Introduction
The human epidermal growth factor receptor 2 gene
(HER2) is located on chromosome 17q12. In 1987, Slamon et
al (1) proposed that the
amplification of HER2 was associated with breast cancer
prognosis. Subsequently, HER2 has been revealed to be
amplified, or HER2 protein overexpressed, in between 20 and 30% of
patients with breast cancer. These patients are generally diagnosed
with high-grade cancer with increased rates of cell proliferation
and a tendency to metastasize to the lymph nodes. Prognosis of
these patients is markedly poorer compared with patients with
breast cancer who do not overexpress HER2 (2–4).
Herceptin/trastuzumab combined with chemotherapy may improve the
quality of life of patients with HER2-positive breast cancer and
prolong their disease-free survival time. Although a limited number
have been described, occasional side effects of Herceptin treatment
do occur, including cardiac toxicity that may weaken cardiac
contractility, leading to cardiac insufficiency (5–9). On this
basis, HER2 status is an important marker for selecting
suitable therapy.
The HER2 test guidelines set out by the
American Society of Clinical Oncology/College of American
Pathologists (ASCO/CAP) were updated in 2013 from the previous 2007
version; the evaluation standards of immunohistochemistry (IHC) and
in situ hybridization (ISH) test results were revised in
these guidelines (10,11). In China, HER2 IHC is extensively
applied as a preliminary screen, whereas ISH is primarily
considered a confirmatory test for HER2 gene amplification,
with the most common ISH test involving double-probe fluorescence
(FISH). Distinctions between the 2013 and 2007 ASCO/CAP evaluation
standards of double-probe FISH results are as follows: i) The
threshold value of HER2 amplification was adjusted to be
≥2.0 (≥2.2 in the 2007 version); ii) in the 2013 version,
HER2 amplification was also defined as HER2/CEP17
<2.0 with mean HER2 copies/nucleus ≥6.0, or HER2/CEP17
≥2.0 with mean HER2 copies/nucleus <4.0. In the 2007
version, these values were considered to represent
non-amplification (HER2/CEP17 <1.8) for patients
identified with simultaneous HER2 and chromosome 17
centromere locus amplification. However, in the 2013 version
HER2 is considered to be amplified in these patients and,
therefore, these patients should be considered for
HER2-targeted therapy. The aim of the present study was to
evaluate the patients that did not exhibit HER2
amplification by 2007 standards, but with potential HER2
amplification by 2013 guidelines.
The selection of control genes for investigations
using double probes is important. A control gene was selected for
chromosome 17 to exclude influences of chromosome 17 polysomy in
cancer cells. A second control gene was selected that is
sufficiently distant from HER2 so as to remain stable when
HER2 is amplified. On the basis of double-probe FISH studies
by Troxell et al (12) and
Varga et al (13), chromosome
enumeration probe 17 (CEP17), tumor protein p53 (TP53) and
retinoic acid receptor (RARA) were selected as controls for
HER2.
In the present study, a retrospective analysis was
performed to review HER2 FISH-analyzed cases and to compare
the 2007 and 2013 ASCO/CAP guidelines. Alterations in HER2
status following the introduction of novel control genes were also
determined. In addition, the effect of amplification or deletion,
or polysomy of CEP17 in screening patients for targeted therapy was
investigated.
Patients and methods
Samples
Specimens from 1518 patients with breast cancer were
previously analyzed by HER2 FISH between February 2011 and
January 2015; samples were collected from 15 hospitals, including
The First Affiliated Hospital of Chongqing Medical University, The
Second Affiliated Hospital of Chongqing Medical University,
Yongchuan Hospital Chongqing Medical University, The Hospital of
Traditional Chinese Medicine of Chongqing, The Fifth People's
Hospital of Chongqing, The Ninth People's Hospital of Chongqing,
The People's Hospital of Chongqing Rongchang, The Centre's Hospital
of Chongqqing Jiangjin, The People' Hospital of Chongqing Bishan,
The People's Hospital of Chongqing Changshou, The People's Hospital
of Chongqing Hechuan, The People's Hospital of Chongqing Qijiang,
The People's Hospital of Chongqing Tongliang, The Centre's Hospital
of Chongqing Fuling. FISH was performed for patients exhibiting
medium to strong HER2 IHC levels prior to Herceptin administration,
according to the ASCO/CAP 2013 criteria (11). From this FISH analysis, 67
specimenswith suspected amplification, polysomy and monosomy of
CEP17 were selected for inclusion in the present study. This
retrospective study was approved by the Chongqing Medical
University ethics committee.
