Introduction
Tooth development is a highly conserved and
coordinated process that controls tooth shape and size. Tooth
morphogenesis and development is regulated by
epithelial-mesenchymal interactions (1). The dentin layer is a type of
mesenchymal-derived calcified tissue, which serves as the
supporting tissue of the tooth. The enamel layer is a unique
epithelium-derived calcified tissue, which covers the crown and
forms the shape of the tooth in vertebrates. Dental enamel is
composed of organized hydroxyapatite crystallites formed by the
differentiation of ameloblasts (AMs), the differential activities
of which may be divided into the initial secretory stage and
maturation stage of amelogenesis (2). A number of factors regulate
enamel-associated gene expression in AMs and dentin-associated gene
expression in human dental pulp stem cells (hDPSCs). These factors
include bone morphogenetic protein 2 (BMP2) (3), distal-less homeobox 3 (4), msh homeobox 2 (5), sonic hedgehog, Wnt (6) and neurogenic locus notch homolog
protein (Notch) (7). It is
meaningful to understand the molecular mechanisms that regulate the
formation of enamel and dentin, which may be applicable to the
prevention and treatment of heritable and acquired enamel diseases,
including caries and enamel fluorosis (8), and to the replacement of enamel with
biomaterials for therapeutic treatment.
The evolutionarily conserved Notch signaling pathway
has a central role in numerous cellular processes during
development and throughout adult life (9). In particular, the Notch pathway
serves an essential role in enamel development (10), which is required for epithelial
stem cell survival and enamel formation during the continuous
growth of the mouse incisor (11).
Protein delta homolog 1 (DLK1) is a transmembrane and secreted
protein belonging to the Notch family of receptors and ligands
(12). DLK1, as a Notch regulator,
has been proven to be essential for normal development (13), which involves processes including
neuroendocrine differentiation (14), hepatocyte and biliary epithelial
cell differentiation (15),
hematopoiesis (16), osteogenesis
(17), skeletal muscle
differentiation (18), and
chondrogenic differentiation (19). DLK1 shares close structural
similarities with periostin, which has additionally been proposed
to serve a role in tooth development (20–22).
A previous study demonstrated that DLK1 regulated the odontoblastic
differentiation of hDPSCs (23).
Therefore, it was hypothesized that DLK1 may exert regulatory
effects on epithelial-mesenchymal interactions during tooth
development. In particular, the present study investigated the
potential roles of DLK1 in the proliferation and differentiation of
AMs.
In the present study, DLK1 expression during enamel
development in the molars of mice was detected by
immunohistochemical analysis. In addition, DLK1 expression during
ameloblast-lineage cell (ALC) differentiation was analyzed by
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) and western blot analyses, and DLK1 was stably
overexpressed in ALCs to determine its effects on AM proliferation
and differentiation.
Materials and methods
Ethics statement
The present study was performed according to an
informed protocol approved by the Ethics Committee of the Ninth
People's Hospital, Shanghai Jiao Tong University School of Medicine
(Shanghai, China).
Immunohistochemical analysis
A total of nine adult C57BL/6J mice (3 male ~22–24 g
and 6 female approximately at 19–21 g; 8-weeks-old) were mated
overnight in a specific pathogen free animal center (~24°C, 60%
humidity, 10 h light/14 h dark cycle, food and water was supplied
ad libtum). The day on which a vaginal plug was observed was
designated as embryonic day 0.5 (E0.5). A total of 3 embryos and 3
postnatal (PN) mice at each enamel developmental stage (E13.5,
E16.5, E18.5, PN2 and PN6) were used in the present study. All the
animals were deeply anesthetized by intraperitoneal injection of
pentobarbital sodium (60 mg/kg) prior to sample collection, and all
procedures and protocols used were in accordance with the
Guidelines for Ethical Care of Experimental Animals (www.nap.edu/readingroom/books/labrats). Samples were
prepared for immunohistochemistry by fixing the mandibles isolated
at each stage in 4% paraformaldehyde overnight at 4°C, followed by
demineralization with 10% EDTA (pH 7.4) for 2–6 days for 2 weeks at
4°C. Following dehydration and embedding in paraffin, the samples
were sectioned to a thickness of 5 µm. The sections were dipped in
xylene to remove the paraffin and rehydrated using a graded alcohol
series (100, 95, 85 and 70%). The sections were incubated in 3%
hydrogen peroxide for 10 min at room temperature to prevent
endogenous peroxidase activity, incubated in 0.01 M citrate for 10
min at 100°C and cooled at room temperature for 20 min. The slides
were subsequently blocked in 5% bovine serum albumin (Shanghai
Weiao Biotechnology Co., Ltd. Shanghai, China) in PBS at 37°C for
30 min. The slides were then incubated with a primary antibody
against DLK1 (1:300; rabbit polyclonal antibody; cat. no. sc-8624;
Santa Cruz Biotechnology, Inc., Dallas, TX, USA) overnight at 4°C.
A subset of slides was incubated with PBS as a negative control.
The slides were washed with PBS and incubated with polymer helper
and poly-horseradish peroxidase-anti-rabbit immunoglobulin G
(OriGene Technologies, Inc., Beijing, China) for 1 h at 37°C.
Following counterstaining with hematoxylin at room temperature for
3 min, the samples were visualized under a light microscope
(magnification ×200 and ×400; Zeiss AG, Oberkochen, Germany).
Cell culture and differentiation
assay
The ameloblast-like cell line ALC was provided by
the Shanghai Research Institute of Stomatology and Shanghai Key
Laboratory of Stomatology, Ninth People's Hospital, Shanghai Jiao
Tong University (Shanghai, China). The cells were maintained in
high-glucose Dulbecco's modified Eagle's medium (Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) containing 100 U/ml
penicillin, 100 mg/l streptomycin and 10% fetal bovine serum
(Gibco; Thermo Fisher Scientific, Inc.) at 37°C in a 5%
CO2 humidified atmosphere. Ameloblastic differentiation
was induced in ALCs as described previously (2). ALCs were seeded at a density of
2×104 cells in six-well plates, and the medium was
replaced with fresh medium supplemented with 20 mg/l retinoic acid
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and 10−7
M dexamethasone (Sigma-Aldrich; Merck KGaA) when the cells reached
~80% confluence. The medium was changed every 3 days. A subset of
induced ALCs was used as a wild-type group in subsequent
assays.
