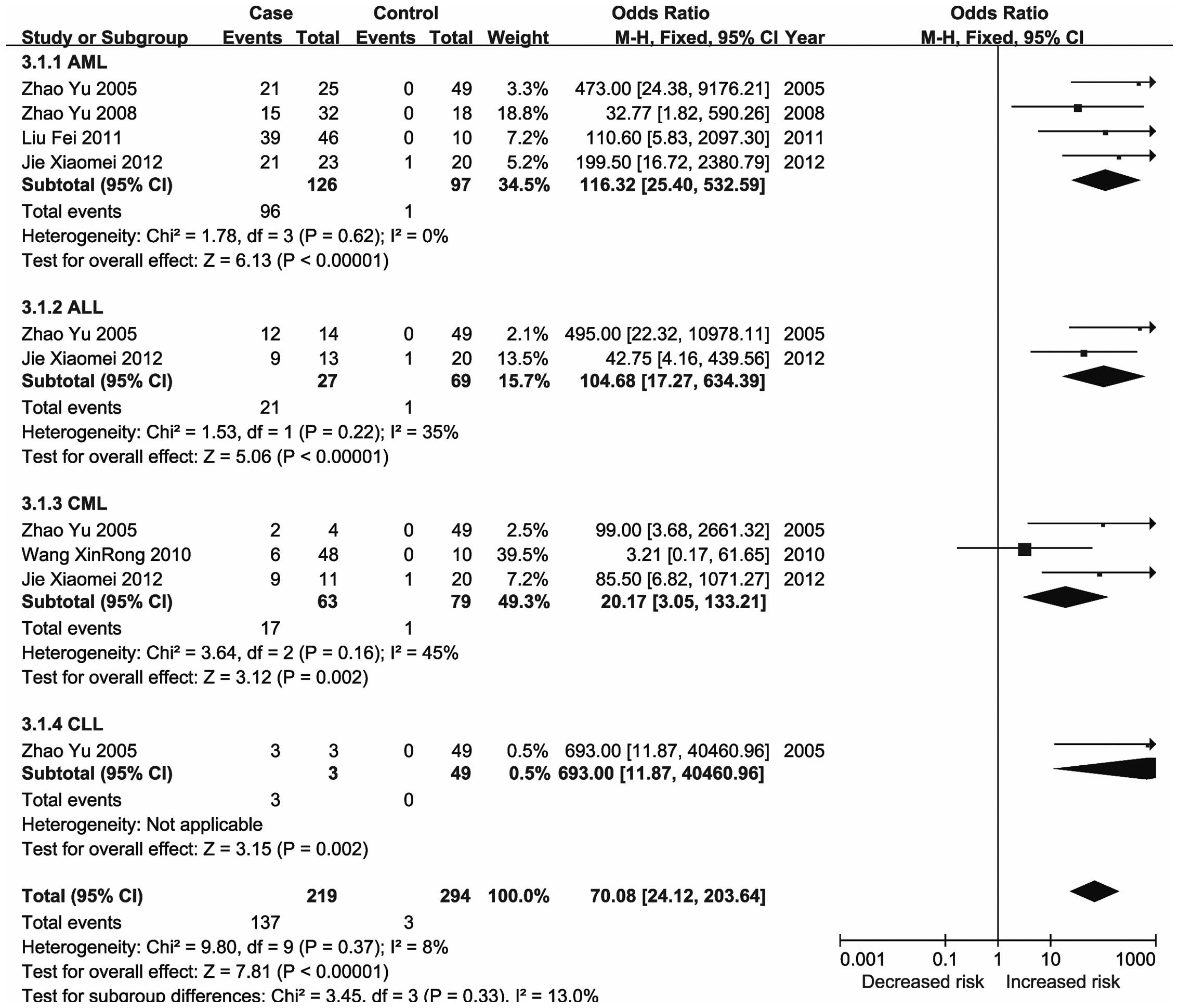
The ID4 protein is a member of the dominant-negative
basic helix-loop-helix transcription factor family that lacks
DNA-binding activity (126) and has
a tumor suppressor function. The promoter of ID4 was
reported to be consistently methylated to various degrees in CLL
and a univariate analysis demonstrated that increased promoter
methylation of ID4 was correlated with shortened patient
survival (127). Previous studies
also reported that ID4 gene promoter hypermethylation was
highly correlated with acute leukemia and may reflect the malignant
degree of AML (128,129). The results of the present
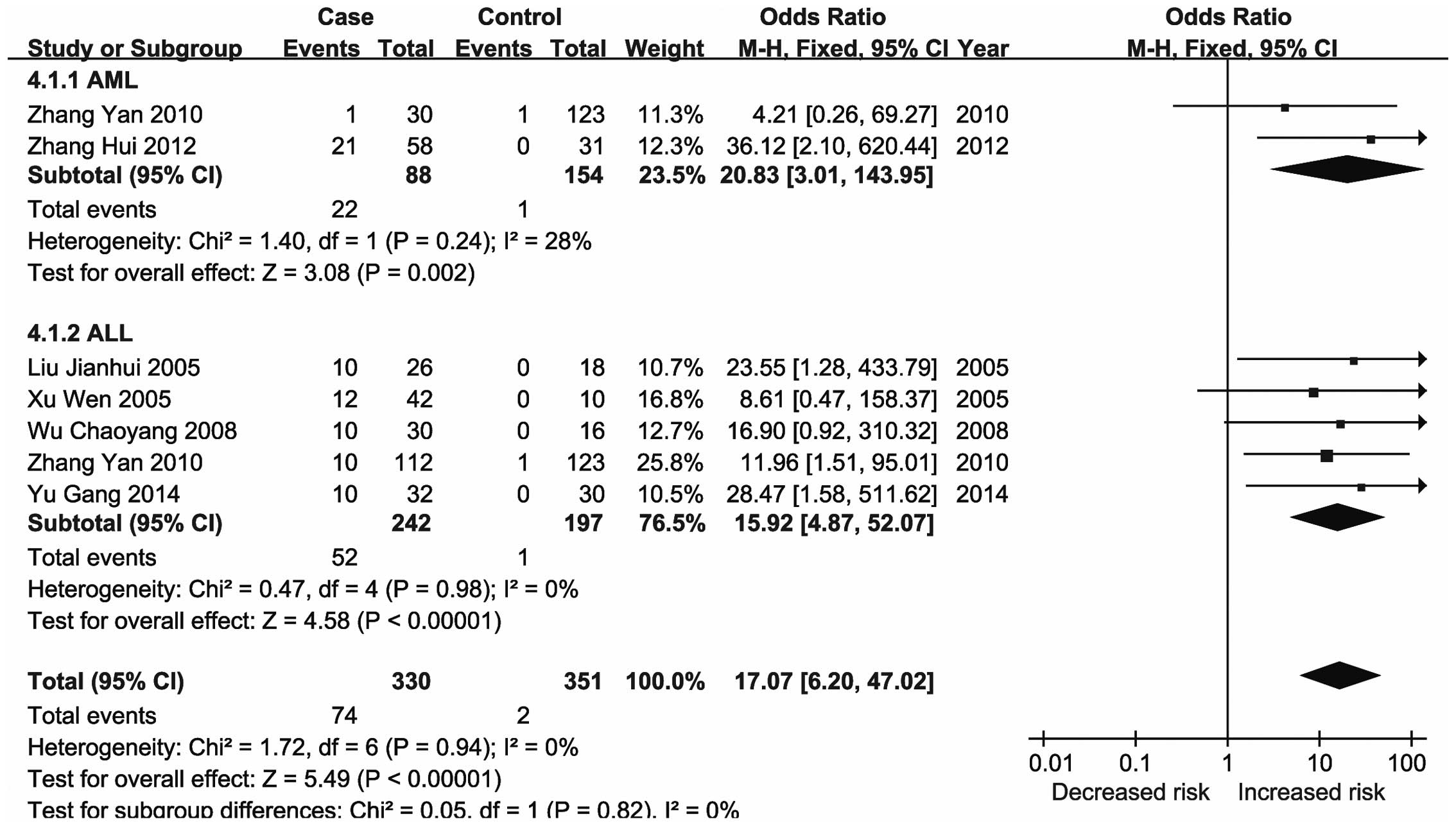
meta-analysis demonstrated that methylation of the ID4 gene
