Introduction
Exosomes are small endosome-derived vesicles that
range between 30 and 100 nm in size, and are actively secreted
through the exocytosis pathway (1).
The major roles of exosomes are intercellular cross talk and
receptor discharge (1–3), and they are typically released from high
viability cells, including cancer cells (1). Previous studies have indicated that
exosomes are capable of modulating intercellular communication and
tumor progression through the transfer of proteins and RNA to
adjacent and distant cells (1,4,5). Additionally, the functions of exosomes
may vary depending on cell type and intracellular contents
(2). In cancer, exosomes are
considered to serve essential roles in tumor metastasis by
regulating complex interactions between tumor cells and their
microenvironment (6,7). However, the regulatory mechanisms of
exosomes in tumor metastasis remain to be elucidated. As exosomes
are carriers of multiple proteins and RNA molecules, the molecules
contained within exosomes may themselves serve key roles in
cell-cell communication (1). Thus,
proteomics profiling and sequencing are promising platforms for
systematically studying exosome components, which may ultimately
improve understanding of exosome function.
At present, hepatocellular carcinoma (HCC) is a
fatal primary malignancy of hepatocytes (8). Emerging diagnostic tools and novel
therapeutic strategies for HCC have substantially improved the
clinical outcomes; however, the long-term survival of patients with
HCC remains relatively poor due to the high possibility of
metastasis and/or recurrence (9).
Whether a tumor is likely to undergo local or distant metastasis is
principally determined by the metastatic potential of tumor cells
and the corresponding microenvironment (10,11). As a
major component of the cellular microenvironment, exosomes secreted
by different tumor cell types are capable of inducing apoptosis of
activated T cells by promoting the expression of cell death ligands
(12–14), inhibiting natural killer cell
functions (15,16), and promoting the generation of
suppressor cells derived from myeloid precursors (13). Additionally, various signaling
pathways and genes are involved in the communication between tumor
cells and their microenvironment (17). For example, a previous study
demonstrated that tumor-activated hepatocytes were capable of
altering the expression profiles of colon cancer cells in order to
support hepatic metastasis (18).
However, despite progress in research regarding the role of
exosomes in cell communication, the mechanism by which exosomes
alter the metastatic potentials of different cell types,
particularly liver cancer cells, still requires further
investigation.
To investigate whether exosomes may alter the
metastatic potential of cancer cells, the present study used two
HCC cell lines with high and low metastatic potential, MHCC97-H and
MHCC97-L, for exosome isolation and characterization. To evaluate
the regulatory effect of exosomes on the mobility of HCC cells,
exosomes from the culture medium of different HCC origins were
isolated for in vitro migration and invasion assays.
Additionally, protein profiling was performed on the exosomes from
different origins to systematically characterize the content of the
exosomes, in order to investigate the regulatory role of exosomes
at the molecular level.
Materials and methods
Cell lines and cell culture
The in-house preserved MHCC97-H and MHCC97-L cell
lines were provided by The Second Military Medical University of
China (Shanghai, China). All cells were cultured in Dulbecco's
modified Eagle medium (DMEM; cat no. C11995500BT; Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA), supplemented with 10%
FBS (cat no. 10100-147-FBS; Gibco; Thermo Fisher Scientific, Inc.),
100 U/ml penicillin and 100 µg/ml streptomycin at 37°C in a 5%
CO2 incubator for 24 h.
Exosome purification
Exosomes were isolated using a total exosome
isolation kit (cat no. 4478359; Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. In
brief, 5×106 cells were seeded in a volume of 15 ml
culture medium at 37°C for 24 h, prior to harvesting the cell
culture medium. The culture medium was centrifuged at 2,000 × g at
4°C for 30 min to remove cells and debris. Subsequently, the
supernatant containing the cell-free culture medium was transferred
to a new tube, and then 15 ml of cell-free culture medium was mixed
with 7.5 ml of total exosome isolation reagent. The culture
medium/reagent was mixed by vortexing until homogenous, and the
samples were incubated at 4°C overnight. Following incubation, the
samples were centrifuged at 10,000 × g for 1 h at 4°C. The
supernatant was aspirated and discarded, and the pelleted exosomes
were resuspended in 1X phosphate-buffered saline (PBS). The
exosomes were then washed with 1X PBS, ultra-filtrated with a
molecular weight cut-off (MWCO) of 100,000 Da, and finally
dissolved in 1X PBS.
