Introduction
Globally, breast cancer (BC) is the most frequently
diagnosed cancer in women and accounts for more than 1 million new
cases yearly (1). Despite the
mortality of breast cancer has been reduced dueing to the early
detection and adjuvant treatments, more than 450,000 deaths still
occur each year (2). As
acknowledged, breast cancer is a complex and heterogeneous disease
which is accompanied with dissimilar cellular origin, etiology,
treatment responses or clinical outcomes (3). Current therapies for breast cancer
which are customized are based on molecular profiles including
luminal, basal-like and human epidermal growth factor receptor-2
positive, and contain chemotherapy, endocrine therapy, selective
estrogen receptor modulators and targeted therapies (4).
Interleukin (IL)-23 is a pro-inflammatory cytokine
which consists of IL-12 p40 and IL-23 p19 subunits. IL-23, which is
primarily secreted by macrophages and dendritic cells, induces
autoimmunity by T-cell-mediated inflammation through impacting T
helper 17 (Th17) cell response (5). Inappropriate expression of IL-23
appeared to coincide with multiple autoimmune disorders (6,7).
Furthermore, previous studies indicate that IL-23/IL-23R is
indispensable for Th17 cell-mediated immune response (8,9), and
IL-23R plays a crucial role in initiating, maintaining and
accelerating IL-23/IL-17 inflammatory signaling pathway (10). The importance of IL-23R during the
development of tumor and the effects on tumor immunity was
previously elucidated (11);
meanwhile, IL-23R was indicated to exert an immunosurveillance
function by CD8+ T-cells and accelerate tumor growth in
a previous study (12).
The correlation between chronic inflammation and
elevated malignancy incidence or the regulatory mechanisms has been
suggested for more than a century (13). The current study aimed to explore
the effects of IL-23/IL-23R on breast cancer, and provide a
therapeutic target for breast cancer.
Materials and methods
Participants
Breast cancer tissues and the adjacent tissues were
gathered from thirty-two patients who were treated in our hospital.
Basic characteristics of patients were presented in Table I. Prior written and informed
consent were obtained from each patient before the beginning of our
research. Current research was approved by the Ethics Committee of
The Third Hospital of Chengde City.
 | Table I.IL-23/IL-23R mRNA level was correlated
with clinicopathological parameters of breast cancer patients. |
Table I.
IL-23/IL-23R mRNA level was correlated
with clinicopathological parameters of breast cancer patients.
| Parameters | Cases | IL-23 | P-value | IL-23R | P-value |
|---|
| Age (years) |
|
| 0.71 |
| 0.54 |
|
<40 | 15 | 5.21±0.67 |
| 48.60±8.04 |
|
| ≥40 | 17 | 5.79±0.74 |
| 52.29±8.97 |
|
| Tumor size (cm) |
|
| 0.04a |
| 0.02a |
| ≥5 | 13 | 5.07±0.72 |
| 48.75±5.97 |
|
|
<5 | 19 | 5.74±0.94 |
| 54.14±6.01 |
|
| Histological
grade |
|
| 0.55 |
| 0.78 |
|
Well-intermediately
differentiation | 11 | 5.36±0.87 |
| 51.35±5.90 |
|
| Poor
differentiation | 21 | 5.14±1.02 |
| 51.98±5.95 |
|
| Invasion depth |
|
| 0.78 |
| 0.93 |
|
T1-T2 | 25 | 5.06±0.90 |
| 51.78±5.90 |
|
|
T3-T4 | 7 | 5.24±1.06 |
| 51.99±6.02 |
|
| Lymph node
metastasis |
|
| 0.89 |
| 0.72 |
| N0 | 3 | 5.17±0.90 |
| 50.87±5.90 |
|
|
N1-N3 | 29 | 5.08±1.06 |
| 51.98±4.96 |
|
| Distant
metastasis |
|
| 0.04a |
| 0.02a |
| M0 | 27 | 5.02±0.68 |
| 50.06±5.99 |
|
| M1 | 5 | 5.75±0.71 |
| 57.17±4.76 |
|
| TNM stage |
|
| 0.01a |
| 0.04a |
|
0–II | 23 | 5.05±0.74 |
| 50.17±5.90 |
|
|
III–IV | 9 | 5.88±0.66 |
| 56.14±5.76 |
|
Cell culture
MCF7 cells were maintained in Dulbecco's modified
Eagle's medium (DMEM)/Ham's F12 medium with 10% fetal bovine serum,
100 U/ml penicillin and 100 µg/ml streptomycin. And incubated in a
humidified atmosphere of 5% CO2 at 37°C.