DLK1 lentiviral transfection
A lentivirus expressing green fluorescent protein
(GFP) and harboring the DLK1 gene was constructed as the
dlk1-overexpression (oe) group, while a lentivirus expressing GFP
without the DLK1 gene was constructed as the control group
(Shanghai GeneChem Co., Ltd., Shanghai, China). ALCs were seeded
into six-well plates and grown to ~50% confluence prior to
transfection. The ALCs were infected with 10 µl lentiviruses
(1×108 U/ml) using polybrene reagent (Sigma-Aldrich;
Merck KGaA) for 24 h, according to the manufacturer's protocol.
Following transfection, 10 µg/ml puromycin (Sigma-Aldrich; Merck
KGaA) was added for 2 weeks to select the positively transfected
cells. A monoclonal population of stably infected cells (termed
dlk1-oe or control) were pooled and used for further experiments,
respectively. The transfected cells were analyzed under a
fluorescence microscope and confirmed by RT-qPCR and western blot
analysis.
Cell proliferation assay
A Cell Counting Kit-8 (CCK-8; Dojindo Molecular
Technologies, Inc., Kumamoto, Japan) assay was used to analyze the
effect of DLK1 on ALC proliferation, according to the
manufacturer's protocol. The control and dlk1-oe groups were seeded
at a density of 2×103 cells/well into 96-well plates (5
wells/group; Corning Incorporated, Corning, NY, USA) and cultured
for 1, 3, 5 and 7 days. The cell medium supplemented with 10% FBS
was replenished daily. Following culturing, the number of cells was
assessed using the CCK-8 assay. Absorbance was measured using a
microplate reader at 450 nm to determine the number of viable cells
in each well. A well containing medium and CCK-8 solution without
cells was used as a blank control. Cell proliferation was
represented as the mean ± standard deviation of the absorbance of
five wells for each group.
RNA isolation and RT-qPCR
analysis
Total RNA was extracted from the wild-type, control
and dlk1-oe groups cultured in ameloblastic induction medium for
different durations (0, 3 and 7 days) using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. cDNA was synthesized using a PrimeScript
RT Reagent kit with gDNA Eraser (Takara Bio, Inc., Otsu, Japan) and
used as a template for PCR. The ameloblastic differentiation of the
cells was monitored by analyzing the expression of the following
AM-related markers: Amelogenin (AMELX), enamelin, kallikrein 4
(KLK4), and matrix metallopeptidase 20 (MMP 20). GAPDH was used to
normalize the expression of the target RNAs (2). The following primer sequences were
used: AMELX forward, 5′-GATGGCTGCACCACCAAATC-3′ and reverse,
5′-CTGAAGGGTGTGACTCGGG-3′; enamelin forward,
5′-TGCAGAAATCCGACTTCTCCT-3′ and reverse, 5′-CATCTGGAATGGCATGGCA-3′;
KLK4 forward, 5′-CCGGATCATACAAGGCCAGG-3′ and reverse,
5′-TGCGGATGCACCAAGACTC-3′; MMP 20 forward,
5′-CACCTCACAAGCCATCTATCC-3′ and reverse,
5′-GAAGCTCCTTTCCCAACATTG-3′; DLK1 forward,
5′-CTCCCTGACTCTTGTTTGG-3′ and reverse, 5′-AACGGTGACAATGACTTGC-3′;
and GAPDH forward, 5′-TGGGTGTGAACCATGAGAAGT-3′ and reverse,
5′-TGAGTCCTTCCACGATACCAA-3′. The PCR reaction was performed with a
Hieff™ qPCR SYBR® Green Master Mix (Shanghai
Qcbio Science & Technologies Co., Ltd., Shanghai, China) in an
ABI 7500 RT-PCR System (Applied Biosystems; Thermo Fisher
Scientific, Inc.). Amplification was performed under the following
conditions: 95°C for 5 min in the holding stage; 40 cycles of 95°C
for 10 sec and 60°C for 30 sec in the cycling stage; and 95°C for
15 sec, 60°C for 1 min and 60°C for 15 sec in the melt curve stage.
Relative gene expression was calculated using the comparative
2−ΔΔCq method (24).
The mean Cq value of the target gene was normalized to the averaged
Cq values of GAPDH to obtain a ΔCq value, which was subsequently
normalized to control samples to obtain a ΔΔCq value. Each
measurement was assessed in triplicate. The gene expression ratio
was presented as the mean ± standard deviation of three independent
experiments.
Western blot analysis
Cells in the control and dlk1-oe groups cultured in
a 6-well plate under ameloblastic induction medium for 0, 3 and 7
days were lysed with a protein extraction kit (Pierce; Thermo
Fisher Scientific, Inc.), and cells were incubated for 15 min at a
temperature of 4°C with gentle agitation every 5 min. The lysate
was collected and transferred to a microcentrifuge tube and
centrifuged at 12,000 × g for 10 min at 4°C. The supernatant was
collected and transferred into a new tube for analysis. Protein
concentration was determined using a bicinchoninic acid protein
assay kit (Pierce; Thermo Fisher Scientific, Inc.). An equal amount
(30 µg per lane) of protein was separated via SDS-PAGE (6 and 10%
gels, respectively) and transferred onto nitrocellulose membranes
(EMD Millipore, Billerica, MA, USA). Following blocking with 5%
milk at room temperature for 1 h, the following primary antibodies
were used overnight at 4°C: Rabbit anti-mouse DLK1 (1:1,000; cat.
no. sc-8624; Santa Cruz Biotechnology, Inc.), rabbit anti-mouse
AMELX (1:1,000; cat. no. sc-365284; Santa Cruz Biotechnology,
Inc.), rabbit anti-mouse MMP 20 (1:1,000; cat. no. ab84737; Abcam,
Cambridge, MA, USA), and rabbit anti-mouse GAPDH (1:5,000; cat. no.
sc-47724; Santa Cruz Biotechnology, Inc.). Following three washes
with TBS containing Tween 20, membranes were incubated with goat
anti-mouse immunoglobulin G (1:10,000; cat. no. 926–32210, LI-COR
Biosciences, Lincoln, NE, USA) and goat anti-rabbit immunoglobulin
G (1:10,000; cat. no. 926-32211, LI-COR Biosciences) secondary
antibodies for 1 h at room temperature with agitation. Subsequent
to the final wash, the membranes were visualized using an Odyssey
LI-COR system (LI-COR Biosciences) and analyzed using an Odyssey
infrared imaging system (LI-COR Biosciences, application software
version 3.0).
Alkaline phosphatase (ALP)
staining
The wild-type, control and dlk1-oe groups were
seeded 5×105 cells/per well in 6-well plates in
triplicate and cultured in ameloblastic induction medium for 7
days. The induction medium was replaced every 3 days. An ALP color
development kit (Beyotime Institute of Biotechnology, Haimen,
China) was used, according to the manufacturer's protocol. The
plates were treated with an ALP staining kit for 30 min at 37°C
following fixation in 4% paraformaldehyde for 15 min at 37°C. The
cells were washed three times with distilled water and observed by
phase-contrast microscopy (magnification ×200).