Transmission electron microscopy
(TEM)
Exosomes isolated from MHCC97-H and MHCC97-L cells
were identified for morphology by transmission electron microscopy
(TEM) as previously described (19).
In brief, exosomes were transferred to a copper grid coated with
0.125% Formvar in chloroform immediately after isolation. Then the
grids were stained with 1% (v/v) uranyl acetate in double-distilled
water right before examination. A Hitachi 7100 transmission
electron microscope was applied for imaging.
Evaluation of HCC cell motile ability
following exosome incubation
MHCC97-H and MHCC97-L cells were freshly cultured in
DMEM supplemented with 10% FBS and incubated with 5% CO2
in air at 37°C for 24 h. Exosomes were isolated from high
metastatic MHCC97-H and low metastatic MHCC97-L cells as described
above. A total of 10 µg pelleted exosomes from each of the MHCC97-H
and MHCC97-L cell lines were individually resuspended in 1 ml
culture medium. The MHCC97-L cells were mixed with the MHCC97-H- or
MHCC97-L-derived exosomes, and the cells were cultured with 5%
CO2 in air at 37°C for 6 h prior to migration and
invasion assays.
Migration and invasion assay
Cell migration was evaluated with a Transwell
migration assay, while the invasion assays were performed using the
Transwell units (Corning Incorporated, Corning, NY, USA) coated
with Matrigel (BD Biosciences, Franklin Lakes, NJ, USA), according
to the manufacturer's protocols. A total of 1×105
MHCC97-H and MHCC97-L cells, and MHCC97-L cells pretreated with
exosomes, were seeded onto the upper chamber of the insert in
serum-free DMEM (cat no. C11995500BT; Gibco; Thermo Fisher
Scientific, Inc.). After 6 h of incubation at 37°C, the membrane of
the insert was fixed in 100% methanol at room temperature for 20
min and stained with crystal violet at room temperature for 20 min.
After washing twice with PBS, tumor cells on the upper surface of
the filters were removed by wiping with cotton swabs. The number of
migrated or invaded cells that had passed through the filter to the
lower surface were counted under an inverted microscope (Axiovert
A1; Zeiss GmbH, Jena, Germany) in ~30 fields of view at ×200
magnification. Mean values were determined from three independent
experiments run in duplicate.
Statistical analysis of in vitro
data
Data are presented as the mean ± standard deviation.
Student's t test and analysis of variance (ANOVA) were used to
determine whether the MHCC97-L and MHCC94-H groups were
statistically significantly different in migratory and invasive
ability. Following ANOVA results, Dunnett's test was used as a post
hoc test. All data were analyzed with GraphPad Prism 6 (GraphPad
Software, Inc., La Jolla, CA, USA) and P<0.05 was considered to
indicate a statistically significant difference.
Protein preparation and isobaric tags
for relative and absolute quantitation (iTRAQ)
For each sample, proteins were precipitated by
ice-cold acetone, and subsequently centrifuged at 10,000 × g at 4°C
for 15 min. A total of 200 µl lysis buffer containing 8 M urea, 2%
SDS and 1X protease inhibitor cocktail (Roche Diagnostics, Basel,
Switzerland) was added to resuspend the precipitate. The protein
concentration of the samples was determined with a bicinchoninic
acid assay (Beijing Transgen Biotech Co., Ltd., Beijing, China)
following the manufacturer's protocol. A total of 5 µl DTT (200 mM)
was then added to the protein samples, and the samples were
incubated at 55°C for 1 h, after which 10 µl iodoacetamide (500 mM)
was added to each sample for 30 min in the dark at room temperature
to alkylate the proteins.