was associated with an increased risk of leukemia, particularly
CML.
Previous studies have reported that the risk of
hematological malignancies varies significantly among different
ethnic groups (9,13,134,135).
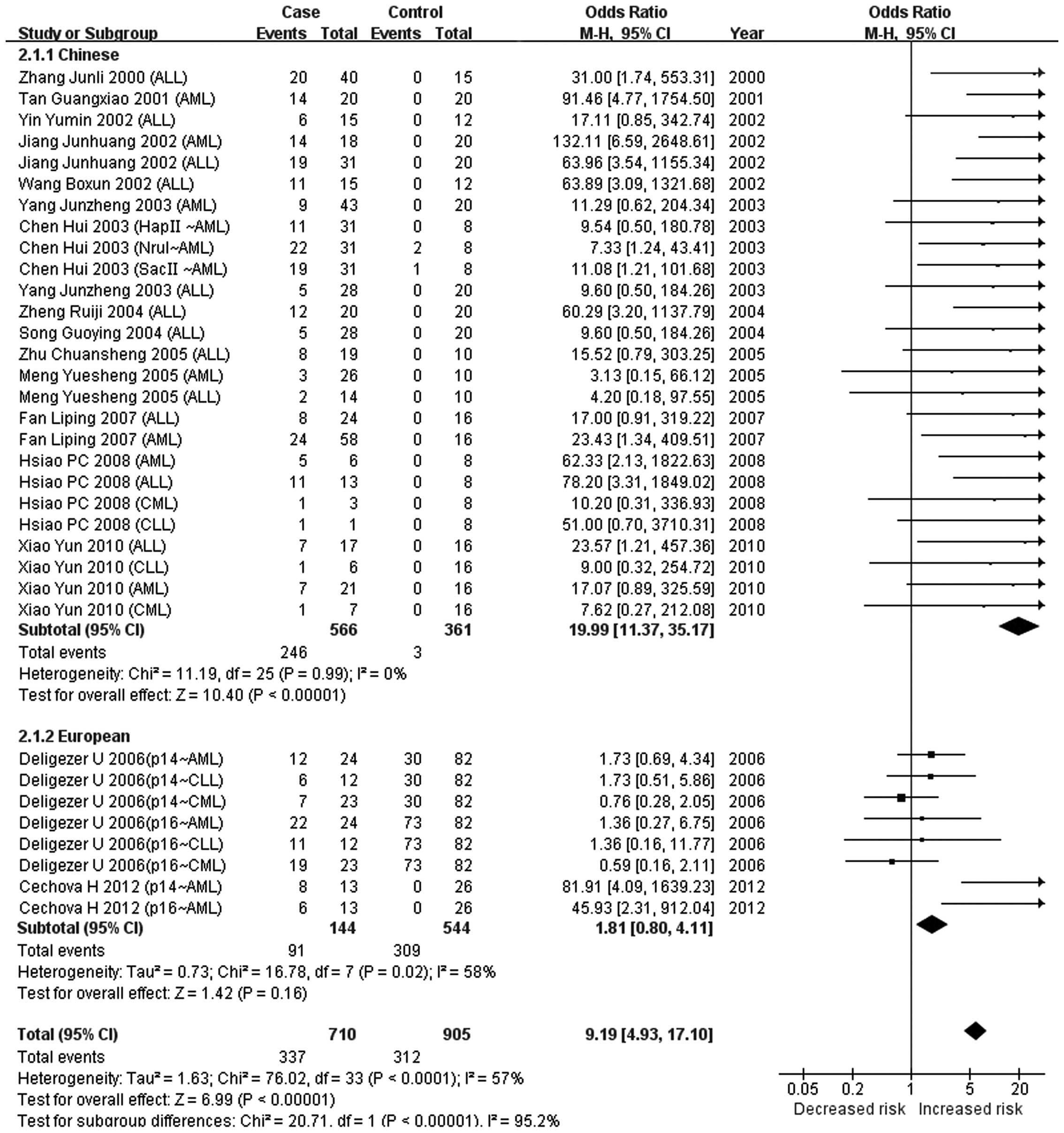
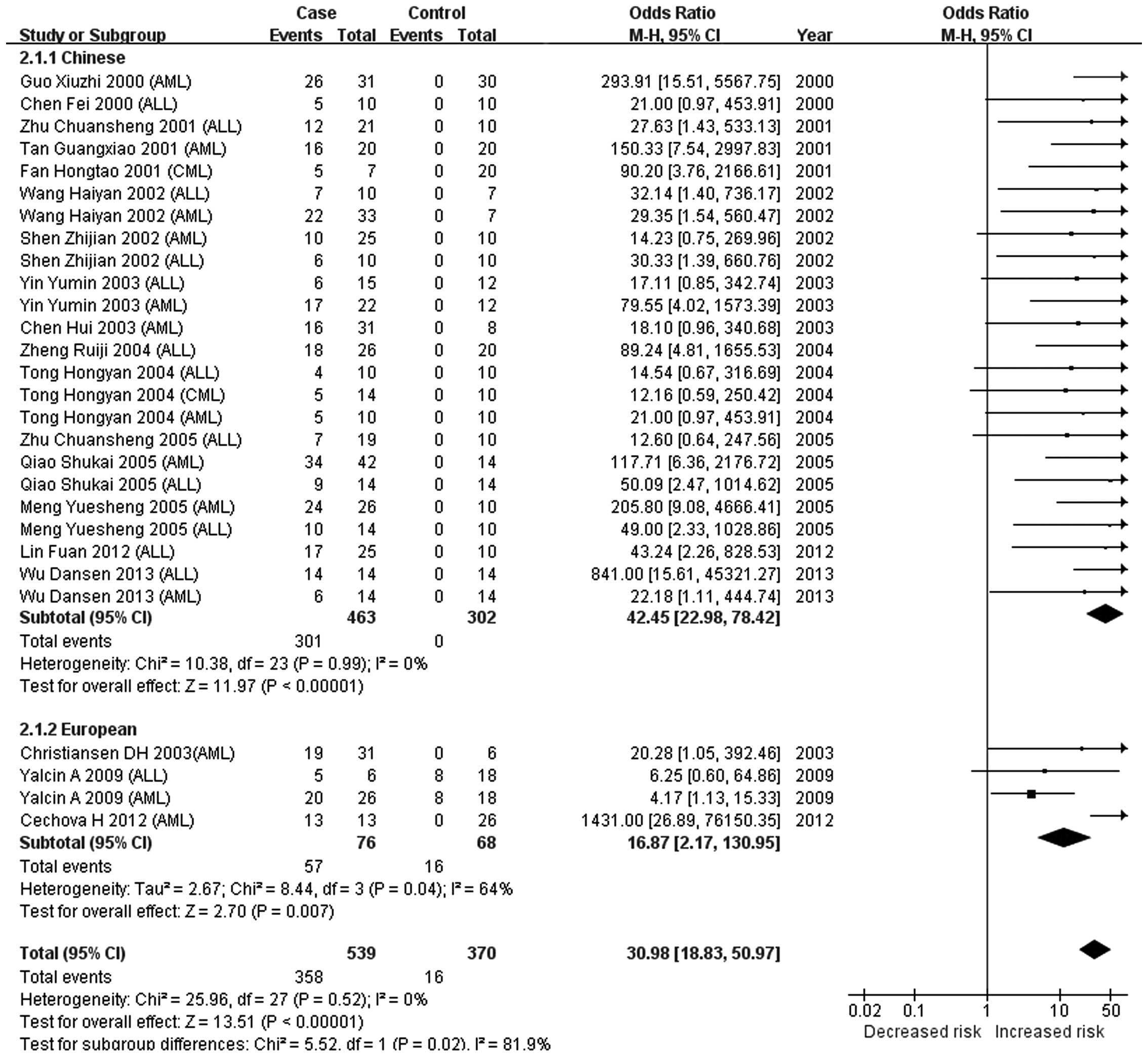
The present meta-analysis indicated that there was no association
between CDKN2A methylation and the risk of leukemia (P=0.16)
in Europeans, while a significant association was observed in
Chinese populations (P<0.00001). A significant difference in the
association of CDKN2A methylation with leukemia was observed
between European and Chinese populations (P<0.00001). This
result may provide molecular evidence to guide future
individualization of chemotherapy for leukemia, although further
research is required to elucidate the precise nature of the ethnic
differences in leukemia.
Of note, the present meta-analysis had certain
limitations. First, the numbers of the studies regarding each gene
and leukemia subtype were uneven. For certain leukemia subtypes,
only a few studies on certain genes were available. The lack of
association of the methylation status of certain genes with several
leukemia subtypes may have been due to a lack of statistical power
of the respective studies, so that the negative results must be
interpreted with caution. Furthermore, a language bias was present,
as only studies written in Chinese and English were included.