FISH
Paraffin-embedded tissue samples (from the 67
selected patients) were fixed in 10% neutral buffered formalin at
room temperature for between 24 and 48 h, and were sectioned at a
thickness of 4 µm. Hematoxylin and eosin staining for 5–10 min at
room temperature was performed to label infiltrating carcinomas,
and observation with an Olympus BX41 microscope (magnification,
×40). FISH for HER2, CEP17, TP53 and RARA was
performed on paraffin sections according to the manufacturer's
instructions (each individual probe of HER2, CEP17,
TP53 and RARA and solid tumor FISH testing protocol
were obtained from Beijing GP Medical Technologies, Ltd.; China
Medical Technologies Inc., Beijing, China). Information about
marker probes is presented in Table
I. Fluorescence signal observation, photography and analysis
were performed using an Olympus BX51 fluorescence microscope
(magnification, ×100) and FISH software (version 2.0; Beijing GP
Medical Technologies, Ltd.; China Medical Technologies Inc.).
HER2 status was interpreted according to the 2007 and 2013
ASCO/CAP HER2 test guidelines as well as the control genes,
TP53 and RARA.
 | Table I.Labeled probes on chromosome 17. |
Table I.
Labeled probes on chromosome 17.
| Gene | Color | Marker site |
|---|
| Human epidermal
growth factor receptor 2 | Red | 17q11.2-q12 |
| Chromosome
enumeration probe 17 | Green | 17p11.1-q11.1 |
| Tumor protein
p53 | Green | 17p13.1 |
| Retinoic acid
receptor α | Red | 17q21.1 |
Results
FISH for CEP17 and HER2, as well as
TP53 and RARA was performed on 67 samples. According
to ASCO/CAP 2007 guidelines, 20 patients exhibited HER2
amplification (29.85%; 16 with CEP17 monosomy and 4 with partial
CEP17 deletion), which was consistent with HER2/CEP17 ≥2.0
(Table II). On this basis,
HER2 was concluded to be amplified. A total of 6 patients
were revealed to be equivocal for HER2/CEP17 (4 patients
with 2.2> HER2/CEP17 >2.0 and 2 patients with 1.8
<HER2/CEP17 <2.0). A total of 41 patients did not
experience HER2 amplification, including 25 with polysomy (6
with CEP17 and HER2 cluster-amplification and 19 with CEP17
and HER2 punctiform-amplification), 15 with monosomy and 1
with suspected monosomy plus co-amplification of HER2 and
CEP17.
 | Table II.Human epidermal growth factor 2 gene
status according to distinct interpretation standards. |
Table II.
Human epidermal growth factor 2 gene
status according to distinct interpretation standards.
|
|
| ASCO/CAP 2013 | Tumor protein p53
or retinoic acid receptor α |
|---|
|
|
|
|
|
|---|
| ASCO/CAP 2007 | n | Non-amplified | Equivocal | Amplified | Non-amplified | Equivocal | Amplified |
|---|
| Amplified | 20 | 0 | 0 | 20 | 4 | 0 | 16 |
| Equivocal | 6 | 0 | 0 | 6 | 0 | 0 | 6 |
| Non-amplified | 41 | 18 | 0 | 23 | 20 | 0 | 21 |
| Total | 67 | 18 | 0 | 49 | 24 | 0 | 43 |
Table II presents
HER2 status according to various interpretation standards
(ASCO/CAP 2007, ASCO/CAP 2013 and reference genes TP53 or
RARA). According to ASCO/CAP 2013 guidelines, 49 patients
were diagnosed with HER2 amplification (73%). The additional
29 patients who were not diagnosed with HER2 amplification
according to the 2007 criteria included 6 patients originally at
the equivocal level but now demonstrating amplification (4 patients
with HER2/CEP17 ≥2.0 and 2 patients with 1.8 <
HER2/CEP17 <2.0 but HER2 ≥6 signals/nucleus), 22
patients originally with polysomy but now exhibiting amplification
(HER2/CEP17 <2, but HER2 ≥6 signals/nucleus) and 1
patient with suspected monosomy plus co-amplification of
HER2 and CEP17 (HER2/CEP17 <2, but HER2 ≥6
signals/nucleus).
The introduction of TP53, RARA and
CEP17 as control genes indicated that HER2 was amplified in
43 patients (64.2%). A total of 6 patients with HER2
amplification according to ASCO/CAP 2013 guidelines did not exhibit
amplification following the introduction of TP53 and
RARA control genes. Among these 6 patients, 4 exhibited
normal TP53 and RARA, partial CEP17 deletion,
HER2/CEP17≥2, but HER2/TP53 <2,
HER2/RARA <2 and HER2 <4
signals/nucleus, and the remaining 2 patients demonstrated
HER2 ≥6 signals/nucleus and HER2/CEP17 <2, but
HER2/TP53 <2 and HER2/RARA <2, on
which basis polysomy was defined. Of the 15 patients with monosomy,
3 patients exhibited normal TP53 and RARA, therefore
the number of monosomic patients was 12.
Using TP53, RARA and CEP17 as control
genes, the incidence of chromosome 17 polysomy in 1,518 patients
was 0.2% (3/1,518) and the incidence of monosomy was 0.8%
(12/1,518). The incidence of co-amplification of HER2 and
CEP12 was 1.4% (21/1518).