For each sample, the proteins were ultra-filtrated
(MWCO, 10,000 Da) and dissolved in 100 µl triethylammonium
bicarbonate (100 mM). The proteins were then digested with
sequence-grade modified trypsin (Promega Corporation, Madison, WI,
USA), and the resultant peptide mixture was labeled using an iTRAQ
reagent kit (Shanghai AB SCIEX, Analytical Instrument Trading Co.,
Shanghai, China). The peptides were labeled with iTRAQ 8-plex
reagent as follows: MHCC97-H and MHCC97-L were labeled with 113 and
114 isobaric tags, respectively; while the peptides from biological
repetitions were labeled with 115 and 117, respectively (MHCC97-H),
or 116 and 118, respectively (MHCC97-L). Equal amounts of labeled
samples (100 µg) were then desalted with the Sep-Pak Vac C18
cartridges and dried in a vacuum centrifuge at 4°C for 2 h.
High pH reverse-phase separation
A total of 400 µg peptide mixture was dissolved in
solution A (5% acetonitrile and 0.1% formic acid in water; pH
adjusted to 10.0 with ammonium hydroxide), and then fractionated by
high pH separation using an Agilent 1260 Infinity System (Agilent
Technologies GmbH, Waldbronn, Germany) connected to a reverse phase
column (Durashell C18, 5 µm, 4.6×250 mm; Bonna-Agela Technologies,
Inc., Tianjin, China). High pH separation was performed using a
linear gradient of solution B [0.1% formic acid in 90% acetonitrile
(ACN); pH adjusted to 10.0 with ammonium hydroxide] from 2 to 40%
over 60 min. The column flow rate was maintained at 700 µl/min and
the column temperature was maintained at 45°C. Following
separation, the column was re-equilibrated at the initial
conditions for 15 min. A total of 40 fractions were collected, and
any two fractions with the same time interval (including, 1 and 21,
2 and 22) were pooled to reduce the fraction numbers. In total, 20
fractions were obtained and dried in a vacuum concentrator at 4°C
for 2 h.
Low pH nano-liquid chromatography-mass
spectrometry (nano-LC-MS)/MS analysis
The fractions were resuspended with 80 µl solution C
(0.1% formic acid in water), separated by nano-LC and analyzed by
electrospray tandem mass spectrometry. The experiments were
performed on a Nano LC1000 system (Thermo Fisher Scientific, Inc.)
connected to a quadrupole-Orbitrap mass spectrometer (Q-Exactive
Plus; Thermo Fisher Scientific, Inc.), equipped with an online
nano-electrospray ion source. A total of 2 µl peptide sample was
loaded onto the trap column (Thermo Fisher Scientific Inc., Acclaim
PepMap C18, 100 µm × 2 cm) with a flow rate of 10 µl/min, and
subsequently separated on the analytical column (Acclaim PepMap
C18, 75 µm × 15 cm; Thermo Fisher Scientific, Inc.), with a linear
gradient of solution D (0.1% formic acid in ACN) between 3 and 35%.
The column flow rate was maintained at 300 nl/min, the column
temperature was maintained at 40°C, the nebulizer pressure of ~15
MPa and an electrospray voltage of 2.8 kV at the inlet of the mass
spectrometer was used. Following the nano-LC separation, the column
was re-equilibrated at the initial conditions for 15 min.
The Q-Exactive Plus mass spectrometer was operated
in the data-dependent mode to switch automatically between MS and
MS/MS acquisition. Survey full-scan MS spectra (m/z
300–1,500) were acquired with a mass resolution of 70 K, followed
by 10 sequential high-energy collisional dissociation MS/MS scans
with a resolution of 17.5 K. In all cases, one microscan was
recorded using a dynamic exclusion of 30 sec.
Mass spectrometry data analysis
The raw files from the Q-Exactive instrument were
searched against the human database provided by the Universal
Protein Resource (http://www.uniprot.org/uniprot, released on 10 April
2014, with 20,264 entries) using Proteome Discoverer (PD) 1.4
(Thermo Fisher Scientific, Inc.). The enzyme specificity of trypsin
and a maximum of two missed cleavages were selected for protease
digestion. PD was used with a parent ion tolerance of 10 parts per
million and a fragment ion mass tolerance of 0.05 Da.