Recombinant human (rh) IL-23 (R&D) was used for
the stimulation of the cells. Effects of rh IL-23 (10 ng/ml) on
cell proliferation were detected after cells were incubated with it
48 h; the influence of pre-treatment of PAb IL-23p19 (Santa Cruz
Biotechnology, Inc., Dallas, TX, USA), a neutralizing antibody
specific for IL-23, on rh IL-23 treated cells were also examined
(14). Therefore, cells were
divided into 3 groups, i.e., control group, IL-23 group (cells
treated with rh IL-23) and PAb IL-23p19 group (cells treated with
PAb IL-23p19 and rh IL-23) to investigate cell behaviors and
correlated protein expression levels.
Real-time PCR analysis of IL-23R
messenger RNA
Tumor tissues and the adjacent tissues of thirty-two
breast cancer patients were extracted and stored at −80°C or
applied for the following experiments. Total RNA was isolated by
TRIzol (Molecular Research Center, Cincinnati, OH, USA). cDNA was
obtained from mRNA with oligo primers and Superscript II
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
based on the manufacturer's protocol. Relative gene level for
IL-23R was quantified by ABI Prism 7000 (Applied Biosystems; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) based on the method of
SYBR-Green. Primers were as followed: IL-23, forward:
TGCTAGGATCGGATATTTTCACAGG; reverse: GAGGCTTGGAATCTGCTGAGTC; IL-23R,
forward: GATATTCCTGATGAAGTAACCTGTGT. C; reverse:
GATACTGTTGCTCTTCTTCTGTCTC; GAPDH, forward: CAGCCTCAAGATCATCAGCA;
reverse: ACAGTCTTCTGGGTGGCAGT, which were as previously reported
study performed (14). Total
volume of PCR reaction was 20 µl which consisted of 0.1 µM
forward/reverse primer, 1× SYBR Premix EX Taq premix (Takara
Biotechnology Co., Ltd., Dalian, China) and 50 ng cDNA. The
conditions were: 95°C for 120 sec, followed by 40 cycles at 95°C
for 15 sec and 60°C for 60 sec. GAPDH was used as an internal
reference gene.
Immunological histological
chemistry
Tissues from patients were first sliced into 4
mm-thick sections. Afterwards, sections were fixed by 7.5% buffered
formalin and embedded with paraffin. Then deparaffinage and antigen
retrieval of slices were performed before incubation with antibody.
Avidin-biotin-peroxidase complex (ABC) method was conducted with
monoclonal IL-23 antibody and IL-23R antibody according to the
manufacturer's protocol. The expression levels of IL-23 and IL-23R
were independently assessed by three senior pathologists in
accordance with the proportion and intensity of positive cells.
MTT assay
Cell proliferation was examined by MTT assay.
Briefly, cells (1×105) were seeded into 12-well plate
and incubated with methamphetamine for 24 h. MTT (5 mg/ml in PBS)
was added into the wells, and cells were incubated for another 1.5
h at 37°C. After extraction and throwing away the supernatant from
the culture medium, dimethylsulfoxide (200 µl) was added to
dissolve the formazan crystals. The optical density (OD) on a
spectrophotometer was measured at 595 nm.
Cell apoptosis analysis
Cells were seeded in 12-well plates and cultured for
48 h in an incubator at 37°C with humidified atmosphere of 5%
CO2. A total of 1×105 cells were collected by
centrifugation (2,000 rpm, 5 min) and washed by ice-cold PBS.
Afterwards, cells were fixed with ice-cold ethanol (70%) and placed
at −20°C overnight. On the next day, cells were stained by Annexin
V-enhanced green fluorescent protein (FITC) and propidium iodide
(PI), then incubated for 15 min at room temperature in dark place.
Assay results were measured by Cell Quest software (BD Biosciences,
Franklin Lakes, NJ, USA). The percentage of cells with apoptotic
nuclei was calculated.
Western blot analysis
Cell were rinsed twice with ice cold PBS and scraped
by RIPA (Beyotime Institute of Biotechnology, Shanghai, China).
Samples were separated by 10% sodium dodecyl sulfate polyacrylamide
gel electrophoresis. Then, gels were transferred onto
polyvinylidene difluoride membranes at 4°C. Membranes reacted with
primary antibody overnight at 4°C, and horseradish
peroxidase-conjugated secondary antibody for 90 min at room
temperature with gentle agitation. Finally, at room temperature,
after washing with Tris Buffered Saline with Tween 20 (TBST) for 10
min, proteins were detected with Super Signal West Pico
Chemiluminescent Substrate (Thermo Fisher Scientific, Inc.).
Statistical analysis
Data were analyzed by SPSS v21.0 (SPSS, Inc.,
Chicago, IL, USA) and presented as mean ± standard error of mean
(SEM). Data were analyzed by t-test. P<0.05 was considered to
indicate a statistically significant difference.