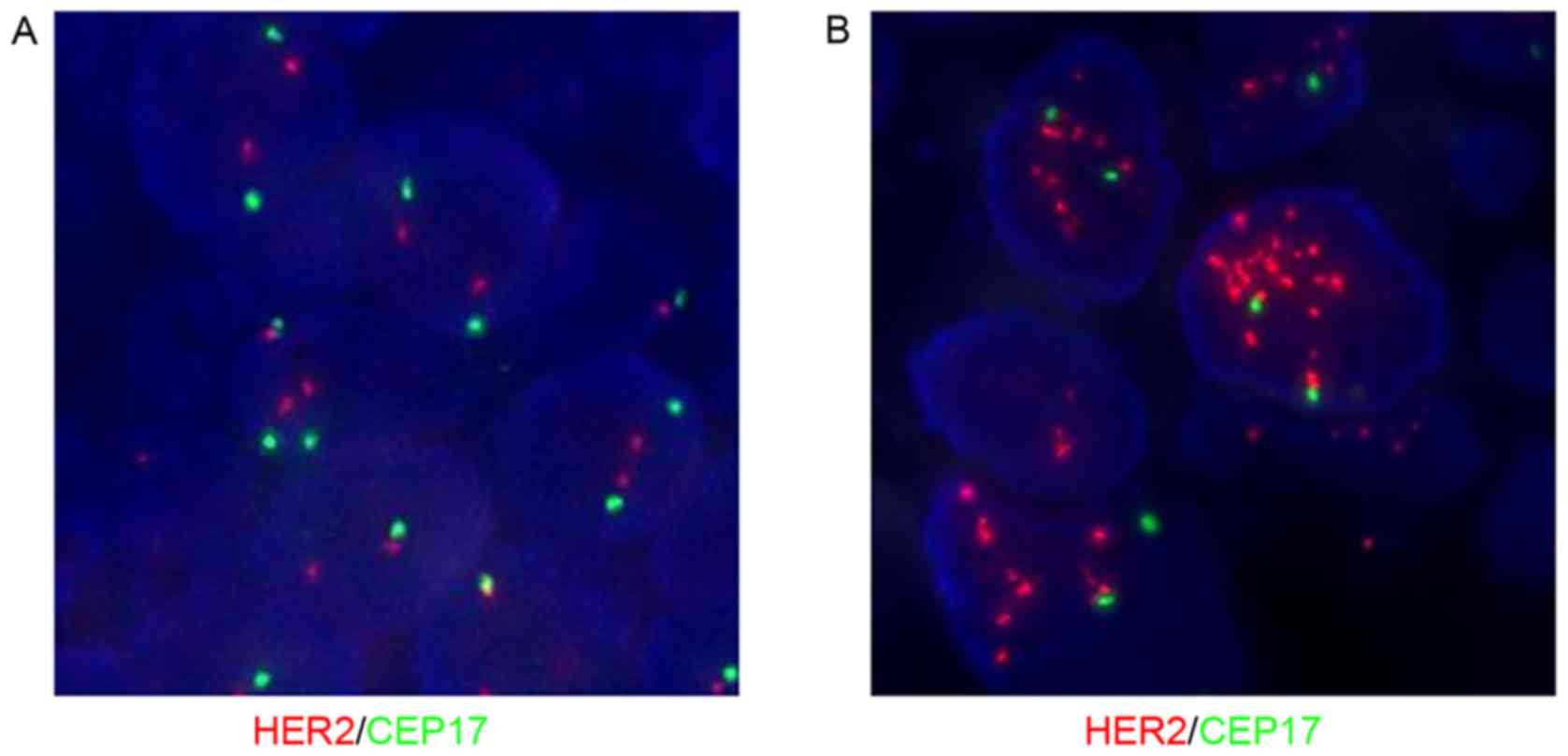
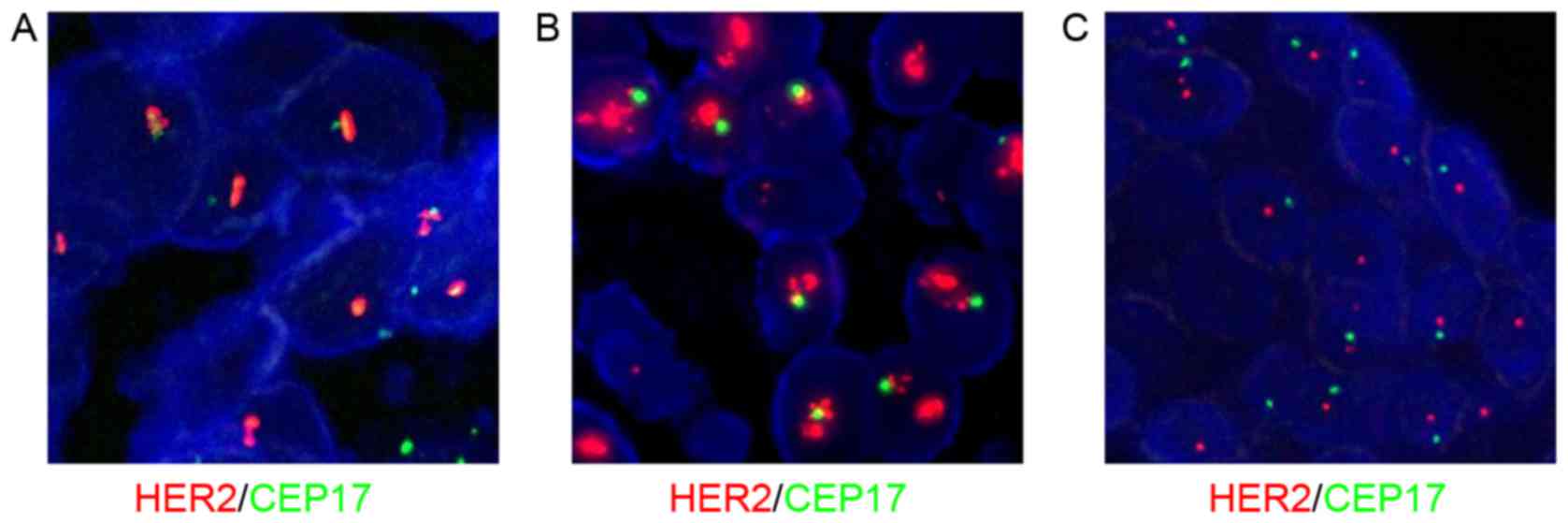
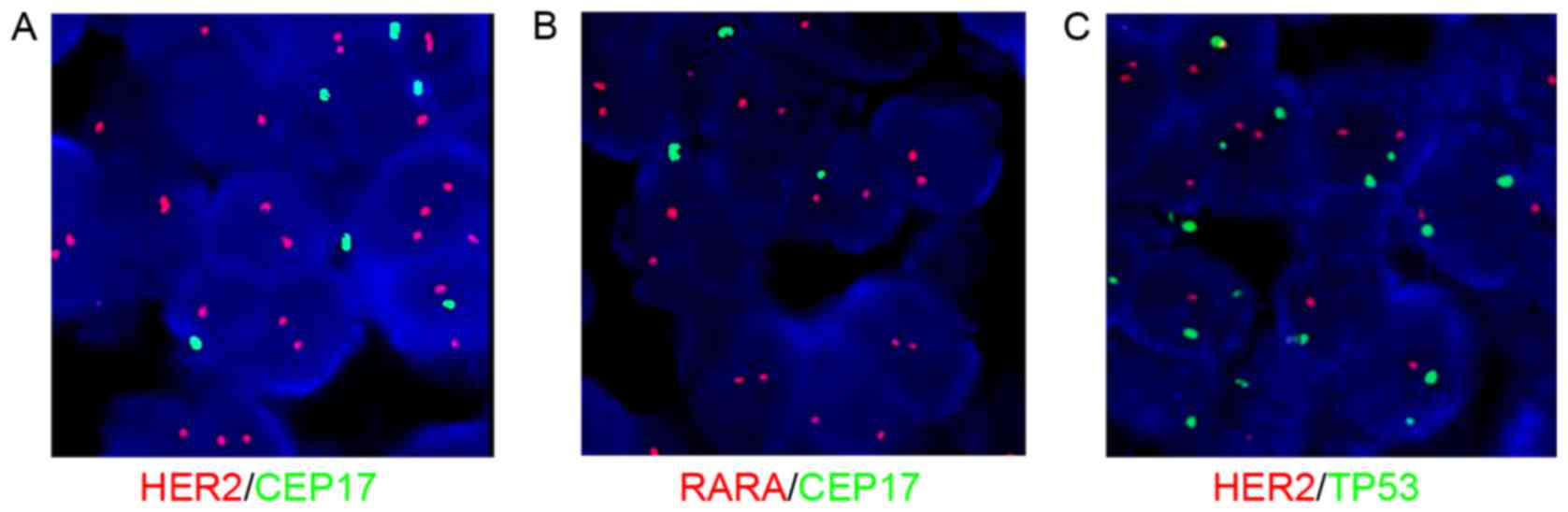
HER2 status was associated with the status of
CEP17 and the reference genes. Fig. 1
demonstrates common HER2 and CEP17 status using FISH.
Fig. 2 reveals co-amplification of
HER2 and CEP17 polysomy. If only applying CEP17,
HER2/CEP17 <2 and therefore HER2 was not amplified
according to the 2007 ASCO/CAP version, but was amplified according
to the 2013 version (HER2 ≥6 signals/nucleus). Fig. 3 reveals that chromosome 17 monosomy
was accompanied by irregular HER2 and CEP17 status. Fig. 4 demonstrates CEP17 deletion by FISH.
If only applying CEP17, HER2/CEP17 ≥2 and therefore
HER2 was amplified according to the 2013 version of ASCO/CAP
guidelines. However, FISH analysis of TP53 and RARA
revealed HER2 to be normal.