Carbamidomethylation of cysteine, as well as iTRAQ modification of
the peptide N-terminus and lysine residues, were set as a fixed
modification; oxidation of methionine and iTRAQ 8-plex labeling of
tyrosine were specified as variable modifications.
A decoy database search strategy was adopted to
estimate the false discovery rate (FDR) for peptide identification.
Scaffold (version 4.3.2, Proteome Software, Inc., Portland, OR,
USA) was used to validate the MS/MS based peptide and protein
identifications. The proteins were assembled using the parsimony
method and accepted if the peptide FDR was <1% and the protein
probability was >99.0%. Proteins containing similar peptides
that could not be distinguished based on MS/MS analysis alone were
grouped to satisfy the principles of parsimony.
Differentially expressed protein
filtering and gene ontology (GO) and kyoto encyclopedia of genes
and genomes (KEGG) pathway analyses
Proteins with expression fold change >2 and
Student's t-tests, P<0.05 were filtered as differentially
expressed proteins between exosomes isolated from MHCC97-H and
MHCC97-L cells. GO and KEGG pathway analyses were conducted using
the R packages GO.db (version 3.4.1), KEGG.db (version 3.23) and
KEGGREST (version 1.16.1). The P-value threshold was set at 0.01 to
filter significantly enriched biological processes and KEGG
pathways.
Identification of significantly
altered subnetworks
Subnetwork identification was conducted with a
heat-diffusion model based on the HotNet2 algorithm. The expression
profile change was used as a heat signal input. The networks used
for this analysis were obtained from the Human Protein Reference
Database (HPRD) (20), iRefIndex
(21) and Multinet (22) as recommended by the algorithm authors.
A subnetwork identification result with P<0.05 and minimum edge
weight threshold δ≥0.0003 were selected for subsequent consensus
subnetwork construction. The consensus subnetworks were derived by
the following steps: Initially, a complete weighted graph combining
all the subnetworks identified from each interaction network was
generated. In the weighted graph, proteins served as vertices and
the edge between any pair of proteins were weighted by the number
of networks, in which HotNet2 reports them in the same subnetwork.
Then, the consensus subnetworks were identified by i) initializing
the consensus subnetworks with connected components with edge
weights ≥2 (connected components were defined as core genes of the
consensus subnetworks), and ii) extending the subnetworks by adding
similar genes to a given subnetwork until all weight one edges
ended in the consensus subnetworks. The construction of the
consensus networks was performed using customized Python scripts
(version 2.7.13, Python Software Foundation, Wilmington, DE,
USA).
Results
Electron microscopy of isolated
exosomes
Exosomes were isolated from the culture media of
MHCC97-H and MHCC97-L cells using a total exosome isolation kit. To
verify that the isolated structures were exosomes, the isolates
were examined by electron microscopy (Fig. 1). The electron images depicted rounded
structures with a size range of 50–100 nm in diameter and a
cup-shaped morphology, which confirmed the successful isolation of
exosomes according to previously described exosome characteristics
(5,23,24).
Exosomes from different origins
significantly alter the migratory and invasive abilities of cancer
cells
The HCC cell lines, MHCC97-H and MHCC97-L, were used
in the present study to assess the potential tumor regulatory role
of exosomes. MHCC97-H cells exhibited a higher motile ability
compared with that of MHCC97-L cells when analyzed by in
vitro migration and invasion assays (Fig. 2A). Statistical analyses confirmed
that, the migratory and invasive capacity of MHCC97-H cells was
~1.5 times higher than that of the MHCC97-L cells (Fig. 2B; P<0.0001 and P<0.005),
respectively, which is in accordance with a previous study
(25).