Statistical analysis
Experiments were performed in triplicate, and data
are presented as the mean ± standard deviation. Data were evaluated
by one-way analysis of variance followed by a Tukey's post hoc test
using SPSS software (version 10.0; SPSS, Inc., Chicago, IL, USA).
P<0.05 was considered to indicate a statistically significant
difference.
Results
Location of DLK1 protein during mouse
molar development
DLK1 protein expression in the front sections of the
first mandibular molar tooth germs in mice was assessed by
immunohistochemical analysis. At the early bud stage (E13.5), DLK1
was strongly expressed in the dental epithelium. Positive
immunolabelling was observed in the basal epithelial cells and
surrounding mesenchymal cells (Fig.
1A). At the early bell stage (E16.5), positive immunolabelling
was observed in the inner enamel epithelium (IEE), particularly in
the cervical loop (CL) and enamel knot (EK) (Fig. 1B). At the late bell stage (E18.5),
DLK1 was expressed at a low level in the dental pulp (DP) cells
located near the odontoblasts (ODs), IEE and outer enamel
epithelium (OEE). However, DLK1 was strongly expressed in the ODs,
stellate intermediate (SI) layer and stellate reticulum (SR)
adjacent to the SI layer (Fig. 1C)
during the late bell stage. At the PN2 stage, DLK1 was located in
the AMs and strongly expressed in DP cells located near the ODs, SI
layer and SR adjacent to the SI layer (Fig. 1D). At the PN6 stage, DLK1 was
strongly expressed in the ODs, AMs and SR adjacent to the AMs
(Fig. 1E). Therefore, positive
expression of DLK1 was observed in the AMs and ODs during
development, particularly in the AMs located near the enamel and
ODs located near the dentin.
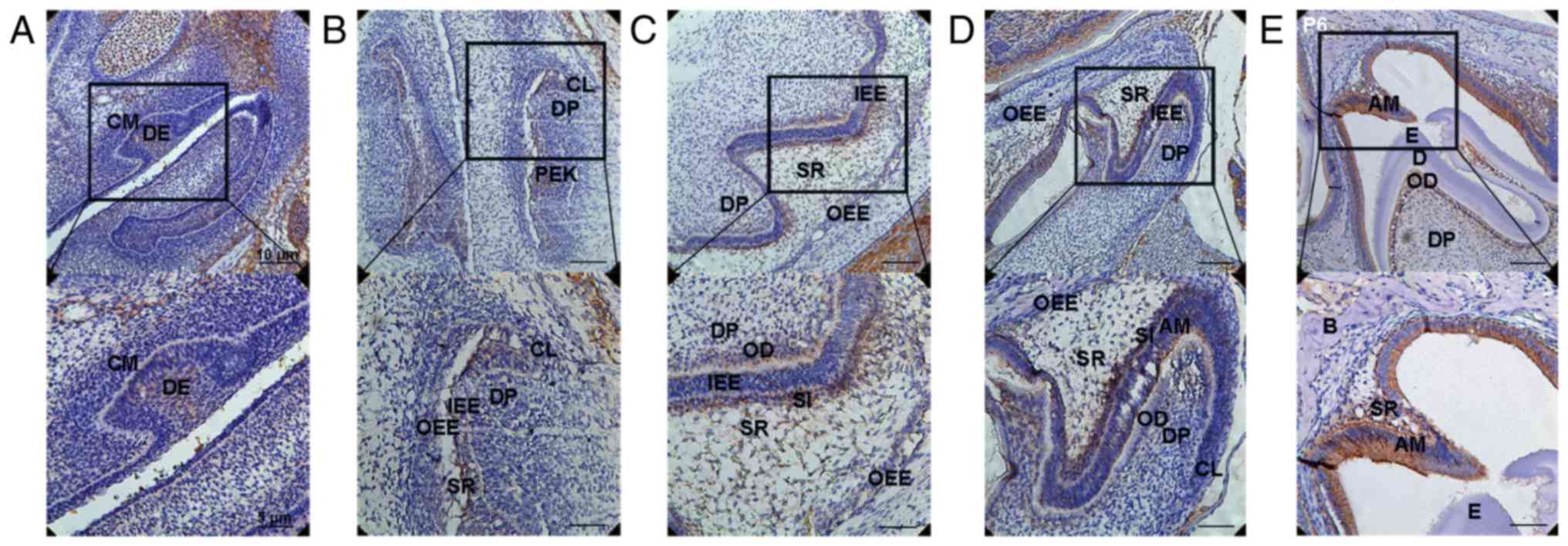 | Figure 1.Expression of DLK1 during enamel
development. (A) DLK1 was strongly expressed in the DE during the
early bud stage (E13.5). Positive immunolabelling was observed in
the basal epithelial cells and surrounding mesenchymal cells. (B)
Positive immunolabelling was also observed in the IEE, particularly
in the CL and EK during the early bell stage (E16.5). (C) At the
late bell stage (E18.5), DLK1 was expressed at a low level in the
DP cells located near the ODs, IEE and OEE. Primarily, DLK1 was
strongly expressed in the ODs, the SI layer and SR adjacent to the
SI layer. (D) During PN2, DLK1 was located in the AMs and strongly
expressed in the DP cells located near the ODs, SI layer and SR
adjacent to the SI layer. (E) During PN6, DLK1 was strongly
expressed in the ODs, AMs and SR adjacent to the AMs. Upper panels,
magnification ×200; lower panels, magnification, ×400. DE, dental
epithelium; IEE, inner enamel epithelium; CL, cervical loop; EK,
enamel knot; OD, odontoblast; OEE, outer enamel epithelium; SI,
stellate intermediate; ST, stellate reticulum; E, embryonic
development stage; DP, dental pulp; DLK1, protein delta homolog 1;
AM, ameloblast; PN, postnatal day. |
DLK1 and AM-associated gene expression
during ameloblastic differentiation of ALCs
In the present study, DLK1 expression was detected
to evaluate its physiological significance during ameloblastic
differentiation. Mineralization-associated genes were measured to
directly verify the functional role of DLK1 in ameloblastic
differentiation. In order to analyze the expression of
AM-associated genes and DLK1 during ameloblastic induction, the
mRNA levels of AMELX, enamelin, MMP 20, KLK4 and DLK1 were detected
in wild-type ALCs by RT-qPCR analysis, and the protein levels of
AMELX, MMP 20 and DLK1 were detected by western blot analysis
(Fig. 2). The mRNA levels of AMELX
(Fig. 2A) [day (D)0 vs. D3,
P=0.029; D0 vs. D7, P=0.0072] and enamelin (Fig. 2B) (D0 vs. D3, P=0.0089; D0 vs. D7,
P=0.0012) were downregulated, whereas those of MMP 20 (Fig. 2C) (D0 vs. D3, P=0.0049; D0 vs. D7,
P=0.0072) and KLK4 (Fig. 2D) (D0
vs. D3, P=0.0031; D0 vs. D7, P=0.0324) were upregulated gradually.