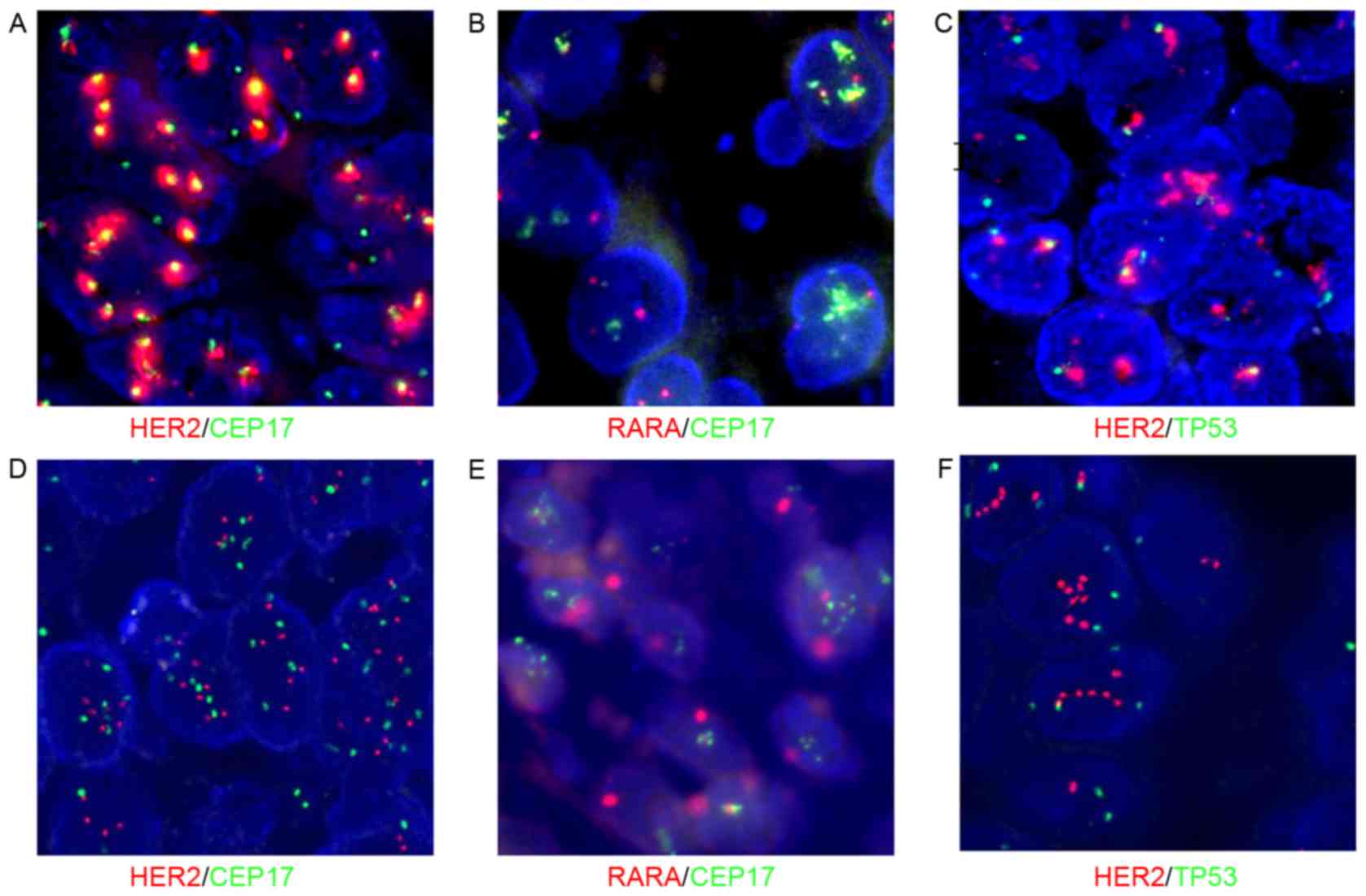 | Figure 2.Co-amplification of HER2 and
CEP17 without polysomy, confirmed using fluorescence in situ
hybridizationfor TP53 and RARA genes. Magnification,
100×10. (A) HER2/CEP17, co-amplification of HER2
(red) and CEP17 (green). (B) RARA/CEP17, normal RARA
(red) and amplification of CEP17 (green). (C)
HER2/TP53, HER2 (red) amplification and normal
TP53 (green). (A-C) Samples from the same case, which
exhibits a high level of co-amplification of HER2 and CEP17.
(D) HER2/CEP17, moderate co-amplification of HER2
(red) and CEP17 (green). (E) RARA/CEP17, CEP17 (green)
amplification and normal RARA (red). (F)
HER2/TP53, HER2 (red) amplification and normal
TP53 (green). (D-F) Samples from the same case, which was
characterized by moderate amplification of HER2 and CEP17.