To investigate the impact of exosomes on the motile
ability of HCC cells, exosomes were individually isolated from
MHCC97-H and MHCC97-L cells and incubated with MHCC97-L cells for 6
h. Following incubation, further migration and invasion assays were
performed to detect changes in the motile abilities of the MHCC97-L
cell groups. As depicted in Fig. 3,
according to the migration and invasion assays, MHCC97-L cells
incubated with MHCC97-H-derived exosomes exhibited higher motile
ability compared with that of MHCC97-L cells incubated with
MHCC97-L-derived exosomes. MHCC97-L cells incubated with
MHCC97-H-derived exosomes demonstrated significantly increased
migratory ability (~2-fold; P<0.05) than those incubated with
MHCC97-L-derived exosomes. For the invasion assay, MHCC97L cells
incubated with MHCC97-H-derived exosomes displayed higher invasive
ability compared with those incubated with MHCC97-L-derived
exosomes (~1.5-fold; P<0.05). Taken together, these results
indicate the ability of exosomes to regulate the motile capacity of
HCC cell lines. It is established that exosomes serve an important
role in cell-cell communication by transferring molecules between
cells (5). Furthermore, a number of
proteins and RNAs have been reported to be enriched in exosomes,
which may be responsible for the regulatory role of exosomes
(1,26). Therefore, the change in motile ability
induced by exosomes in the present study was the basis for
subsequent analysis of the molecular content of exosomes.
Mass spectrum analysis of exosomal
proteins from different origins
Using a quantitative MS-based discovery strategy,
the overall proteomes of exosomes extracted from MHCC97-H and
MHCC97-L cells were investigated to characterize the molecular
mechanism by which exosomes regulate the motile ability of cells.
Exosomes were extracted from the supernatant of the HCC cell lines,
and eluted proteins were digested, separated by high pH
reversed-phase LC and analyzed by iTRAQ 2D LC-MS/MS. In total,
three biological repeats from MHCC97-H and MHCC97-L exosomes were
labeled and processed for quantitative analysis. The representative
MS/MS result is depicted in Fig. 4.
Reporter ion intensities of representative peptides derived from
MHCC97-H and MHCC97-L exosomes are depicted in the inset. Peptides
from the MHCC97-H exosomes were labeled with 113, 115 and 117
isobaric reagents, respectively, while peptides from the MHCC97-L
exosomes were labeled with 114, 116 and 118 isobaric reagents,
respectively. The Scaffold software quantified a total of 129
proteins in the exosome samples isolated from MHCC97-H and MHCC97-L
cells.
GO and KEGG enrichment analyses of
differentially expressed exosomal proteins in MHCC97-H and MHCC97-L
cells
Among the identified genes, adenylyl
cyclase-associated protein 1 (CAP1) had a significantly altered
expression pattern in exosomes isolated from MHCC97-H cells
compared with those from MHCC97-L cells (fold change ≥2; P≤0.05).
Other differentially expressed genes in the MHCC97-H-derived
exosomes included peptidylprolyl isomerase A (PPIA), keratin, type
I cytoskeletal 20 (KRT20) and inter-α-trypsin inhibitor heavy chain
family member 4 (ITIH4; fold-change ≥1.5; P≤0.05). The
differentially expressed genes information is presented in Table I. As the aim was to elucidate the
roles of exosomes in different biological processes and pathways,
GO and KEGG enrichment analyses were performed for all the proteins
identified by MS-based discovery. The 129 identified proteins were
over-represented in 316 biological processes and 6 KEGG pathways.
The top 10 filtered biological processes were ‘response to
stimulus’ (GO:0050896), ‘response to stress’ (GO:0006950), ‘wound
healing’ (GO:0042060), ‘response to wounding’ (GO:0009611),
‘regulation of body fluid levels’ (GO:0050878), ‘blood coagulation’
(GO:0007596), ‘coagulation’ (GO:0050817), ‘hemostasis’
(GO:0007599), ‘platelet activation’ (GO:0030168) and ‘platelet
degranulation’ (GO:0002576), as depicted in Fig. 5. Significantly enriched KEGG pathways
were glycolysis/gluconeogenesis, focal adhesion, extracellular
matrix-receptor interaction, and complement and coagulation
cascades (Table II; P<0.001).