Results
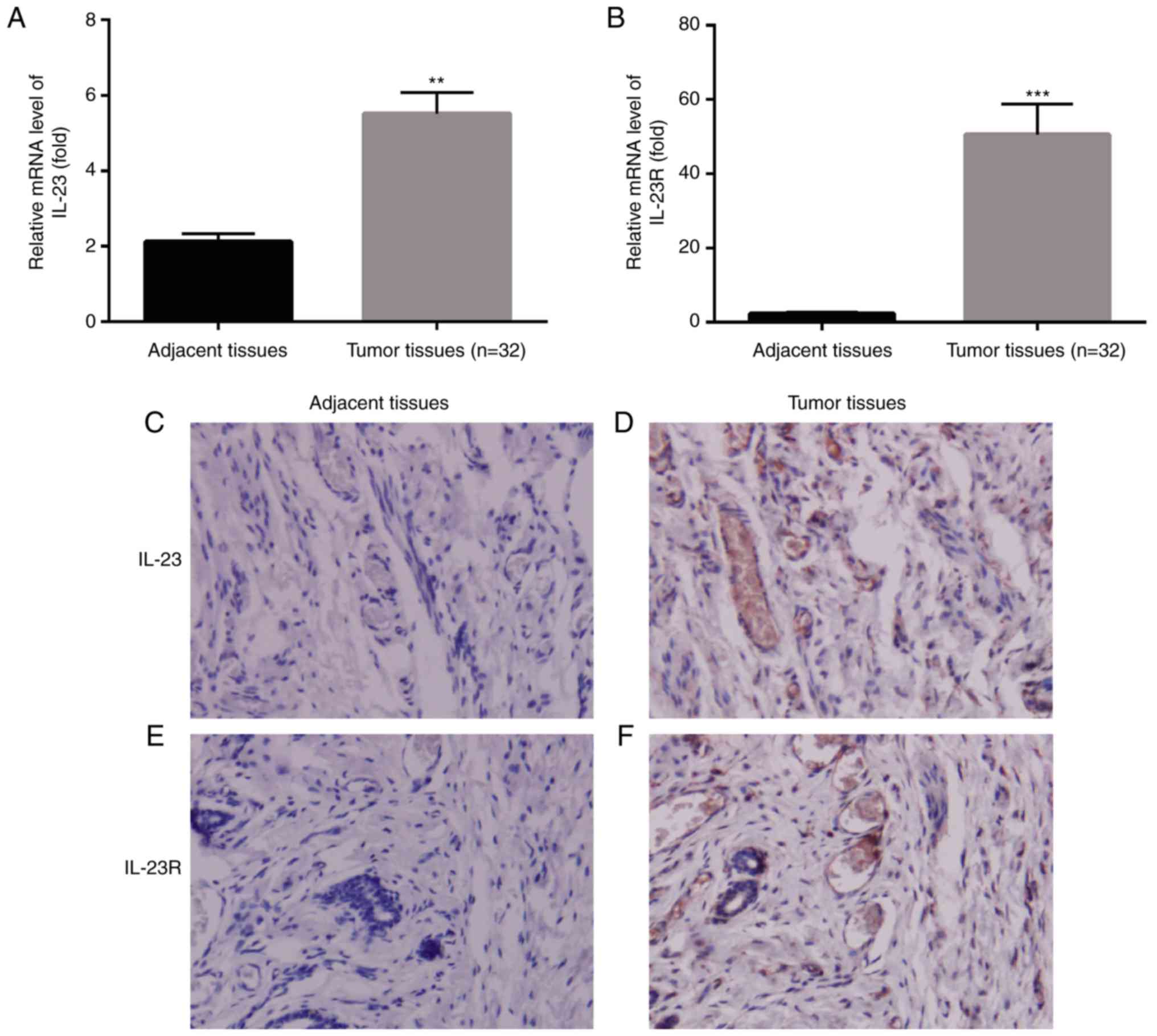
mRNA level of IL-23/IL-23R is higher
in breast cancer tissues than the adjacent tissues
Previous study has reported the differences of IL-23
mRNA expression level between tumor tissues and the adjacent normal
tissues in numerous types of organs, including conlon, ovarian,
lung, stomach and breast, among all of which there was significant
up-regulation of IL-23 mRNA level in tumor tissues than in the
normal tissues (12). In our
present study, we also first explored the differences of IL-23 mRNA
level between breast cancer tissue and the adjacent normal tissue
by RT-PCR. Results indicated that, IL-23 mRNA level was also
significantly higher in tumor tissue in comparison with the normal
tissue (P<0.01, n=32, Fig. 1A),
which was in consistent with the previous reported study (12). Moreover, IL-23 was considered as a
prognostic factor in breast cancer patients which was reported by
Gangemi et al (15).
Therefore, we put forward a hypothesis that IL-23-mediated
responses might be crucial for the promotion of tumor.