HER2, human epidermal growth factor receptor 2; CEP17,
chromosome enumeration probe 17; TP53, tumor protein p53;
RARA, retinoic acid receptor α. |
Discussion
Samples without HER2 amplification according
to the ASCO/CAP 2007 HER2 test guidelines may be classified
as with HER2 amplification according to the revised 2013
HER2 test guidelines, particularly in contentious
co-amplified specimens. This suggests that these patients may
benefit from HER2-targeted medicine. Therefore, in the
present study, FISH results from 1,518 patients were reviewed and
67 patients were identified with abnormal CEP17 signals, including
suspicious co-amplification, depletion, polysomy and monosomy.
The incidence rate of co-amplification of
HER2 and CEP17 was 1.4% (21/1518), which demonstrates
distinction from previous studies. Troxell et al (12) identified that 7/858 patients with
cancer exhibited abnormal HER2 and CEP17 (6 with breast
cancer and 1 with ovarian carcinoma); the incidence rate of CEP17
amplification was 0.8%, whereas no HER2 amplification was
revealed in 3/7 patients. On this basis, the incidence rate of
co-amplification was 0.47%. Varga et al (13) identified that 14 patients were
diagnosed with co-amplification of >5,000 patients with breast
cancer who underwent FISH analysis between 1999 and 2009, on the
basis of which, the co-amplification incidence rate was 0.3%. Press
(14) observed co-amplification in
2/2,600 patients with breast cancer, on the basis of which the
co-amplification incidence rate was 0.08%. Gunn et al
(15) selected 20 patients who
exhibited unclear HER2 status following routine FISH and IHC
investigations, and identified HER2 status through
array-based comparative genomic hybridization (aCGH).
Co-amplification of HER2 and CEP17 was observed in 3/20
patients, for which the co-amplification rate was 15% in patients
suspected to be positive for HER2; there was a tendency for
a false negative result if based only on the HER2/CEP17
ratio. Marchio et al (16)
randomly selected 18 patients (~8% of all cases) with a mean CEP17
>3 signals/nucleus to perform an aCGH test and identified that
17q containing the centromere locus was amplified in 11 patients,
17q excluding the centromere locus was amplified in 1 patient and
was combined with true polysomy in 1 other patient, whereas
amplification of only the centromere locus was identified in 5
patients. Therefore, the co-amplification incidence rate was 61.1%
(11/18). On this basis, the overall co-amplification rate was 4.9%.
Tse et al (17) selected 171
patients with a mean CEP17 signals/nucleus of >2.6 to analyze
HER2 FISH results from 5,683 patients. Novel control genes
were introduced into the interpretation standards, RARA and
TP53. Following the introduction of these control genes,
HER2 of 58 patients (43.9%) was defined to be amplified in
132 patients previously identified as non-amplified (on the 2007
ASCO/CAP criteria of HER2/CEP17). HER2 gene
amplification was identified in 13/14 patients at the threshold
value. The ratio of HER2/CEP17 was at the threshold value of
1.8–2.2 or HER2 gene copy 4.0–6.0. Additionally, HER2
status continued to be defined as amplified in 25 patients in whom
amplification was classified previously. The results observed a
limited number of patients with polysomy, and the co-amplification
rate was 1.8% [(58+13+25)/5863]. Egervari et al (18) investigated chromosome 17 polysomy and
observed, using FISH, that 5/405 patients with breast cancer
presented CEP17 ≥3 alongside HER2 amplification, on the
basis of which the co-amplification incidence was 1.23%. At the
same time, Egervari et al (18) proposed that a pseudomorph of
chromosome 17 polysomy was induced by CEP17 centromere locus
amplification and therefore the incidence of chromosome 17 polysomy
may be less.
Distinctions were observed in the incidence rates of
co-amplification between the results of the present study and the
aforementioned previous studies. A total of 22/1518 patients,
analyzed using FISH in the present study, were observed to exhibit
co-amplification, all of whom presented with medium to strong
levels of HER2 IHC and excluded HER2 negative and weak specimens.
If counting these negative or weak specimens, the incidence rate of
co-amplification was ~0.55% (22/4016).
Currently, the definitions of polysomy and monosomy
are as follows, polysomy occurs when an entire chromosome is
duplicated one or more times, whereas monosomy is the result of
complete deletion of a chromosome (11). With the inclusion of the control genes
TP53 and RARA in the present study, the incidence
rate of polysomy was ~0.2% (3/1518), suggesting that true polysomy
was less common than what was previously observed in the
literature. In cases where increased levels of polysomy are
detected, it may have occurred due to CEP17 amplification, as
suggested by Zeng et al (19),
whereas decreases in polysomy incidence rate may be caused by the
section thickness being less than the diameter of cells (20,21).
Chromosome 17 polysomy may indicate poor efficacy of cytotoxic
medicines, leading to tumor metastasis (22,23), on
the basis of which Herceptin and/or anthracyclines may be more
suitable. However, whether patients with breast cancer who exhibit
chromosome 17 polysomy should receive Herceptin therapy is
disputed. Moelans et al (24)
recommended not using the term ‘polysomy 17’ when in actuality a
‘CEP17 copy number increase’ was meant. Hanna et al
(25) suggested that mean HER2
copies/cell should replace the HER2/CEP17 ratio to evaluate
HER2 status.