In conclusion, the present meta-analysis revealed
that aberrant DNA methylation of the promoters of 47 genes was
associated with leukemia. Further subgroup meta-analysis revealed 5
hypermethylated genes (CDKN2A, CDKN2B, ID4,
GliPR1 and p73) in various leukemia subtypes. In
addition, a difference in the association of CDKN2A and
CDKN2B hypermethylation with leukemia was identified between
Chinese and European populations. The results of the present study
may enhance the current understanding of the association of DNA
methylation with leukemia in the Chinese population.
The present study was supported by grants from
National Natural Science Foundation of China (nos. 31100919 and
81371469), Natural Science Foundation of Zhejiang Province (no.
LR13H020003), K.C. Wong Magna Fund at Ningbo University and Ningbo
Social Development Research Projects (nos. 2010C50019 and
2012C50032).
|
1
|
Davis AS, Viera AJ and Mead MD: Leukemia:
An overview for primary care. Am Fam Physician. 89:731–738.
2014.PubMed/NCBI
|
|
2
|
Zhong S, Chen Z, Yu X, Chen W, Lv M, Ma T
and Zhao J: Tea consumption and leukemia risk: A meta-analysis.
Tumour Biol. 35:5205–5212. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Greim H, Kaden DA, Larson RA, et al: The
bone marrow niche, stem cells, and leukemia: Impact of drugs,
chemicals, and the environment. Ann NY Acad Sci. 1310:7–31. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Laurier D, Grosche B, Auvinen A, et al:
Childhood leukaemia risks: From unexplained findings near nuclear
installations to recommendations for future research. J Radiol
Prot. 34:R53–R68. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Malinge S, Chlon T, Doré LC, et al:
Development of acute megakaryoblastic leukemia in Down syndrome is
associated with sequential epigenetic changes. Blood. 122:e33–e43.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wakeford R: The risk of childhood
leukaemia following exposure to ionising radiation - a review. J
Radiol Prot. 33:1–25. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Woo JS, Alberti MO and Tirado CA:
Childhood B-acute lymphoblastic leukemia: A genetic update. Exp
Hematol Oncol. 3:162014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bhatia S: Disparities in cancer outcomes:
Lessons learned from children with cancer. Pediatr Blood Cancer.
56:994–1002. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Carranza C, Granados L, Morales O, et al:
Frequency of the ETV6-RUNX1, BCR-ABL1, TCF3-PBX1, and MLL-AFF1
fusion genes in Guatemalan pediatric acute lymphoblastic leukemia
patients and their ethnic associations. Cancer Genet. 206:227–232.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Goggins WB and Lo FF: Racial and ethnic
disparities in survival of US children with acute lymphoblastic
leukemia: Evidence from the SEER database 1988–2008. Cancer Causes
Control. 23:737–743. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kennedy AE, Kamdar KY, Lupo PJ, et al:
Genetic markers in a multi ethnic sample for childhood acute
lymphoblastic leukemia risk. Leuk Lymphoma. 56:169–174. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Savage SA: Genomic clues to ethnic
differences in ALL. Blood. 123:2440–2442. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lim JY, Bhatia S, Robison LL and Yang JJ:
Genomics of racial and ethnic disparities in childhood acute
lymphoblastic leukemia. Cancer. 120:955–962. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Adkins RM, Krushkal J, Tylavsky FA and
Thomas F: Racial differences in gene-specific DNA methylation
levels are present at birth. Birth Defects Res A Clin Mol Teratol.
91:728–736. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xia YY, Ding YB, Liu XQ, et al:
Racial/ethnic disparities in human DNA methylation. Biochim Biophys
Acta. 1846:258–262. 2014.PubMed/NCBI
|
|
16
|
Baylin SB and Jones PA: A decade of
exploring the cancer epigenome - biological and translational
implications. Nat Rev Cancer. 11:726–734. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Plass C, Pfister SM, Lindroth AM, et al:
Mutations in regulators of the epigenome and their connections to
global chromatin patterns in cancer. Nat Rev Genet. 14:765–780.
2013. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jiang D, Hong Q, Shen Y, et al: The
diagnostic value of DNA methylation in leukemia: A systematic
review and meta-analysis. PLoS One. 9:e968222014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Higgins JP, Thompson SG, Deeks JJ and
Altman DG: Measuring inconsistency in meta-analyses. BMJ.
327:557–560. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bax L, Ikeda N, Fukui N, Yaju Y, Tsuruta H
and Moons KG: More than numbers: The power of graphs in
meta-analysis. Am J Epidemiol. 169:249–255. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pluta A, Nyman U, Joseph B, Robak T,
Zhivotovsky B and Smolewski P: The role of p73 in hematological
malignancies. Leukemia. 20:757–766. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cahill N and Rosenquist R: Uncovering the
DNA methylome in chronic lymphocytic leukemia. Epigenetics.
8:138–148. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Esteller M: CpG island hypermethylation
and tumor suppressor genes: A booming present, a brighter future.
Oncogene. 21:5427–5440. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhao Y, Guo J, Zhang X, et al:
Downregulation of p21 in myelodysplastic syndrome is associated
with p73 promoter hypermethylation and indicates poor prognosis. Am
J Clin Pathol. 140:819–827. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Volodko N, Gordon M, Salla M, Ghazaleh HA
and Baksh S: RASSF tumor suppressor gene family: Biological
functions and regulation. FEBS Lett. 588:2671–2684. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Benezra R, Davis RL, Lockshon D, Turner DL
and Weintraub H: The protein Id: A negative regulator of
helix-loop-helix DNA binding proteins. Cell. 61:49–59. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bodoor K, Haddad Y, Alkhateeb A, et al:
DNA hypermethylation of cell cycle (p15 and p16) and apoptotic
(p14, p53, DAPK and TMS1) genes in peripheral blood of leukemia
patients. Asian Pac J Cancer Prev. 15:75–84. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chmelařová M, Křepinská E, Spaček J, Laco
J, Nekvindová J and Palička V: Methylation analysis of tumour
suppressor genes in ovarian cancer using MS-MLPA. Folia Biol
(Praha). 58:246–250. 2012.PubMed/NCBI
|
|
29
|
Cechova H, Lassuthova P, Novakova L, et
al: Monitoring of methylation changes in 9p21 region in patients
with myelodysplastic syndromes and acute myeloid leukemia.