Studies found that genetic variants of IL-23R might
contribute to the pathological development of hepatitis to HCC
(16); while hepatitis B virus
could induce hepatitis by elevating IL-23 expression and lead to
liver damage via IL-23/IL-17 axis (17). IL-23R plays a crucial role in
initiating, maintaining and accelerating IL-23/IL-17 inflammatory
signaling pathway (10). Since
IL-23 mRNA level was upregulated in breast cancer tissue than in
the adjacent normal tissue, we were eager to know the changes of
IL-23R between tumor tissue and the normal tissue. RT-PCR results
exhibited that, there was much higher mRNA level of IL-23R in
breast cancer tissue compared to the normal tissue (P<0.001,
n=32, Fig. 1B).
Meanwhile, we detected the protein levels of IL-23
and IL-23R in the adjacent tissues and tumor tissues from patients
by immunohistochemistry. We found that, both IL-23 and IL-23R
exhibited much more expression level in tumor tissues in comparison
with the adjacent tissues (Fig.
1C-F).
Correlation between IL-23/IL-23R and
patients' characteristics
Both IL-23 and IL-23R were positively correlated
with patients' tumor size, TNM stage and metastasis. Patients were
divided into two groups according to the age, there were 17
patients aged ≥40 and 15 patients aged <40, IL-23 and IL-23R in
patients aged ≥40 years old were higher than in patients aged
<40 years old, however, there was no significant difference; as
for histological grade, there were 11 patients showed
well-intermediately differentiation while 21 patients showed poor
differentiation, IL-23/IL-23R in poor differentiation was a little
lower than in well-intermediately differentiation, whereas, there
was no significant difference; regarding invasion depth, there were