Currently, compared with polysomy, investigations
into monosomy are rare. Following the inclusion of TP53 and
RARA control genes in the present study, the number of
patients with monosomy was decreased from 15 to 12. The 3
discrepant cases experienced CEP17 deletion rather than true
monosomy, leading to HER2 false positives (HER2/CEP17
≥2). Those patients with HER2 amplification induced by true
monosomy were not sensitive to targeted therapy and prognosis was
unsatisfactory (26).
In the present study, no TP53 or RARA
amplification was identified in breast cancer cells. Therefore,
TP53 and RARA may be considered as control genes of
HER2, suitable for the diagnosis of suspected HER2
and CEP17 co-amplification. However, TP53 and RARA
only represent part of, not the whole of, chromosome 17.
Previous studies indicate that gene sequencing may
be carried out directly on chromosome 17 based on aCGH (16). Observation using aCGH of whether
HER2 was amplified was the optimal method to evaluate gene
status, which was expensive. It was reported that when chromosome
17 was in a complex gene status, whole gene tests were recommended
as positive FISH results were consistent with results of aCGH tests
(16).
In conclusion, HER2 was previously determined
to not be amplified in 29 patients but was revealed, through
retrospective analysis in the present study, to be amplified
according to ASCO/CAP 2013 HER2 test guidelines. HER2
in 23 patients which had previously been judged to not be
amplified, was revealed to be amplified following the inclusion of
RARA and TP53 control genes. The distinction of
HER2 status is important as it enables patients to receive
targeted medicine. ASCO/CAP 2013 HER2 test guidelines are
more accurate than 2007 guidelines. In addition, RARA and
TP53 may be considered suitable control genes to evaluate
HER2 status.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 81100443), Chongqing
Yuzhong District Science and Technology Plan projects (grant no.
20120214) and Chongqing Municipal Health Bureau Scientific Research
Project (grant no. 20132151).
References
|
1
|
Slamon DJ, Clark GM, Wong SG, Levin WJ,
Ullrich A and McGuire WL: Human breast cancer: Correlation of
relapse and survival with amplification of the HER-2/neu oncogene.
Science. 235:177–182. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pauletti G, Dandekar S, Rong H, Ramos L,
Peng H, Seshadri R and Slamon DJ: Assessment of methods for
tissue-based detection of the HER-2/neu alteration in human breast
cancer: A direct comparison of fluorescence in situ hybridization
and immunohistochemistry. J Clin Oncol. 18:3651–3664. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ross JS, Fletcher JA, Bloom KJ, Linette
GP, Stec J, Clark E, Ayers M, Symmans WF, Pusztai L and Hortobagyi
GN: HER-2/neu testing in breast cancer. Am J Clin Pathol. 120
Suppl:S53–S71. 2003.PubMed/NCBI
|
|
4
|
Winston JS, Ramanaryanan J and Levine E:
HER-2/neu evaluation in breast cancer: Are we there yet? Am J Clin
Pathol. 121 (Suppl):S33–S49. 2004.PubMed/NCBI
|
|
5
|
Slamon DJ, Leyland-Jones B, Shak S, Fuchs
H, Paton V, Bajamonde A, Fleming T, Eiermann W, Wolter J, Pegram M,
et al: Use of chemo therapy plus a monoclonal antibody against HER2
for metastatic breast cancer that overexpresses HER2. N Engl J Med.
344:783–792. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Vogel CL, Cobleigh MA, Tripathy D, Gutheil
JC, Harris LN, Fehrenbacher L, Slamon DJ, Murphy M, Novotny WF,
Burchmore M, et al: Efficacy and safety of trastuzumab as a single
agent in first-line treatment of HER2-overexpressing metastatic
breast cancer. J Clin Oncol. 20:719–726. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cobleigh MA, Vogel CL, Tripathy D, Robert
NJ, Scholl S, Fehrenbacher L, Wolter JM, Paton V, Shak S, Lieberman
G and Slamon DJ: Multinational study of the efficacy and safety of
humanized anti-HER2 monoclonal antibody in women who have
HER2-overexpressing metastatic breast cancer that has progressed
after chemotherapy for metastatic disease. J Clin Oncol.
17:2639–2648. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Romond EH, Perez EA, Bryant J, Suman VJ,
Geyer CE Jr, Davidson NE, Tan-Chiu E, Martino S, Paik S, Kaufman
PA, et al: Trastuzumab plus adjuvant chemotherapy for operable
HER2-positive breast cancer. N Engl J Med. 353:1673–1684. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Piccart-Gebhart MJ, Procter M,
Leyland-Jones B, Goldhirsch A, Untch M, Smith I, Gianni L, Baselga
J, Bell R, Jackisch C, et al: Trastuzumab after adjuvant
chemotherapy in HER2-positive breast cancer. N Engl J Med.