Neoplasma. 59:168–174. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Deligezer U, Erten N, Akisik EE and Dalay
N: Methylation of the INK4A/ARF locus in blood mononuclear cells.
Ann Hematol. 85:102–107. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hsiao PC, Liu MC, Chen LM, et al: Promoter
methylation of p16 and EDNRB gene in leukemia patients in Taiwan.
Chin J Physiol. 51:27–31. 2008.PubMed/NCBI
|
|
32
|
Zheng R, Shen S, Shen J and Ma X: Studies
on p16, p15, p18 and p19 methylation of INK4 series cancer
inhibitive gene in leukemia. J Fujian Med Univ. 38:257–260.
2004.
|
|
33
|
Xiao Y, Liu Y, Jiang L and Ran L: Study on
the deletion point mutation and promoter methylation analysis of
p16 in leukemia. Guizhou Med J. 34:784–787. 2010.
|
|
34
|
Yang J, Li X, Sun L, Song G, Chen H and
Liu Y: Expression of p16 gene in adult acute leukemia. J Zhengzhou
Univ (Med Sci Ed.). 38:33–36. 2003.
|
|
35
|
Song G, Yang J, Sun H, Liu C, Chen H and
Li X: Deletion and methylation of p16 gene in an adult acute
leukemia. Henan J Oncol. 17:229–231. 2004.
|
|
36
|
Tan G and Guo X: Methods of CpG islands
methylation detection. J Jinan Univ. 22:24–26. 2001.
|
|
37
|
Zhu C, Xu W, Liu D, Wang Y and Bi K: Study
of the methylation of p15 and p16 gene in acute leukemia. Shandong
Med J. 45:4–5. 2005.
|
|
38
|
Zhang J, Wu J and Tan Y: The relationship
between P16 gene inactivation and expression of mRNA in ALL. J
Wenzhou Med Coll. 30:5–7. 2000.
|
|
39
|
Fan L, Shen J, Ye B, et al: Detection of
p16 gene methylation status in adult patients with acute leukemia
by using n-MSP. J Exp Hematol. 15:258–261. 2007.
|
|
40
|
Jiang J, Shen J, Yan J, et al: The value
of p15 gene methylation in minimal residual disease of childhood
acute leukemia by MSP. China Child Blood. 7:1–3. 2002.
|
|
41
|
Meng Y, Yu H, Guo C, et al: Quantitative
analysis of p15 and p16 gene methylation and mRNA expression levels
in acute leukemia patients. Chin J Hematol. 26:434–435. 2005.
|
|
42
|
Wang B: The clinical significance of p16
gene methylation in leukemia and other hematologic malignancies.
Master's degree dissertation, China Medical University. 2002.
|
|
43
|
Yin Y: The clinical significance of p15
gene methylation in leukemia and other hematologic malignancies.
Master's degree dissertation, China Medical University. 2002.
|
|
44
|
Chen H, Sun H, Liu C, Liu Y, Song G and Li
X: Methylation of p16/p15 exon 2 in adult acute myeloid leukemia. J
Zhengzhou Univ (Med Sci Ed.). 38:722–724. 2003.
|
|
45
|
Lin F, Ye B, Shen J, et al: Detection of
p15 methylation and deletion status in acute lymphoblastic leukemia
using hn-MSP. J Leuk Lymphoma. 21:208–212. 2012.
|
|
46
|
Zhu C, Bi K and Liu X: Homozygous deletion
and methylation of P15 gene in acute laukemia. Shandong Med J
Shandong Yiyao. 41:10–11. 2001.
|
|
47
|
Shen Z, Zhang J, Yu K and Jiang S:
Evaluation of methylation-specific polymerase chain reaction and
its potential clinical value in detecting minimal residual disease
of adult acute leukemia with P15. J Wenzhou Med Coll. 32:230–232.
2002.
|
|
48
|
Fan H, Guo X, Tan G, Wu Q, Zhou T and Guo
Q: Study on methylation of p15 INK4B in myelodysplastic syndrome.
Chin J Pathophysiol. 17:349–352. 2001.
|
|
49
|
Tong H and Lin M: Methylation of p15INK4B
gene and its mechanism in patients with myelodysplastic syndrome
and leukemia. Chin J Pathophysiol. 20:2257–2260. 2004.
|
|
50
|
Guo X, Fan H, Wu Q, et al: Study on
methylation of p15INK4B gene in acute myeloid leukemia and chronic
myeloid leukemia. J Exp Hematol. 8:257–260. 2000.
|
|
51
|
Qiao S, Xu S, Guo X and Wang Y: Clinical
significance of the expression of DNA methyltransferase genes
(DNMT) in acute leukemia patients. J Exp Hematol. 13:260–265.
2005.
|
|
52
|
Chen F and Zeng X: Detection of p15 gene
hypermethylation in acute leukemia by MSP. Chin J Hematol.
21:2502000.
|
|
53
|
Yin Y, Li Y and Liu D: The clinical
significance of p15 gene methylation in leukemia and other
hematologic malignancies. Chin J Hematol. 24:383–384. 2003.
|
|
54
|
Wu D: p15INK4b gene DNA methylation and
histone modification mechanism of leukemia cell mediated by
p15-piRNAs. PhD dissertation, Fujian Medical University. 2013.
|
|
55
|
Wang H: The study of p15 gene methylation
in malignancies. Master's degree dissertation, Qingdao University.