25 patients at T1-T2 and 7 patients at T3-T4, we found that,
IL-23/IL-23R levels were higher in T3-T4 group than in T1-T2 group,
however, there was no significant difference (Table I).
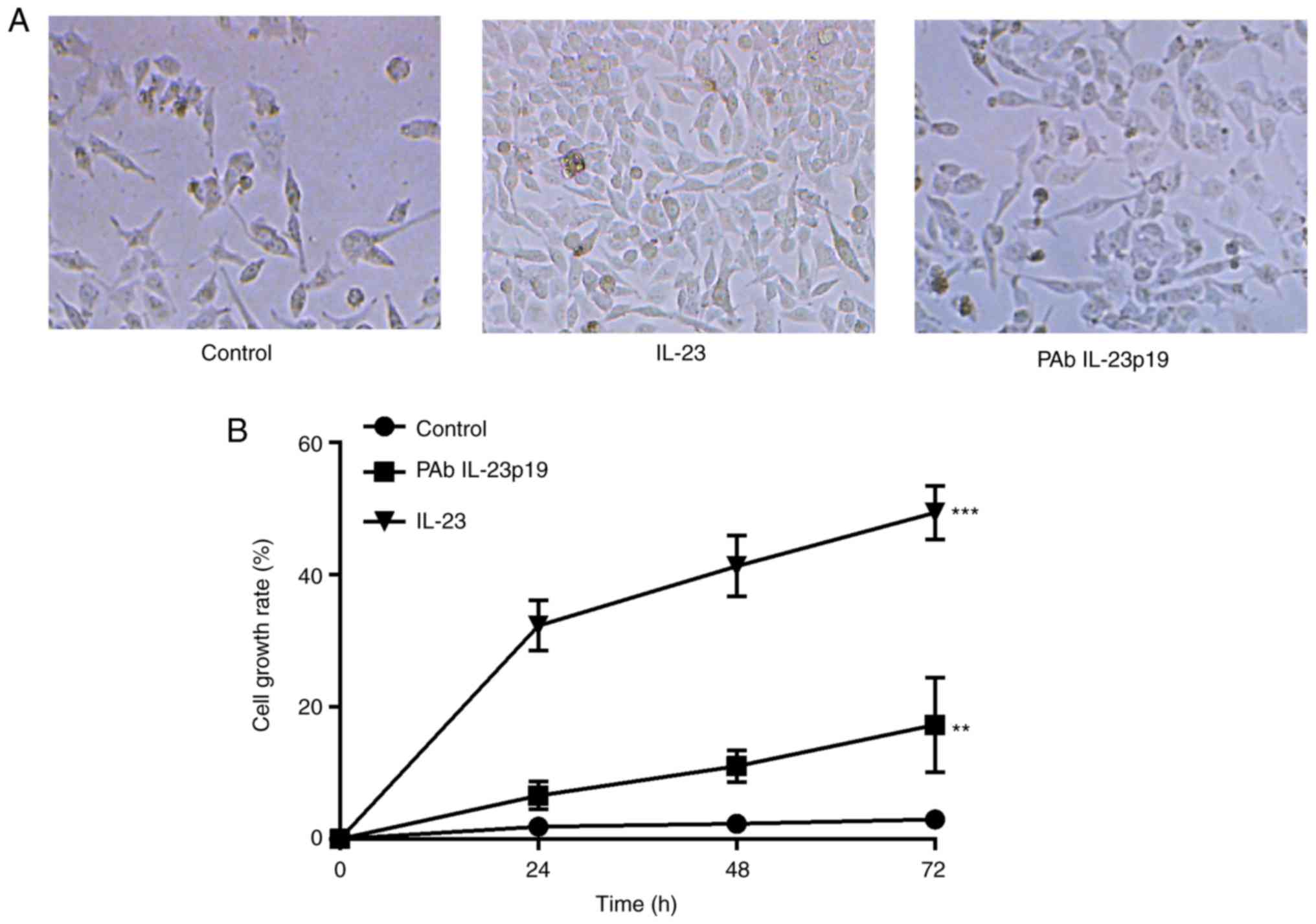
Pre-treatment of PAb IL-23p19
repressed IL-23 induced cell proliferation
To investigate whether IL-23 could induce cell
proliferation of breast cancer cells, cell proliferation assay was
performed. Cell proliferation rate revealed to be distinctly
increased by treatment with 10 ng/ml rhIL-23 in comparison with the
untreated control (P<0.001, n=3, Fig. 2A and B). Cells were further treated
with IL-23 at the presence of PAb IL-23p19, PAb IL-23p19 abolished
the induction of cell proliferation by IL-23 (P<0.01, n=3,
Fig. 2A and B). Taken together,
these results demonstrated that, the promotion of cell
proliferation was an effect of IL-23 in breast cancer cell line,
which could be distinctly inhibited by the pre-treatment of PAb
IL-23p19.
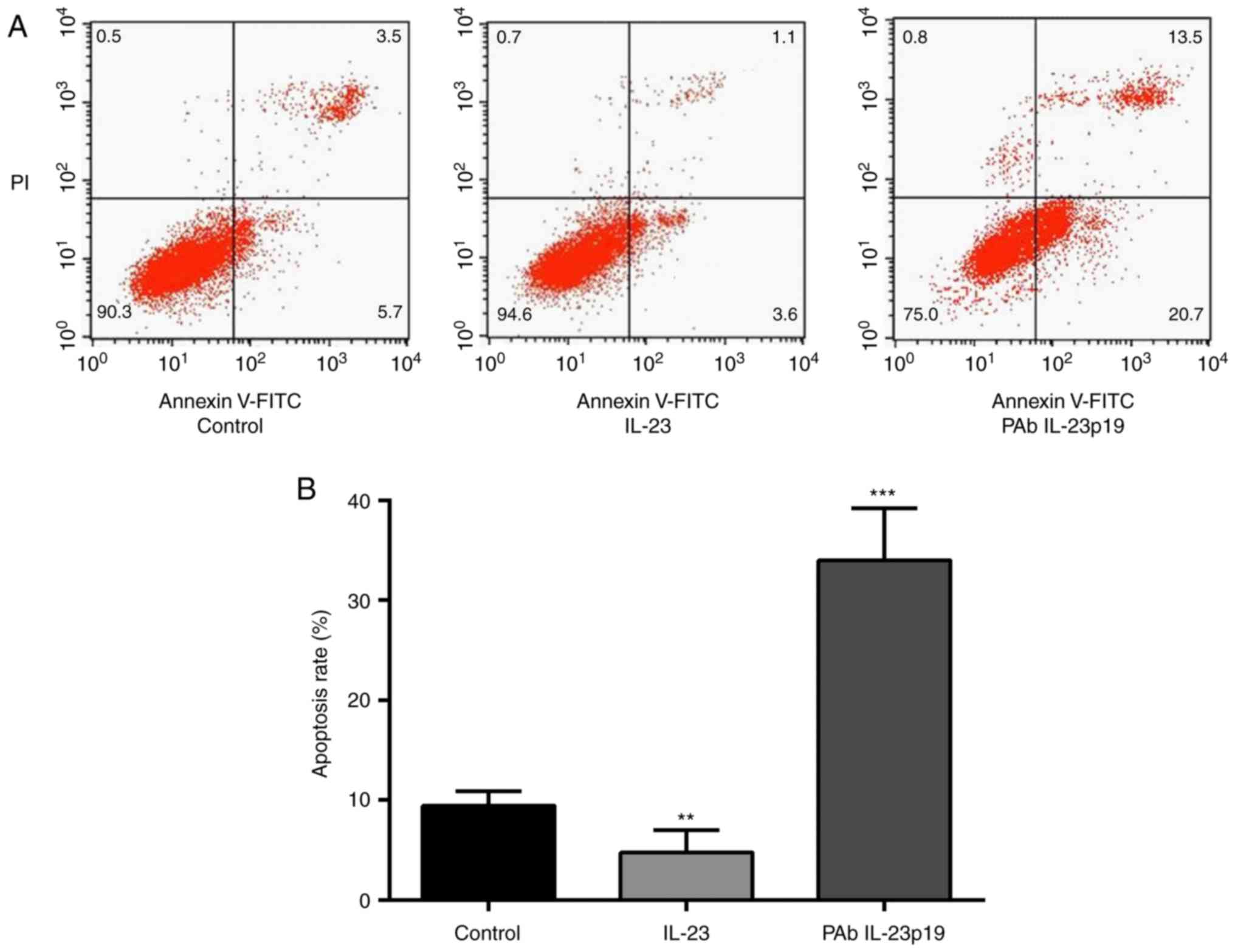
Pre-treatment of PAb IL-23p19 induced
IL-23 repressed cell proliferation
To investigate whether IL-23 could inhibit cell
apoptosis of breast cancer cells, FCM assay was carried out. Cell
apoptosis rate revealed to be distinctly decreased by 10 ng/ml
rhIL-23 in comparison with control group (P<0.01, n=3, Fig. 3A and B). Cells were further treated
with IL-23 at the presence of PAb IL-23p19, PAb IL-23p19 abolished
the inhibition of cell apoptosis by IL-23 (P<0.001, n=3,
Fig. 3A and B). Taken together,
these results suggested that, the inhibition of cell apoptosis is
also an effect of IL-23 in breast cancer cell line, which could be
abolished by the pre-treatment of PAb IL-23p19.