The protein levels of AMELX (Fig.
2G) (D0 vs. D3, P=0.0369; D0 vs. D7, P=0.0352) and MMP 20
(Fig. 2H) (D0 vs. D3, P=0.0273; D0
vs. D7, P=0.0002) followed the same trends as their mRNA expression
during ameloblastic induction (Fig.
2F-H). Additionally, the mRNA and protein levels of DLK1 were
upregulated during ameloblastic differentiation, as presented in
Fig. 2E (D0 vs. D3, P=0.0119; D0
vs. D7, P=0.0022), and Fig. 2F and
I (D0 vs. D3, P=0.0072; D0 vs. D7, P=0.0033).
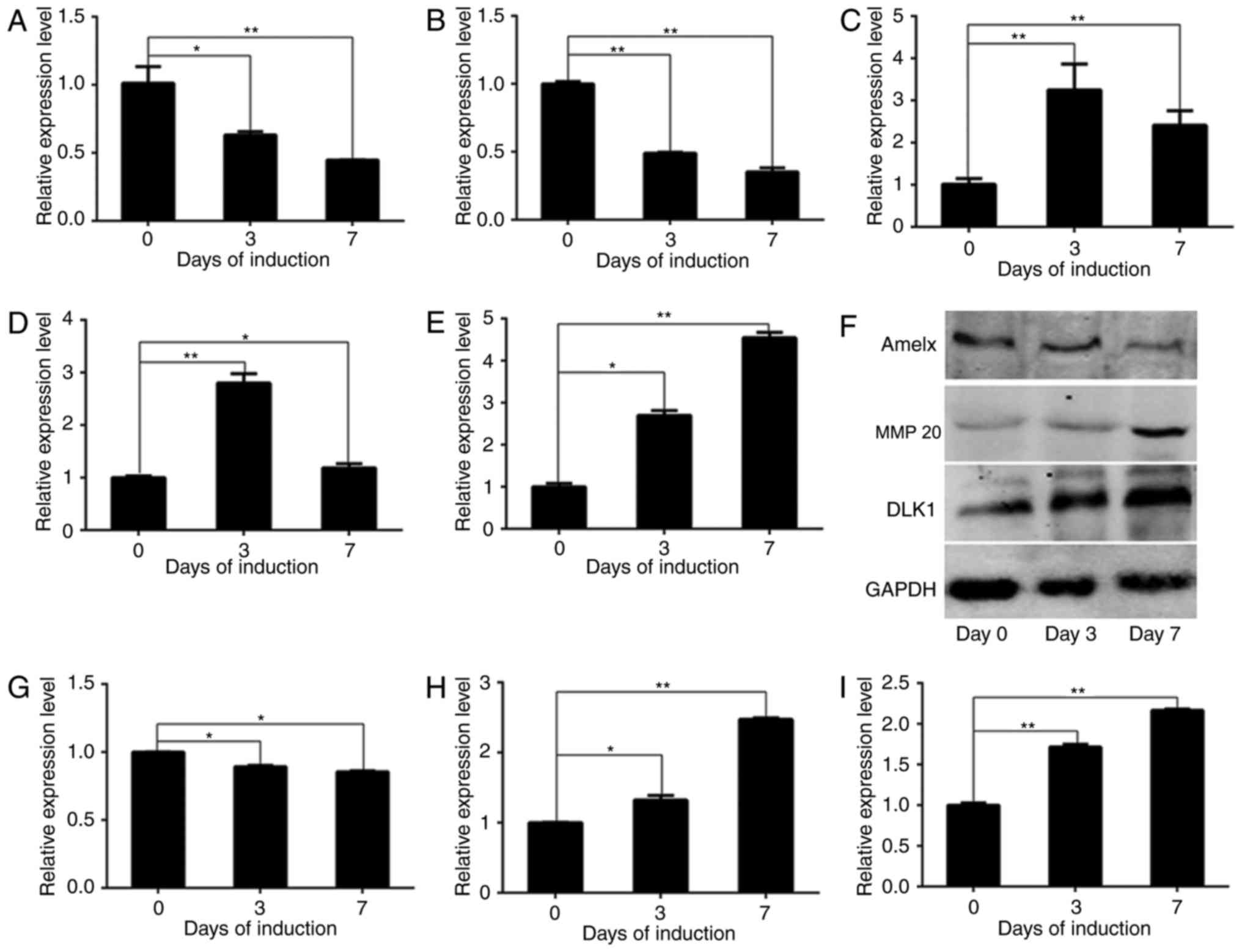 | Figure 2.DLK1 expression during the
ameloblastic differentiation of ameloblast-lineage cells. The mRNA
levels of AMELX, enamelin, MMP 20, KLK4 and DLK1 are presented, in
addition to the protein levels of AMELX, MMP 20 and DLK1 during
ameloblastic induction. Quantified results of the western blot
analysis based on measurements of optical density are presented,
and the data represent the mean ± standard deviation of three
independent experiments. The mRNA levels of (A) AMELX and (B)
enamelin decreased significantly, whereas those of (C) MMP 20 (D)
KLK4 increased significantly. (E) The mRNA levels of DLK1 increased
gradually. The protein levels of AMELX and MMP 20 exhibited the
same trends as their mRNA levels, as demonstrated by (F) western
blot analysis. The results of the western blot analysis of (G)
AMELX, (H) MMP 20 and (I) DLK1 are presented. These results
demonstrated that ameloblastic differentiation was successfully
induced in vitro. *P<0.05; **P<0.01. DLK1, protein
delta homolog 1; AMELX, amelogenin; MMP 20, matrix metallopeptidase
20; KLK4, kallikrein 4. |
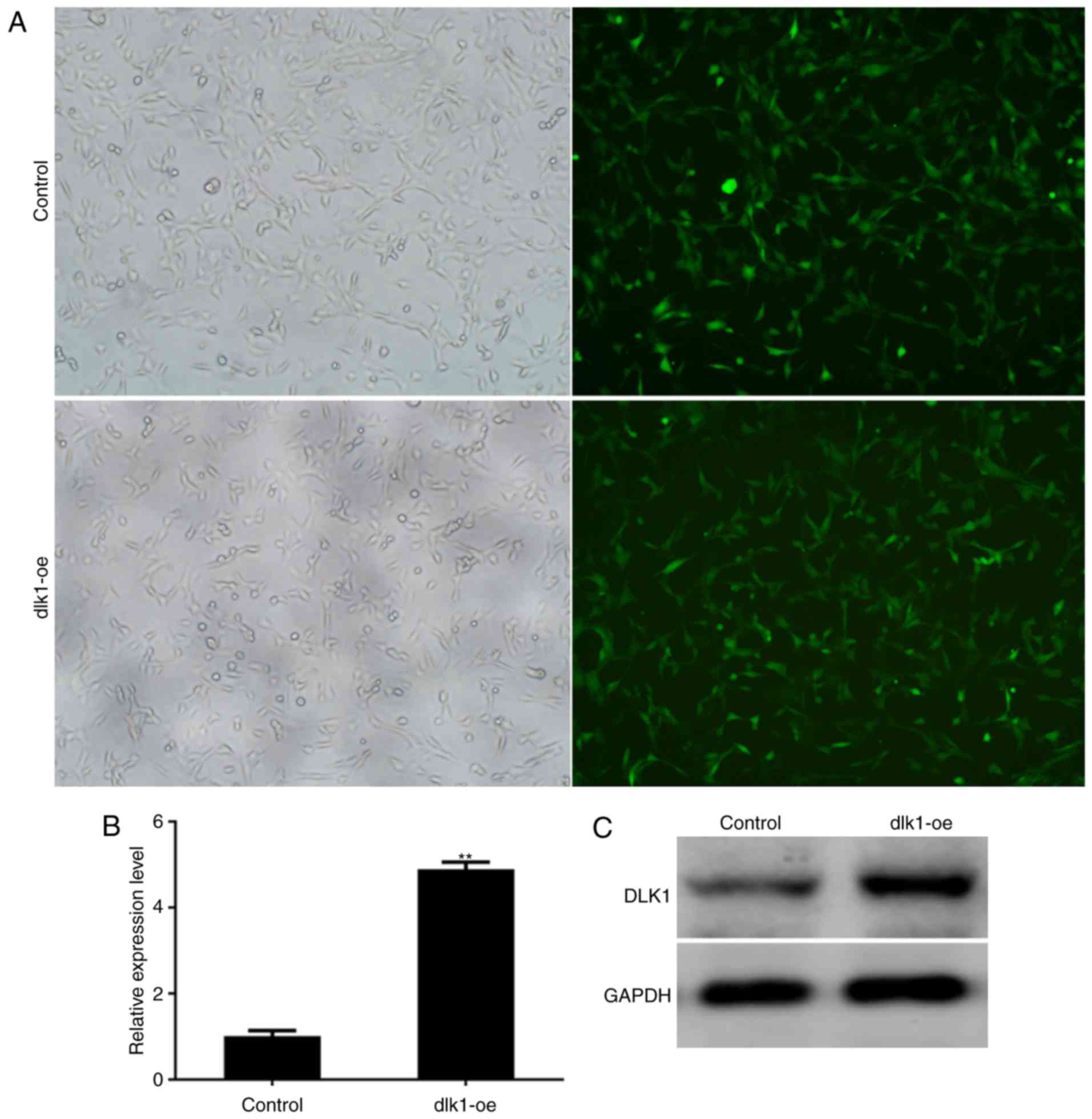
Stable overexpression of DLK1 in
ALCs
ALCs were transfected with a lentiviral vector
expressing GFP alone (control group) or a lentiviral vector
expressing DLK1 and GFP (dlk1-oe group) in vitro, and
exhibited GFP expression within 3 days of transduction (Fig. 3A). The expression of DLK1 in stably
transfected cells was subsequently assessed. The mRNA and protein
levels of DLK1 were markedly increased in the dlk1-oe group
compared with the control group following infection (Fig. 3B and C; P=0.0019). These results
demonstrated that stable overexpression of DLK1 was successfully
established in ALCs following lentiviral transfection.