These results indicated enrichments in proteins involved in cell
microenvironment construction and cell responses to environmental
stress, thus suggesting the regulatory role of exosomes within the
cell microenvironment.
 | Table I.Differentially expressed exosomal
proteins in MHCC97-H and MHCC97-L cells. |
Table I.
Differentially expressed exosomal
proteins in MHCC97-H and MHCC97-L cells.
| Protein name | Fold-change | P-value |
|---|
| CAP1 | 4.047908066 | 0.035 |
| PPIA | 1.827357238 | 0.028 |
| KRT20 | 1.583989312 | 0.041 |
| ITIH4 | 1.944871172 | 0.048 |
 | Table II.Significantly enriched Kyoto
Encyclopedia of Genes and Genomes pathways associated with proteins
identified by mass spectrum-based discovery. |
Table II.
Significantly enriched Kyoto
Encyclopedia of Genes and Genomes pathways associated with proteins
identified by mass spectrum-based discovery.
| ID | Description | Count | P-value |
|---|
| hsa00010 |
Glycolysis/gluconeogenesis | 10 |
4.16×10−8 |
| hsa04510 | Focal adhesion | 16 |
4.69×10−8 |
| hsa04512 | ECMa-receptor interaction | 11 |
5.38×10−8 |
| hsa04610 | Complement and
coagulation | 10 |
7.51×10−8 |
Subnetwork identification based on a
heat-diffusion-like model
To elucidate the potential influences of
differentially expressed proteins, a heat-diffusion-like model was
constructed based on the HotNet2 algorithm (27). This model used the different
expression profiles between exosomes from MHCC97-H and MHCC97-L
cells as stimulated directed heat signals radiated from the
corresponding protein to its interacting partner. Subsequently,
diffusion was evaluated in the genome-scale interaction networks to
identify significantly altered gene subnetworks, which revealed a
combination of proteins across different pathways and complexes.
The three interaction networks were extracted from the HPRD
(20), iRefIndex (21) and Multinet (22). A single subnetwork was significantly
altered in the exosomes of MHCC97-H cells when compared with
MHCC97-L cells (P=0.0001), as depicted in Fig. 6. This subnetwork contained 38 proteins
(Presented in Table III), of which
the core components principally belonged to three different groups
according to their interactions. The first group contained
apolipoprotein E (APOE), low density lipoprotein receptor-related
protein 1 (LRP1), talin 1 (TLN1), filamin A (FLNA), lamin A,
β-actin (ACTB) and CAP1. These core components were indicated to
directly interact and were thus associated. Furthermore, it has
been reported that these proteins are associated with cell-cell
interactions in cell migration indicating their relevance to cancer
metastasis (28). Proteins with close
interactions to the core components of the subnetwork (PYGL, G6PD,
LUM and TKT) were associated with glucose metabolization, which
likely provides the energy required for cell transformation and
migration. CAP1, the expression of which was significantly altered
between MHCC97-H and MHCC97-L exosomes, was a core component and
placed centrally in the subnetwork, implicating it as a key linker
gene. The other two highly interactive groups contained laminin
subunit γ1, nidogen 1 and fibulin 1 (FBLN1), and actinin α1 (ACTN1)
and glycogen phosphorylase B, respectively.