2002.
|
|
56
|
Zhao Y, Wang QS, Li HH, et al:
[Significance of id4 promoter methylation in monitoring AML
patients with completely remission]. J Exp Hematol. 16:476–478.
2008.
|
|
57
|
Wang X, Kang H, Cen J, Li Y, Wang L and Yu
L: Methylation Status of id4 gene promoter in patients with chronic
myeloid leukemia. J Exp Hematol. 18:1402–1404. 2010.
|
|
58
|
Liu F, Xu R, Cui X, Zhang X and Wang Y:
ID4 promoter methylation in acute myeloid leukemia. J Exp Hematol.
19:582–584. 2011.
|
|
59
|
Jie X: WT1, ID4, GLIPR1 aberrant gene
promoter methylation and mRNA expression in leukemia patients.
Master's degree dissertation, Guangdong Medical College. 2012.
|
|
60
|
Zhao Y: The significance of Id4 gene
methylation in hematological malignancies. PhD dissertation, PLA
Postgraduate Medical School. 2005.
|
|
61
|
Xiao YH, Li XH, Tan T, et al:
Identification of GLIPR1 tumor suppressor as methylation-silenced
gene in acute myeloid leukemia by microarray analysis. J Cancer Res
Clin Oncol. 137:1831–1840. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Liang T, Tan T, Xiao Y, et al: Methylation
and expression of glioma pathogenesis-related protein 1 gene in
acute myeloid leukemia. J Cent South Univ (Med Sci Ed.).
34:388–394. 2009.
|
|
63
|
Zhang Y, Gao Y, Wang X, et al: The
relationship between p73 gene methylation and childhood acute
leukemia. Proceedings of the Ninth National Conference on
Environmental and Occupational Medicine Graduate (Shanghai, China).
254–256. 2010.
|
|
64
|
Zhang H, Mo W and Yang Z: The expression
of WWOX and p73 in acute myeloid leukemia. Guangdong Med J.
33:1567–1570. 2012.
|
|
65
|
Wu C, Jin J, Ji Y, et al: Abnormal
expression and significance of p73 gene in adult acute
lymphoblastic leukemia. Jiangsu Med J. 34:339–341. 2008.
|
|
66
|
Liu J, Jiang Y, Xu W and Liang X:
Alterations of p53 and p73 genes in lympho-plasmacytic diseases.
Pract Clin Med. 6:9–13. 2005.
|
|
67
|
Xu W: Study on methylation of the p73 gene
in acute lymphoblastic leukemia and its clinical significances.
Chin J Pract Int Med. 25:905–906. 2005.
|
|
68
|
Yu G: Abnormal expression of p73 gene in
adult acute lymphoblastic leukemia. Med Innov Chin. 2:121–122.
2014.
|
|
69
|
Xie X, Chen Z, Zhang X and Huang M:
Detecting methylation of the calcitonin gene in monitoring
treatment and disease evolution for myelogenous leukemia. Chin J
Cancer. 22:616–619. 2003.
|
|
70
|
Tang Y, Deng C, Du Q and Li G: Study on
hypermethylation of the calcitonin gene in malignant hematological
disorders. J West Chin Univ Med Sci. 32:86–88. 2001.
|
|
71
|
Wang S, Hong W and Peng A: The study of CT
gene methylation in chronic myeloid leukemia. Chin J Hematol.
19:321998.
|
|
72
|
Qian J, Yao D, Lin J, et al: Alteration of
methylation status of death-associated protein kinase (dapk) gene
promoter in patients with acute myeloid leukemia. J Exp Hematol.
18:1390–1394. 2010.
|
|
73
|
Niu Y, Wang P, Wang Y, Wang Y, Cai D and
Li Y: Expression of death-associated protein kinase gene and
methylation status of promoter region in acute leukimia. J Exp
Hematol. 22:30–34. 2014.
|
|
74
|
Zhao W: DAPK gene promoter methylation,
expression and biological function in acute leukemia. PhD
dissertation, Southern Medical University. 2009.
|
|
75
|
Qian J, Wang YL, Lin J, Yao DM, Xu WR and
Wu CY: Aberrant methylation of the death-associated protein kinase
1 (DAPK1) CpG island in chronic myeloid leukemia. Eur J Haematol.
82:119–123. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Lin W, Ye Z and Tan J: Gene methylation
abnormalities in chronic lymphocytic leukemia patients by PCR. J
Qiqihar Med Coll. 31:2376–2377. 2010.
|
|
77
|
Wu B, Chen E and Zhu B: Detecting the
aberrant methylation of progesterone receptor B gene in human
chronic lymphocytic leukemia by MSP. Chin J Hemorheol. 18:140–141.
2008.
|
|
78
|
Zhang X: The experimental study of PR
promoter CpG island methylation in leukemia. Master's degree
dissertation, Third Military Medical University. 2003.
|
|
79
|
Shi X, Chang N, Fan Y, et al: Detection
and clinical significance of SFRP2 promoter methylation status in
patients with acute myeloid leukemia. Chin J Clin, Electron Ed.