DLK 1 promotes the proliferation of
ALCs
Cell proliferation was measured using a CCK-8 assay,
according to the manufacturer's protocol. The results indicated
that the rate of cell proliferation in the dlk1-oe group was
markedly increased compared with that of the control group on days
1 (P=0.031), 3 (P=0.042), 5 (P=0.0053) and 7 (P=0.018) (Fig. 4A). These results demonstrated that
cell proliferation was promoted in the dlk1-oe group.
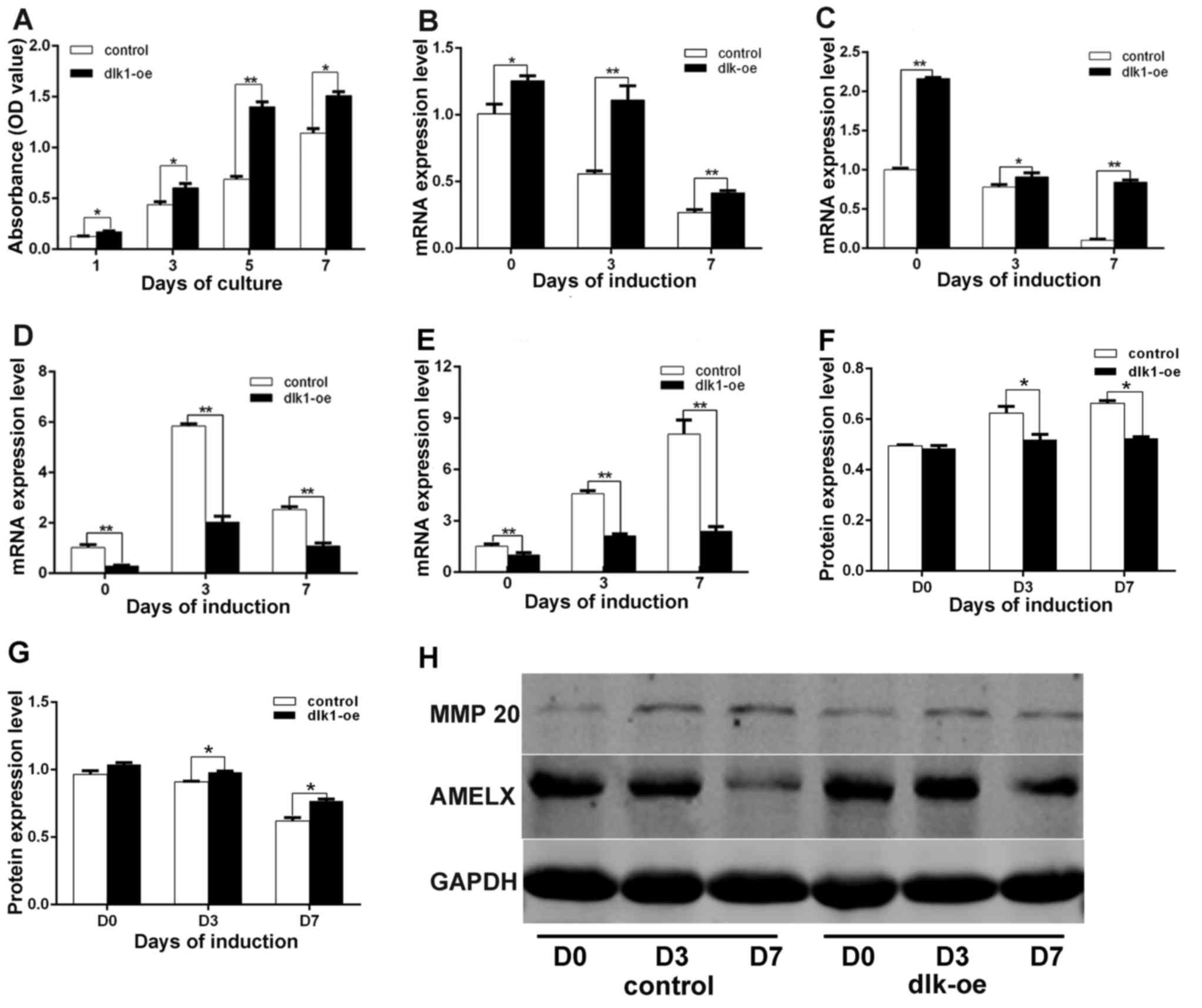 | Figure 4.Effects of DLK1 on ALC proliferation
and differentiation. (A) The stimulatory effect of DLK1 on ALC
proliferation. A CCK-8 assay indicated that the rate of cell
proliferation in the dlk1-oe group was markedly increased compared
with the control group on days 1, 3, 5 and 7. The mRNA levels of
(B) AMELX, (C) enamelin, (D) MMP 20 and (E) KLK4 were measured
following DLK1 overexpression in ALCs during ameloblastic
differentiation, which indicated that DLK1 overexpression
upregulated AMELX and enamelin mRNA, while downregulating the mRNA
levels of MMP 20 and KLK4. The quantified protein expression data
for (F) MMP 20 and (G) AMELX were calculated based on measurements
of the optical density value. The protein levels of AMELX and MMP
20 followed the same trend as their mRNA levels. (H) The protein
levels of MMP 20 and AMELX were measured by western blot analysis.
All data are presented as the mean ± standard deviation of at least
three independent experiments. *P<0.05; **P<0.01. DLK1,
protein delta homolog 1; AMELX, amelogenin; MMP 20, matrix
metallopeptidase 20; KLK4, kallikrein 4; ALC, ameloblast-lineage
cell; D, day; oe, overexpression. |
Inhibition of ameloblastic
differentiation in ALCs following DLK1 overexpression
The mRNA levels of AMELX (Fig. 4B; P=0.0413, P=0.0028 and P=0.0091)
and enamelin (Fig. 4C; P=0.0013,
P=0.048 and P=0.0001) in the dlk1-oe group were significantly
increased compared with those in the control group following 7 days
of culture in induction medium. In addition, the mRNA levels of MMP
20 (Fig. 4D; P=0.0029, P=0.0068
and P=0.0054) and KLK4 (Fig. 4E;
P=0.0093, P=0.0056 and P=0.0027) in the dlk1-oe group were
significantly decreased compared with those in the control group.
Similar trends in the protein expression levels of MMP 20 (Fig. 4F; P=0.0073, P=0.025 and P=0.039)