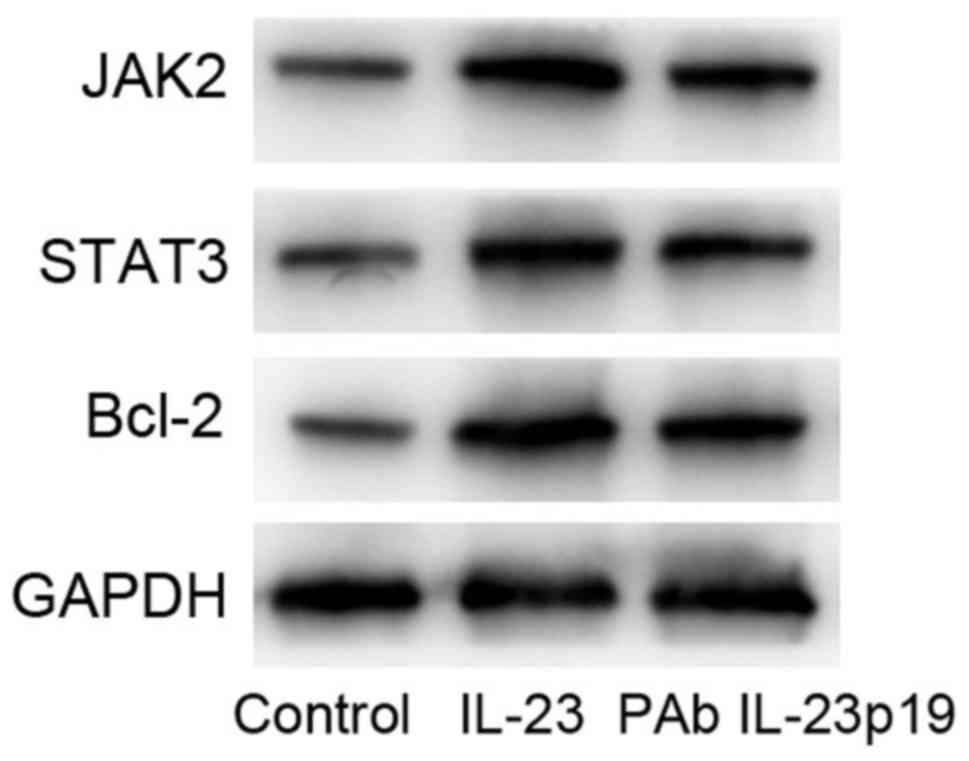
Pre-treatment of PAb IL-23p19
inhibited the activation of JAK2/STAT3 signaling pathway induced by
IL-23
Recent evidence indicated that IL-23-mediated
responses were crucial in promoting tumor progression of various
tissues. IL-23 signaling occurred by activation of the JAK/STAT
(specifically STAT3) pathway, thus resulting in up-regulation of
proteins which were linked with proliferation, survival (bcl-x),
metastasis (VEGF, HIF1α, MMP2, MMP9), angiogenesis and
immunosuppression, etc (18).
Consequently, we tested the expression levels of STAT3, JAK2 and
bcl-2 in MCF-7 cells by western blot. Results showed that, the
activation levels of STAT3 and JAK2 were induced by IL-23, and
pre-treatment of PAb IL-23p19 inhibited the activation of STAT3 and
JAK2. Meanwhile, bcl-2 protein expression was also upregulated by
IL-23, which was downregulated by pre-treatment of PAb IL-23p19
(Fig. 4). These results suggested
that, pre-treatment of PAb IL-23p19 abolished IL-23-mediated
aberrant cell proliferation and apoptosis by inhibiting the
activation of JAK2/STAT3 signaling pathway and reduced the
expression of bcl-2.