5:1962–1966. 2011.
|
|
80
|
Wang H, Zhang G, Yao K, et al: The gene
methylation and protein expression of SFRP5 in leukemic cells. J
China Med Univ. 41:2012.
|
|
81
|
Xu C, Shen J, Shen S, Fu H, Zhu Y and Chen
L: The significance of methylation status of secreted
frizzled-related protein gene promoter in acute leukemia. Chin J
Intern Med. 49:769–771. 2010.
|
|
82
|
Li M: A preliminary study of inactivation
of IGSF4 gene methylation in leukemia cell. PhD dissertation, PLA
Postgraduate Medical School. 2004.
|
|
83
|
Chen W, Huang C, Zhan X, et al: RASSF1A
gene methylation and its clinical significance in leukemia
patients. Lab Med. 27:554–556. 2012.
|
|
84
|
Song J, Li Y, Yang Z, Yang L and Chen J:
Hypermethylation in SFRP2 promoter in human leukemia K562 cells and
bone marrow specimen with chronic myeloid leukemia. Acad J Third
Med Coll PLA. 33:2583–2586. 2011.
|
|
85
|
Dou L: The study of LRP15 gene methylation
in blood diseases. Master's degree dissertation, PLA Postgraduate
Medical School. 2004.
|
|
86
|
Yao C, Fang J, Gao L, Ding J, Shu G and Yu
W: Study on the relationship between the RIZ1 gene expression and
methylation status of the gene promoter in acute myeloid leukemia.
J Southeast Univ (Med Sci Ed.). 29:119–122. 2010.
|
|
87
|
Cai F: RIZ1 gene expression and promoter
methylation status in acute childhood leukemia. PhD dissertation,
Huazhong University Science and Technology. 2012.
|
|
88
|
Shi X, Fan Y, Zhou C, et al: Study of
abnormal methylation in the promoter region of SFRP4 gene in acute
myeloid leukemia. Chin J Clin, Electron Ed. 5:990–995. 2011.
|
|
89
|
Dou LP, Liu JH, Wang C, et al: [Study on
the involvement of ZO-1 gene in leukemogenesis]. Chin J Hematol.
30:473–476. 2009.
|
|
90
|
Wang C, Wang G, Tan Y, Li W, Liu C and Yu
L: The methylation pattern and clinical significance of Zonula
occludens-1 gene promoter in acute leukemia. Chin J Intern Med.
47:111–113. 2008.
|
|
91
|
Chim CS, Wong SY, Pang A, et al: Aberrant
promoter methylation of the retinoic acid receptor alpha gene in
acute promyelocytic leukemia. Leukemia. 19:2241–2246. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Wang G, Liu L, Chen J, et al: Promoter
methylation profile of androgen receptor gene in leukemic cells. J
Third Mil Med Univ. 29:932–934. 2007.
|
|
93
|
Wang X, Li J, Fu B, Guo L, Zhang J and
Huang S: Methylation Status of JunB and CDH13 gene promoter in CD34
CD38−chronic myelogenous leukemia cells. J Exp Hematol.
17:1405–1408. 2009.
|
|
94
|
Liu J: CDH13 gene promoter methylation and
expression in acute myeloid leukemia. Master's degree dissertation,
Guangzhou Medical University. 2013.
|
|
95
|
Wang Y: The study of DDIT3 gene promoter
methylation in hematological tumors. Master's degree dissertation,
Jiangsu University. 2009.
|
|
96
|
Zhu X, Zhu C, Wan Y, et al: Analysis of
methylation of the Dickkopf1 (DKK-1) gene in acute leukemia. J
Shandong Univ (Health Sci). 50:84–87. 2012.
|
|
97
|
Yuan X: The study of EDNRB gene promoter
methylation in acute leukemia. Master's degree dissertation,
Zhengzhou University. 2010.
|
|
98
|
Yu Y: The study of FANCF methylation in
patients with acute myeloid leukemia and myelodysplastic syndrome.
Master's degree dissertation, China Medical University. 2008.
|
|
99
|
Deng N: The study of FANCF/FANCD2
expression in patients with acute myeloid leukemia and
myelodysplastic syndrome. Master's degree dissertation, China
Medical University. 2009.
|
|
100
|
Qian Z: The significance of GRAF gene
promoter methylation in hematopoietic tumors. Master's degree
dissertation, Jiangsu University. 2010.
|
|
101
|
Chen Q: HAGE gene promoter hypomethylation
in myeloid tumors. Master's degree dissertation, Jiangsu
University. 2012.
|
|
102
|
Li Y, Wang Y, Zhou J, Zhou S, Fang G and
Liu X: Analysis and clinical significance of hPer3 promoter
methylation status for CML monitoring. Chin J Pathophysiol.
27:2111–2115. 2011.
|
|
103
|
Wang Y, Zhou J, Zhou S, et al: Promoter
methylation status of hPer3 gene in AML patients and the in
vitro effect of decitabine on the status. Chin J Hematol.
32:317–321. 2011.
|
|
104
|
Wang N: MiR-34b expression, methylation
regulation and clinical significance in acute childhood leukemia.
Master's degree dissertation, Suzhou University. 2013.
|
|
105
|
Chai H: RAGE-1 gene promoter
hypomethylation in myeloid tumors. Master's degree dissertation,
Shanxi Medical University. 2013.
|
|
106
|
Lin D, Fan R and Liu X: Significance of
DNA methylation status of runx3 gene promoter region in acute
leukemia. J Exp Hematol. 16:263–266. 2008.
|
|
107
|
Chim CS, Wong AS and Kwong YL: Epigenetic
dysregulation of the Jak/STAT pathway by frequent aberrant
methylation of SHP1 but not SOCS1 in acute leukaemias. Ann Hematol.
83:527–532. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Wang Y, Zhu CS, Bi KH, Xu WW, Dong L and
Hou M: [Study of WIF-1 promoter methylation with expressions of
β-catenin in acute leukemia]. Chin Med J. 91:2858–2860. 2011.
|
|
109
|
Liu W, Xu C, Guan M and Zhang Y: Aberrant
promoter hypermethylation of AKAP12 gene in acute lymphoblastic
leukemia in children. Lab Med. 23:344–348. 2008.
|
|
110
|
Gao F, Li Y, Liu W, et al: Studies on gene
expression and the 5′ CpG islands methylation status of E-cadherin
in acute myeloid leukemia. Chin J Hematol. 27:25–27. 2006.
|
|
111
|
Yao DM, Qian J, Lin J, et al: Aberrant
methylation of CCAAT/enhancer binding protein zeta promoter in
acute myeloid leukemia. Leuk Res. 35:957–960. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Guan M, Xu C, Liu W and Chen B:
Identification of aberrant methylation of DLC-1 by pyrosequencing
in acute lymphoblastic leukemia in children. Chin J Lab Med.