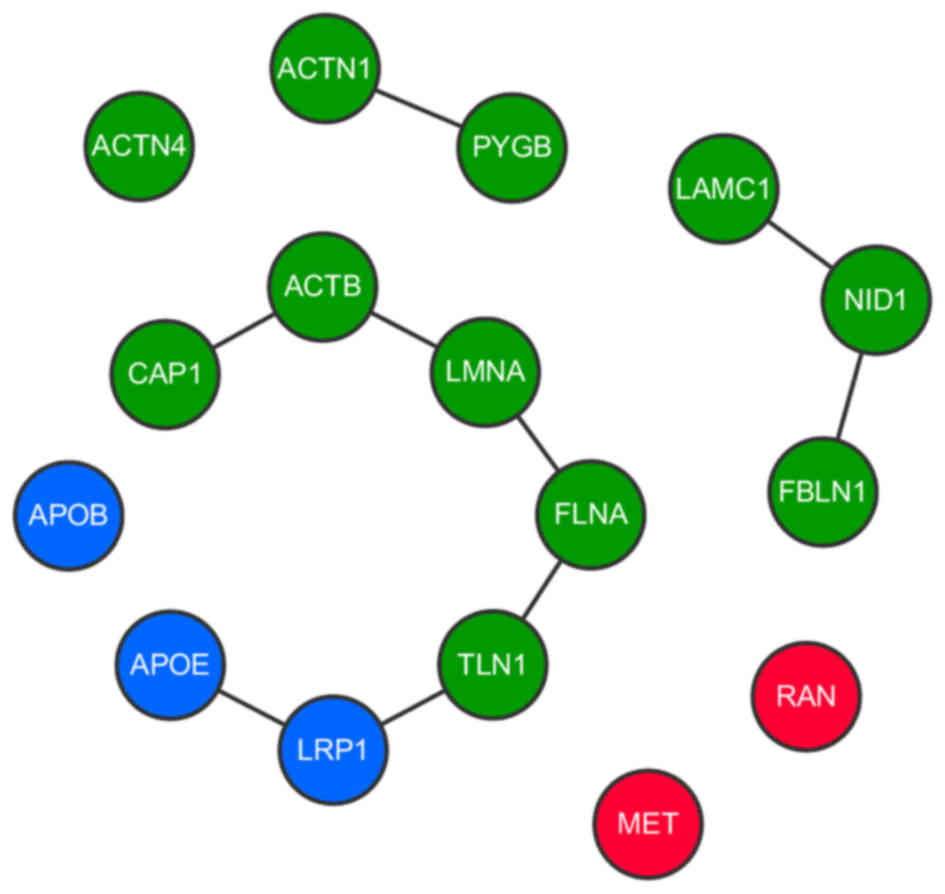 | Figure 6.Interactions between core proteins in
the subnetwork. Interactions between proteins in the subnetwork
determined from three different databases. Each type of protein is
depicted as a different color node (blue represents
lipoprotein-associated proteins; green represents cell
microenvironment and extracellular matrix formation-associated
proteins; red represents oncogene encoded proteins). CAP1, adenylyl
cyclase-associated protein 1; ACTN1/4, actinin α1/4; PYGB, glycogen
phosphorylase B; ACTB, β-actin; LMNA, lamin A; FLNA, filamin A;
TLN1, talin 1; LRP1, low density lipoprotein receptor-related
protein 1; APOB, apolipoprotein B; APOE, apolipoprotein E; LAMC,
laminin subunit γ1; NID1, nidogen 1; FBLN1, fibulin 1; RAN,
ras-related nuclear protein. |
 | Table III.Consensus subnetworks identified by
HotNet2 algorithm. |
Table III.
Consensus subnetworks identified by
HotNet2 algorithm.
| Group | Component | Size |
|---|
| 1 | APOB, APOE, FLNA,
LRP1, PYGL, TLN1, UBA1, FBLN1, LAMC1, NID1, ACTN1, ACTN4, PYGB,
LMNA, ACTB, ANXA2, CAP1, WDR1, G6PD, LUM, PGK1, PPIA, TKT, MET,
RAN | 25 |
| 2 | C1S, ITIH4, ITIH3,
ALDOA, LDHB, MYH9, NRP2, NAMPT, PSMD3, UGDH, SDC4, MYL6, PLEC | 13 |
All the core components were classified based on
function, as the majority were either associated with lipoprotein
activity (APOB, APOE and LRP1, depicted as blue nodes in Fig. 6), or with the cell microenvironment
and extracellular matrix formation (including FLNA, TLN1, FBLN1,
ACTN1/4 and ACTB, depicted as green nodes in Fig. 6). The other core components that did
not belong to an interactive group were MET and ras-related nuclear
protein (RAN; depicted as red nodes in Fig. 6), indicating that these genes exert a
distinct influence in the subnetwork while remaining indirectly
associated with the CAP1-centered core. MET and RAN are established
cancer-associated genes, particularly regarding cancer development
and migration (29,30). Thus, taken together these results
indicated an altered fraction of genes that may serve crucial
functions in cancer cell-derived exosomes.