and AMELX (Fig. 4G; P=0.0063,
P=0.0045 and P=0.0024) were observed in the dlk1-oe and control
groups via western blot analysis, and the western blot results are
illustrated in Fig. 4H.
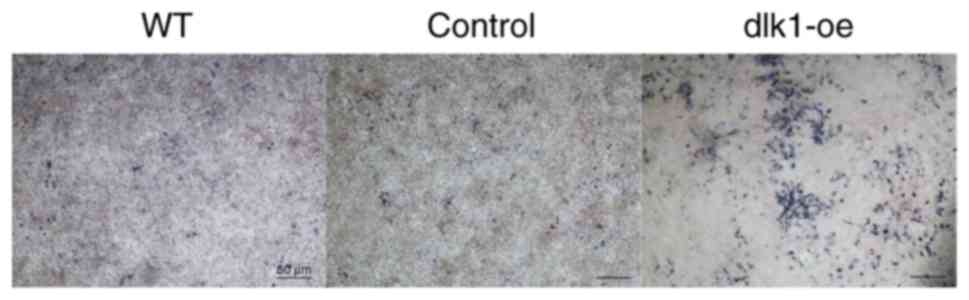
Additionally, ALP staining was markedly reduced in the wild-type
and control groups on day 7 when compared with the dlk1-oe group
(Fig. 5). These results indicated
that DLK1 overexpression inhibited the ameloblastic differentiation
of ALCs.
Discussion
Tooth development requires continuous interaction
and cross-talk between various epithelial and mesenchymal cells,
which involves different growth factors and signaling molecules.
DLK1 exerts regulatory effects on cell differentiation and controls
various signaling pathways during normal development. Numerous
heritable defects, including growth retardation, obesity and
skeletal malformations, have been observed in DLK1-null mice, and
symptoms of maternal uniparental disomy syndrome have been
associated with DLK1 deficiency in humans (25,26).
DLK1 is a negative regulator of a number of differentiation
processes, including neuroendocrine differentiation (14), osteogenesis, chondrogenesis
(19) and muscle regeneration
(17). However, the in vivo
and in vitro effects of DLK1 on ameloblastic proliferation
and differentiation remain unclear.
During embryonic development, DLK1 is expressed at a
high level in a number of tissues, including the lungs, adrenal
cortex, proximal tubules of the kidneys and mesoderm-derived
tissues, including chondroblast tissue (19) and skeletal myotubes (14). A previous study demonstrated that
DLK1 may regulate the development of mesoderm-derived dentin tissue
in mice (23). In the present
study, DLK1 expression was upregulated between E13.5 and E18.5 in
the inner and outer enamel epithelium, and was markedly expressed
in the AMs at stage PN6 following enamel formation. These results
demonstrated that DLK1 may serve a regulatory role in the
development of epithelium-derived enamel tissue, ultimately
suggesting that DLK1 serves important roles in tooth development
through epithelial-mesenchymal interactions. Therefore, it is
useful to analyze the function of DLK1 during AM proliferation and
differentiation.