Discussion
IL-23 was identified as a cancer-related cytokine in
a recent study. The expression of IL-23 was significantly increased
in majority of human tumors from various organs compared to normal
adjacent tissues (12). As
acknowledged, multiple cancers arose from chronic inflammation and
inflammatory mediators were synthesized by tumors (19,20).
The ability to respond to IL-23 is determined by the expression
level of IL-23R in cells (21).
In our study, we indicated that mRNA and protein
levels of IL-23 and its receptor IL-23R were constitutively
co-expressed in breast cancer tissues, and IL-23/IL-23R was
increased in breast cancer tissues in comparison with the adjacent
tissues (Fig. 1), which was in
consistent with a previous reported research (12). Taken together, the results verified
the potential of IL-23 as a prognosis factor in breast cancer.
Thereafter, we determined the effects of IL-23 on
breast cancer cell line MCF-7 behaviors, and studied whether a
neutralizing antibody specific for IL-23, PAb IL-23p19 could
attenuate the effects of IL-23.
To investigate whether IL-23 could induce cell
proliferation of breast cancer cells, cell proliferation assay was
carried out. Cell proliferation rate revealed to be distinctly
increased by treatment with rhIL-23 in comparison with control;
cells were further treated with IL-23 at the presence of PAb
IL-23p19, and PAb IL-23p19 abolished IL-23 induced cell
proliferation (Fig. 2A and B).
We also investigate whether IL-23 could inhibit cell
apoptosis of breast cancer cells, FCM assay was carried out. Cell
apoptosis rate revealed to be distinctly decreased by rhIL-23 in
comparison with control; cells were further treated with IL-23 at
the presence of PAb IL-23p19, PAb IL-23p19 abolished the inhibition
of cell apoptosis by IL-23 (Fig. 3A
and B).
Taken together, these results suggested that, the
inhibition of cell apoptosis and promotion of cell proliferation by
IL-23 could be abolished by pre-administration of PAb IL-23p19.
Whereas, it was unclear which signaling pathway did IL-23 depend on
to function.
IL-23-mediated responses were indicated to be
crucial in promoting tumor progression. IL-23 signaling occurred by
activating JAK/STAT3 pathway, up-regulating proteins that were
correlated with proliferation and survival (bcl-x) (18). The JAK-STAT signaling cascade
consisted of three main components: One Janus kinase (JAK, a cell
surface receptor) and two signal transducer and activator of
transcription (STAT) proteins; JAK-STAT signaling pathway
transmitted information from extracellular chemical signals to
nucleus, generating DNA transcription and expression of genes which
were correlated with proliferation, apoptosis and oncogenesis
(22). Disruption or dysregulation
of JAK-STAT functionality lead to immune deficiency syndromes and
cancers (22). Activated JAKs
phosphorylated tyrosine residues on the receptor, creating binding
sites for proteins which possessed SH2 domains; SH2 domain
containing STATs were recruited to the receptor and
tyrosine-phosphorylated (activated) by JAKs followed by forming
hetero- or homodimers and translocating to the cell nucleus to
induce target gene transcription (23). Moreover, activation of JAK2/STAT3
pathway contributed to the poor outcomes of metastatic breast
cancers (24,25).
Consequently, we tested the expression levels of
STAT3, JAK2 and bcl-2 in MCF-7 cells by western blot. Results
showed that, the activation levels of STAT3 and JAK2 were induced
by IL-23, and pre-treatment of PAb IL-23p19 inhibited the
activation of STAT3 and JAK2. Meanwhile, bcl-2 protein expression
was also upregulated by IL-23, which was downregulated by
pre-treatment of PAb IL-23p19 (Fig.
4). These results suggested that, pre-treatment of PAb IL-23p19
abolished IL-23-mediated aberrant cell proliferation and apoptosis
by inhibiting the activation of JAK2/STAT3 signaling pathway and
reduced the expression of bcl-2.
In conclusion, blocking the function of IL-23
inhibited the proliferative activity and induced the apoptotic
activity of tumor cells via inhibiting the activation of JAK2/STAT3
pathway in MCF-7 cells. We provide a potential therapeutic target
for breast cancer.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar
|
|
3
|
Cardoso F, Harbeck N, Barrios CH, Bergh J,
Cortés J, EI Saghir N, Francis PA, Hudis CA, Ohno S, Partridge AH,
et al: Research needs in breast cancer. Ann Oncol. 28:208–217.
2017.PubMed/NCBI
|
|
4
|
Harbeck N and Gnant M: Breast cancer.
Lancet. 389:1134–1150. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ahern PP, Izcue A, Maloy KJ and Powrie F:
The interleukin-23 axis in intestinal inflammation. Immunol Rev.