353:1659–1672. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wolff AC, Hammond ME, Schwartz JN, Hagerty
KL, Allred DC, Cote RJ, Dowsett M, Fitzgibbons PL, Hanna WM, Langer
A, et al: American Society of Clinical Oncology/College of American
Pathologists guideline recommendations for human epidermal growth
factor receptor 2 testing in breast cancer. J Clin Oncol.
25:118–145. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wolff AC, Hammond ME, Hieks DG, Dowsett M,
McShane LM, Allison KH, Allred DC, Bartlett JM, Bilous M,
Fitzgibbons P, et al: Recommendations for human epidermal growth
factor receptor 2 testing in breast cancer: American Society of
Clinical Oncology/College of American Pathologists clinical
practice guideline update. J Clin Oncol. 31:3997–4013. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Troxell ML, Bangs CD, Lawce HJ, Galperin
IB, Baiyee D, West RB, Olson SB and Cherry AM: Evaluation of
Her-2/neu status in carcinomas with amplified chromosome 17
centromere locus. Am J Clin Pathol. 126:709–716. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Varga Z, Tubbs RR, Wang Z, Sun Y, Noske A,
Kradolfer D, Bosshard G, Jochum W, Moch H and Öhlschlegel C:
Co-amplification of the HER2 gene and chromosome 17 centromere: A
potential diagnostic pitfall in HER2 testing in breast cancer.
Breast Cancer Res Treat. 132:925–935. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Press MF: How Is Her-2/neu Status
Established When Her-2/neu gene and chromosome 17 centromere are
both amplified? Am J Clin Pathol. 126:673–674. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gunn S, Yeh IT, Lytvak I, Tirtorahardjo B,
Dzidic N, Zadeh S, Kim J, McCaskill C, Lim L, Gorre M and Mohammed
M: Clinical array-based karyotyping of breast cancer with equivocal
HER2 status resolves gene copy number and reveals chromosome 17
complexity. BMC Cancer. 10:3962010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Marchiò C, Lambros MB, Gugliotta P, Di
Cantogno LV, Botta C, Pasini B, Tan DS, Mackay A, Fenwick K, Tamber
N, et al: Does chromosome 17 centromere copy number predict
polysomy in breast cancer? A fluorescence in situ hybridization and
microarray-based CGH analysis. J Pathol. 219:16–24. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tse CH, Hwang HC, Goldstein LC, Kandalaft
PL, Wiley JC, Kussick SJ and Gown AM: Determining true HER2 gene
status in breast cancers with polysomy by using alternative
chromosome 17 reference genes: Implications for Anti-HER2 targeted
therapy. J Clin Oncol. 29:4168–4174. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Egervari K, Kosa C and Szollosi Z: Impact
of chromosome 17 centromere region assessment on HER2 status
reported in breast cancer. Pathol Res Pract. 207:468–471. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zeng X, Liang ZY, Wu SF, Gao J, Zhou WX
and Liu TH: HER2 status in breast cancer of Chinese women: A study
of 1,170 cases using fluorescence in-situ hybridization. Zhonghua
Bing Li Xue Za Zhi. 37:594–598. 2008.(In Chinese). PubMed/NCBI
|
|
20
|
Orsaria M, Khelifa S, Buza N, Kamath A and
Hui P: Chromosome 17 polysomy: Correlation with histological
parameters and HER2NEU gene amplification. J Clin Pathol.
66:1070–1075. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jiang H, Bai X, Zhao T, Zhang C and Zhang
X: Fluorescence in situ hybridization of chromosome polysomy
in breast cancer using thin tissue sections causes the loss of
CEP17 and HER2 signals. Oncol Rep. 32:1889–1896. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jiang H, Bai X, Meng F, Zhang C and Zhang
X: Evaluation of chromosome 17 polysomy in breast cancer by FISH
analysis of whole nuclei, and its clinicopathological significance.
Oncol Lett. 7:1954–1958. 2014.PubMed/NCBI
|
|
23
|
Krishnamurti U, Hammers JL, Atem FD,
Storto PD and Silverman JF: Poor prognostic significance of
unamplified chromosome 17 polysomy in invasive breast carcinoma.
Mod Pathol. 22:1044–1048. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Moelans CB and van Diest PJ: CEP17 copy
number increase does not indicate polysomy 17. J Clin Pathol.
67:454–455. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hanna WM, Rüschoff J, Bilous M, Coudry RA,
Dowsett M, Osamura RY, Penault-Llorca F, van de Vijver M and Viale
G: HER2 in situ hybridization in breast cancer: Clinical
implications of polysomy 17 and genetic heterogeneity. Mod Pathol.
27:4–18. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Risio M, Casorzo L, Redana S and
Montemurro F: HER2 gene-amplified breast cancers with monosomy of
chromosome 17 are poorly responsive to trastuzumab-based treatment.
Oncol Rep. 13:305–309. 2005.PubMed/NCBI
|