Mouse ALCs, based on a previous study (2), which express secretory-stage and
maturation-stage-associated genes, were selected to verify the
effects of DLK1 on ameloblastic differentiation and proliferation
in vitro. Proteins of the enamel matrix (enamelin, AMELX,
MMP 20 and KLK4) are essential for enamel formation and may be used
as mineralization markers to assess the ameloblastic
differentiation of ALCs (2). In
the present study, enamelin and AMELX were downregulated during the
ameloblastic differentiation of ALCs, while MMP 20 and KLK4 were
upregulated. The variations in these markers during the present
experiments exhibited the same trends as in previous reports
(2), and demonstrated that the
ALCs underwent differentiation into a more mature stage during the
course of the study; additionally, the mRNA and protein levels of
DLK1 were significantly upregulated during this process.
Lentiviral vectors were used to achieve efficient
gene overexpression in vitro. Results of the CCK-8 assay
demonstrated that cell proliferation in the Dlk1-oe group was
significantly upregulated. No significant differences were observed
between the wild-type and control groups in terms of proliferation
and differentiation. These findings indicated that recombinant
lentiviral infection did not disturb cellular properties. Notably,
DLK1 overexpression promoted the proliferation of ALCs, an effect
that has previously been documented in lung epithelial cell- and
pluripotent stem cell-derived neural progenitors (27,28).
In addition, the ameloblastic differentiation
markers enamelin, AMELX, MMP 20 and KLK4 were observed to be
regulated in the dlk1-oe group, as demonstrated by RT-qPCR and
western blot analyses. The mRNA levels of enamelin and AMELX, and
the protein levels of AMELX, were upregulated in the dlk1-oe group
during ameloblastic differentiation compared with the control
group. By contrast, the protein and mRNA levels of MMP 20, and the
mRNA level of KLK4, were downregulated in the dlk1-oe group. These
results confirmed that DLK1 overexpression inhibited the
ameloblastic differentiation of ALCs. Immunohistochemistry of DLK1
expression during tooth germ development demonstrated that DLK1 was
highly expressed in the EK, which serves as a signaling center
during the formation and regulation of tooth shape and size
(29), at stage E16.5. DLK1 was
additionally highly expressed in the AMs following enamel formation
at PN6. These results suggested that DLK1 may inhibit AM
differentiation to prevent excessive enamel formation.
DLK1, a Notch regulator, is able to activate Notch
signaling, which serves important roles in the differentiation of
odontoblasts and osteoblasts, calcification of hard tooth tissues,
formation of cusp patterns and generation of tooth roots (30). Additionally, DLK1 activates human
stem cells by interrupting the sonic hedgehog signaling pathway
(31). DLK1 additionally activates
the mitogen-activated protein kinase (MAPK) and MAPK kinase
(MEK)/extracellular signal-regulated kinase (ERK) pathways to
inhibit adipogenesis and chondrogenesis (19,32,33).
In addition, DLK1 regulates osteogenesis and chondrogenesis through
phosphatidylinositol 3-kinase (PI3K)/RAC-α serine/threonine-protein
kinase (AKT) signaling (19).
Notably, tooth development involves a number of overlapping
pathways, including the MAPK (34), MEK/ERK (34) and PI3K/AKT (35) pathways, which are associated with
epithelial-mesenchymal interactions. The development of enamel and
dentin involves processes similar to those in osteogenesis and
chondrogenesis. Therefore, it was hypothesized that DLK1 may be
involved in enamel development. Further intensive studies are
required to elucidate the molecular cross-talk and signaling
pathways associated with DLK1 in dentin and enamel development.
In conclusion, the present study detected the
expression of DLK1 during the entire process of tooth development.
DLK1 overexpression resulted in the increased proliferation
capacity of AMs and regulated the expression of enamel
mineralization-associated genes and proteins. Thus, DLK1 may serve
a role in enamel development.
Acknowledgements
The present study was supported by grants from the
National Natural Science Foundation of China (grant nos. 81500806
and 31271341), the China Postdoctoral Science Foundation (grant no.
2015M581633), and the Science Foundation of Shanghai Health and
Family Planning Commission (grant no. 20144Y0257).
References
|
1
|
Mina M and Kollar EJ: The induction of
odontogenesis in non-dental mesenchyme combined with early murine
mandibular arch epithelium. Arch Oral Biol. 32:123–127. 1987.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sarkar J, Simanian EJ, Tuggy SY, Bartlett
JD, Snead ML, Sugiyama T and Paine ML: Comparison of two mouse
ameloblast-like cell lines for enamel-specific gene expression.
Front Physiol. 5:2772014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Guo F, Feng J, Wang F, Li W, Gao Q, Chen
Z, Shoff L, Donly KJ, Gluhak-Heinrich J, Chun YH, et al: Bmp2
deletion causes an amelogenesis imperfecta phenotype via regulating
enamel gene expression. J Cell Physiol. 230:1871–1882. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Price JA, Wright JT, Walker SJ, Crawford
PJ, Aldred MJ and Hart TC: Tricho-dento-osseous syndrome and
amelogenesis imperfecta with taurodontism are genetically distinct
conditions. Clin Genet. 56:35–40. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhou YL, Lei Y and Snead ML: Functional
antagonism between Msx2 and CCAAT/enhancer-binding protein alpha in
regulating the mouse amelogenin gene expression is mediated by
protein-protein interaction. J Biol Chem. 275:29066–29075. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu F, Chu EY, Watt B, Zhang Y, Gallant
NM, Andl T, Yang SH, Lu MM, Piccolo S, Schmidt-Ullrich R, et al:
Wnt/beta-catenin signaling directs multiple stages of tooth
morphogenesis. Dev Biol. 313:210–224. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mitsiadis TA, Graf D, Luder H, Gridley T
and Bluteau G: BMPs and FGFs target Notch signalling via jagged 2
to regulate tooth morphogenesis and cytodifferentiation.
Development. 137:3025–3035. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Riksen EA, Kalvik A, Brookes S, Hynne A,
Snead ML, Lyngstadaas SP and Reseland JE: Fluoride reduces the
expression of enamel proteins and cytokines in an
ameloblast-derived cell line. Arch Oral Biol. 56:324–330. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Artavanis-Tsakonas S, Rand MD and Lake RJ:
Notch signaling: Cell fate control and signal integration in
development. Science. 284:770–776. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mitsiadis TA, Regaudiat L and Gridley T:
Role of the Notch signalling pathway in tooth morphogenesis. Arch
Oral Biol. 50:137–140. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Felszeghy S, Suomalainen M and Thesleff I:
Notch signalling is required for the survival of epithelial stem
cells in the continuously growing mouse incisor. Differentiation.