226:147–159. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Langrish CL, Chen Y, Blumenschein WM,
Mattson J, Basham B, Sedgwick JD, McClanahan T, Kastelein RA and
Cua DJ: IL-23 drives a pathogenic T cell population that induces
autoimmune inflammation. J Exp Med. 201:233–240. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lee E, Trepicchio WL, Oestreicher JL,
Pittman D, Wang F, Chamian F, Dhodapkar M and Krueger JG: Increased
expression of interleukin 23 p19 and p40 in lesional skin of
patients with psoriasis vulgaris. J Exp Med. 199:125–130. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen Z, Laurence A and O'Shea JJ: Signal
transduction pathways and transcriptional regulation in the control
of Th17 differentiation. Semin Immunol. 19:400–408. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Volpe E, Servant N, Zollinger R, Bogiatzi
SI, Hupé P, Barillot E and Soumelis V: A critical function for
transforming growth factorbeta, interleukin 23 and proinflammatory
cytokines in driving and modulating human T(H)-17 responses. Nat
Immunol. 9:650–657. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cho JH and Weaver CT: The genetics of
inflammatory bowel disease. Gastroenterology. 133:1327–1339. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Le Gouvello S, Bastuji-Garin S, Aloulou N,
Mansour H, Chaumette MT, Berrehar F, Seikour A, Charachon A, Karoui
M, Leroy K, et al: High prevalence of Foxp3 and IL17 in MMR
proficient colorectal carcinomas. Gut. 57:772–779. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Langowski JL, Zhang X, Wu L, Mattson JD,
Chen T, Smith K, Basham B, McClanahan T, Kastelein RA and Oft M:
IL-23 promotes tumor incidence and growth. Nature. 442:461–465.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Balkwill F and Mantovani A: Inflammation
and cancer: Back to Virchow? Lancet. 357:539–545. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fukuda M, Ehara M, Suzuki S and Sakashita
H: Expression of interleukin-23 and its receptors in human squamous
cell carcinoma of the oral cavity. Mol Med Rep. 3:89–93.
2010.PubMed/NCBI
|
|
15
|
Gangemi S, Minciullo P, Adamo B, Franchina
T, Ricciardi GR, Ferraro M, Briguglio R, Toscano G, Saitta S and
Adamo V: Clinical significance of circulating interleukin-23 as a
prognostic factor in breast cancer patients. J Cell Biochem.
113:2122–2125. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xu Y, Liu Y, Pan S, Liu L, Liu J, Zhai X,
Shen H and Hu Z: IL-23R polymorphisms, HBV infection and risk of
hepatocellular carcinoma in a high-risk Chinese population. J
Gastroenterol. 48:125–131. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang Q, Zhou J, Zhang B, Tian Z, Tang J,
Zheng Y, Huang Z, Tian Y, Jia Z, Tang Y, et al: Hepatitis B virus
induces IL-23 production in antigen presenting cells and causes
liver damage via the IL-23/IL-17 axis. PLoS Pathog. 9:e10034102013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yu H, Kortylewski M and Pardoll D:
Crosstalk between cancer and immune cells: Role of STAT3 in the
tumour microenvironment. Nat Rev Immunol. 7:41–51. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Coussens LM and Werb Z: Inflammation and
cancer. Nature. 420:860–867. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Balkwill F, Charles KA and Mantovani A:
Smoldering and polarized inflammation in the initiation and
promotion of malignant disease. Cancer Cell. 7:211–217. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Parham C, Chirica M, Timans J, Vaisberg E,
Travis M, Cheung J, Pflanz S, Zhang R, Singh KP, Vega F, et al: A
receptor for the heterodi-meric cytokine IL-23 is composed of
IL-12Rbeta1 and a novel cytokine receptor subunit, IL-23R. J
Immunol. 168:5699–5708. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Aaronson DS and Horvath CM: A road map for
those who don't know JAK-STAT. Science. 296:1653–1655. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hebenstreit D, Horejs-Hoeck J and Duschl
A: JAK/STAT-dependent gene regulation by cytokines. Drug News
Perspect. 18:243–249. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Marotta LL, Almendro V, Marusyk A,
Shipitsin M, Schemme J, Walker SR, Bloushtain-Qimron N, Kim JJ,
Choudhury SA, Maruyama R, et al: The JAK2/STAT3 signaling pathway
is required for growth of CD44+CD24 stem cell-like
breast cancer cells in human tumors. J Clin Invest. 121:2723–2735.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hennessy BT, Gonzalez-Angulo AM,
Stemke-Hale K, Gilcrease MZ, Krishnamurthy S, Lee JS, Fridlyand J,
Sahin A, Agarwal R, Joy C, et al: Characterization of a naturally
occurring breast cancer subset enriched in
epithelial-to-mesenchymal transition and stem cell characteristics.
Cancer Res. 69:4116–4124. 2009. View Article : Google Scholar : PubMed/NCBI
|