31:389–393. 2008.
|
|
113
|
Yu L, Wang Q, Gu Q, Shi Z and Jin H: Study
on aberrant DNA methylation of dopamine D4 receptor gene in acute
myeloid leukemia. Acad J Chin PLA Postgrad Med Sch. 21:62–64.
2000.
|
|
114
|
Tao YF, Xu LX, Lu J, et al:
Metallothionein III (MT3) is a putative tumor suppressor gene that
is frequently inactivated in pediatric acute myeloid leukemia by
promoter hypermethylation. J Transl Med. 12:1822014. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Zheng Y: Abnormal methylation of the
promoter region of the p53 gene in acute leukemia. Master's degree
dissertation, Sun Yat-sen University. 2007.
|
|
116
|
Li Y, Qian J, Chen Q, et al: Expression
pattern and promoter hypermethylation of PDLIM4 gene in patients
with chronic myeloid leukemia. J Jiangsu Univ (Med Ed.). 23:17–21.
2013.
|
|
117
|
Song JJ, Liu Q, Li Y, et al: Epigenetic
inactivation of PLCD1 in chronic myeloid leukemia. Int J Mol Med.
30:179–184. 2012.PubMed/NCBI
|
|
118
|
Yao D, Qian J, Lin J, Li Y, Chen Q and
Chen X: Alteration of methylation status of PRAME gene promoter in
patients with chronic myeloid leukemia. J Pract Med. 29:3495–3498.
2013.
|
|
119
|
Yan W, Hu W and Yang W: Expression and
methylation study of PRDX2 gene in acute myeloid leukemia. J China
Med Univ. 41:1047–1049. 2012.
|
|
120
|
Yang J, Du W, He Y, Liu J, Zheng J and
Huang S: Research on PTEN expression and its promoter methylation
in B-ALL patients. J Intern Intens Med. 13:141–143. 2007.
|
|
121
|
Du C, Mao P and Wang Y: The clinical
significance of the RIL gene methylation levels in acute myeloid
leukemia. China J Mod Med. 23:46–51. 2013.
|
|
122
|
Jiao X, Chen Q, Lin J, et al: SALL4 genes
methylation changes and clinical significance in acute myeloid
leukemia patients. J Jiangsu Univ (Med Ed.). 23:532–535. 2013.
|
|
123
|
Zhuang Y, Cheng Y, Wang L, Dou H, Zhu Q
and Hu J: The methylation of SOCS-1 gene in adult acute myeloid
leukemia. Prog Mod Biomed. 18:3417–3420. 2011.
|
|
124
|
Deng G, Li ZQ, Zhao C, et al: WNT5A
expression is regulated by the status of its promoter methylation
in leukaemia and can inhibit leukemic cell malignant proliferation.
Oncol Rep. 25:367–376. 2011.PubMed/NCBI
|
|
125
|
Iacobucci I, Ferrari A, Lonetti A, et al:
CDKN2A/B alterations impair prognosis in adult BCR-ABL1-positive
acute lymphoblastic leukemia patients. Clin Cancer Res.
17:7413–7423. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Hu HB and Hu Q: ID4 methylation patterns
in childhood T line and B line lymphocytic leukemia. Chin J Contemp
Pediatr. 12:940–942. 2010.(In Chinese).
|
|
127
|
Chen SS, Claus R, Lucas DM, et al:
Silencing of the inhibitor of DNA binding protein 4 (ID4)
contributes to the pathogenesis of mouse and human CLL. Blood.
117:862–871. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Liu Y, Kang HY, Wang LL, Lu XC, Zhu HL and
Yu L: Establishment of methylation-specific quantitative PCR system
for ID4 gene in acute leukemia cells and its specificity and
sensitivity. J Exp Hematol. 22:269–274. 2014.(In Chinese).
|
|
129
|
Liu F and Xu RR: Study on the correlation
between Chinese medical syndrome types and ID4 gene promoter
methylation in human acute myeloid leukemia. Chin J Integr Tradit
West Med. 32:471–473. 2012.(In Chinese).
|
|
130
|
Awasthi A, Woolley AG, Lecomte FJ, et al:
Variable expression of GLIPR1 correlates with invasive potential in
melanoma cells. Front Oncol. 3:2252013. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Capalbo G, Mueller-Kuller T, Koschmieder
S, et al: Endoplasmic reticulum protein GliPR1 regulates G protein
signaling and the cell cycle and is overexpressed in AML. Oncol
Rep. 30:2254–2262. 2013.PubMed/NCBI
|
|
132
|
He Z, Liu H, Agostini M, et al: p73
regulates autophagy and hepatocellular lipid metabolism through a
transcriptional activation of the ATG5 gene. Cell Death Differ.
20:1415–1424. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Toska E and Roberts SG: Mechanisms of
transcriptional regulation by WT1 (Wilms tumour 1). Biochem J.
461:15–32. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Dores GM, Devesa SS, Curtis RE, Linet MS
and Morton LM: Acute leukemia incidence and patient survival among
children and adults in the United States, 2001–2007. Blood.
119:34–43. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Shirley MH, Sayeed S, Barnes I, Finlayson
A and Ali R: Incidence of haematological malignancies by ethnic
group in England, 2001-7. Br J Haematol. 163:465–477. 2013.
View Article : Google Scholar : PubMed/NCBI
|