80:241–248. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Traustadóttir GÁ, Jensen CH, Thomassen M,
Beck HC, Mortensen SB, Laborda J, Baladrón V, Sheikh SP and
Andersen DC: Evidence of non-canonical NOTCH signaling: Delta-like
1 homolog (DLK1) directly interacts with the NOTCH1 receptor in
mammals. Cell Signal. 28:246–254. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Falix FA, Aronson DC, Lamers WH and
Gaemers IC: Possible roles of DLK1 in the Notch pathway during
development and disease. Biochim Biophys Acta. 1822:988–995. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Floridon C, Jensen CH, Thorsen P, Nielsen
O, Sunde L, Westergaard JG, Thomsen SG and Teisner B: Does fetal
antigen 1 (FA1) identify cells with regenerative, endocrine and
neuroendocrine potentials? A study of FA1 in embryonic, fetal, and
placental tissue and in maternal circulation. Differentiation.
66:49–59. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tanimizu N, Nishikawa M, Saito H,
Tsujimura T and Miyajima A: Isolation of hepatoblasts based on the
expression of Dlk/Pref-1. J Cell Sci. 116:1775–1786. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Moore KA, Pytowski B, Witte L, Hicklin D
and Lemischka IR: Hematopoietic activity of a stromal cell
transmembrane protein containing epidermal growth factor-like
repeat motifs. Proc Natl Acad Sci USA. 94:pp. 4011–4016. 1997;
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Abdallah BM, Jensen CH, Gutierrez G,
Leslie RG, Jensen TG and Kassem M: Regulation of human skeletal
stem cells differentiation by Dlk1/Pref-1. J Bone Miner Res.
19:841–852. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Andersen DC, Petersson SJ, Jørgensen LH,
Bollen P, Jensen PB, Teisner B, Schroeder HD and Jensen CH:
Characterization of DLK1+ cells emerging during skeletal muscle
remodeling in response to myositis, myopathies, and acute injury.
Stem Cells. 27:898–908. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen L, Qanie D, Jafari A, Taipaleenmaki
H, Jensen CH, Säämänen AM, Sanz ML, Laborda J, Abdallah BM and
Kassem M: Delta-like 1/fetal antigen-1 (Dlk1/FA1) is a novel
regulator of chondrogenic cell differentiation via inhibition of
the Akt kinase-dependent pathway. J Biol Chem. 286:32140–32149.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li H, Marijanovic I, Kronenberg MS, Erceg
I, Stover ML, Velonis D, Mina M, Heinrich JG, Harris SE, Upholt WB,
et al: Expression and function of Dlx genes in the osteoblast
lineage. Dev Biol. 316:458–470. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hassan MQ, Saini S, Gordon JA, van Wijnen
AJ, Montecino M, Stein JL, Stein GS and Lian JB: Molecular switches
involving homeodomain proteins, HOXA10 and RUNX2 regulate
osteoblastogenesis. Cells Tissues Organs. 189:122–125. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ma D, Zhang R, Sun Y, Rios HF, Haruyama N,
Han X, Kulkarni AB, Qin C and Feng JQ: A novel role of periostin in
postnatal tooth formation and mineralization. J Biol Chem.
286:4302–4309. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Qi S, Yan Y, Wen Y, Li J, Wang J, Chen F,
Tang X, Shang G, Xu Y and Wang R: The effect of delta-like 1
homolog on the proliferation and odontoblastic differentiation in
human dental pulp stem cells. Cell Prolif. 50:2017. View Article : Google Scholar
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cheung LY, Rizzoti K, Lovell-Badge R and
Le Tissier PR: Pituitary phenotypes of mice lacking the notch
signalling ligand delta-like 1 homolog. J Neuroendocrinol.
25:391–401. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mortensen SB, Jensen CH, Schneider M,
Thomassen M, Kruse TA, Laborda J, Sheikh SP and Andersen DC:
Membrane-tethered delta-like 1 homolog (DLK1) restricts adipose
tissue size by inhibiting preadipocyte proliferation. Diabetes.
61:2814–2822. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Weng T, Gao L, Bhaskaran M, Guo Y, Gou D,
Narayanaperumal J, Chintagari NR, Zhang K and Liu L: Pleiotrophin
regulates lung epithelial cell proliferation and differentiation
during fetal lung development via beta-catenin and Dlk1. J Biol
Chem. 284:28021–28032. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Surmacz B, Noisa P, Risner-Janiczek JR,
Hui K, Ungless M, Cui W and Li M: DLK1 promotes neurogenesis of
human and mouse pluripotent stem cell-derived neural progenitors
via modulating Notch and BMP signalling. Stem Cell Rev. 8:459–471.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mustonen T, Tümmers M, Mikami T, Itoh N,
Zhang N, Gridley T and Thesleff I: Lunatic fringe, FGF, and BMP
regulate the Notch pathway during epithelial morphogenesis of
teeth. Dev Biol. 248:281–293. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cai X, Gong P, Huang Y and Lin Y: Notch
signalling pathway in tooth development and adult dental cells.
Cell Prolif. 44:495–507. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Han Z, Yu C, Tian Y, Zeng T, Cui W, Mager
J and Wu Q: Expression patterns of long noncoding RNAs from
Dlk1-Dio3 imprinted region and the potential mechanisms of Gtl2
activation during blastocyst development. Biochem Biophys Res
Commun. 463:167–173. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kim KA, Kim JH, Wang Y and Sul HS: Pref-1
(preadipocyte factor 1) activates the MEK/extracellular
signal-regulated kinase pathway to inhibit adipocyte
differentiation. Mol Cell Biol. 27:2294–2308. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kim SW, Muise AM, Lyons PJ and Ro HS:
Regulation of adipogenesis by a transcriptional repressor that
modulates MAPK activation. J Biol Chem. 276:10199–10206. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Greenblatt MB, Kim JM, Oh H, Park KH, Choo
MK, Sano Y, Tye CE, Skobe Z, Davis RJ, Park JM, et al: p38α MAPK is
required for tooth morphogenesis and enamel secretion. J Biol Chem.
290:284–295. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sakoda K, Nakajima Y and Noguchi K: Enamel
matrix derivative induces production of vascular endothelial cell
growth factor in human gingival fibroblasts. Eur J Oral Sci.
120:513–519. 2012. View Article : Google Scholar : PubMed/NCBI
|