Discussion
The interactions between cancer cells and their
microenvironment are important in tumor development (16). In recent studies on HCC, it has been
demonstrated that exosomes secreted by cancer cells serve as
messengers in cell-cell communication by conveying molecular
information between tumor and adjacent cells (31,32).
However, while exosomes are a key component of the cellular
microenvironment, whether they regulate the migration and invasion
of HCC cells remains unresolved. Additionally, the molecular
contents of exosomes derived from HCC cells remain largely unclear,
and the regulatory role of exosomes regarding metastatic potential
requires further elucidation. The present study has indicated that
horizontal transfer of exosomes between cells of different origin
may lead to changes in the receiver cell metastatic potential,
which thus provides insight into how exosomes may be involved in
cancer development. This alteration in metastatic ability is
unlikely to result from a change in the direct expression of
several individual proteins, but rather from changes in complex
mechanisms consisting of multiple biological pathways (33).
A number of proteins are enriched in exosomes,
including membrane trafficking proteins (Rab proteins and
Annexins), adhesion molecules (lactadherin) and signal transduction
proteins (protein kinases) (1,26,34). Following validation of the regulatory
role of exosomes regarding cell mobility, the present study
conducted protein profiling of exosomes from different origins to
systematically investigate the differentially expressed proteins
within the exosomes. Among the differentially expressed proteins,
CAP1 was revealed to be significantly upregulated within exosomes
from high metastatic HCC cells. CAP1 has been demonstrated to be
significantly overexpressed in human HCCs and correlated with HCC
metastasis, and thus was suggested to be a potential independent
prognostic factor in patients with liver cancer (35). Loss of CAP1 expression may lead to
cell polarity defects and altered distributions of actin filaments
and mRNA determinants during development (36). Meanwhile, a previous study
demonstrated that CAP1 was overexpressed in pancreatic cancer, and
indicated an involvement of CAP1 in aggressive pancreatic cancer
cell behavior (37).
The present study identified that CAP1 expression
was relatively higher in the exosomes of MHCC97-H cells compared
with those of MHCC97-L cells, which indicates a regulatory role of
CAP1 in cell mobility alterations associated with secreted
exosomes. However, a limitation of the present study is that
RNA-sequencing screening was not performed to compare the genetic
components within exosomes of different origins. It has previously
been indicated that the exchange of genetic materials, including
mRNA and microRNA, is an alternative way of exosome-mediated
intercellular communication that may reprogram the recipient cells
(5,38). Thus, future studies should focus on
the potential regulatory functions of genetic materials within
exosomes. Additionally, the level of CAP1 within exosomes and its
association with the regulation of the motile ability of cancer
cells requires further characterization.
By constructing a heat-diffusion-like model, a
fraction of genes were identified that was significantly altered
between exosomes isolated from MHCC97-H and MHCC97-L cells. Genes
within this group were divided into several classes. A major part
of this subnetwork consisted of lipoprotein genes, which is
consistent with the origin of exosomes, as cell-derived vesicles
with membranous structure. These lipoproteins are frequently
present in different classes of extracellular vesicles, including
microvesicles (39), microparticles
(40,41) and exosomes (42,43).
Lipoproteins are key components of exosome structure
and participate in exosome synthesis, transport and interactions
with cells. It has also been reported that APOE and APOB were
associated with hepatocyte-derived exosomes (44), while other proteins of this group
(including CLIC1 and NRP2) have been associated with the cell
microenvironment and extracellular structure (45,46), thus
indicating that these proteins may serve key roles in the exosomal
regulation of cell metastatic ability. Additionally, it has been
reported that tumor cells may modulate the surrounding
microenvironment to enhance their metastatic potential through the
secretion of exosomes (47). In the
present study, the altered expression of exosome lipoproteins may
have resulted from differences in the status of the origin cell, as
the cell lines possessed different motile abilities. Indeed,
several of these proteins (including APOB and APOE) have previously
been identified to be associated with cancer (48,49).
FLNA, which crosslinks actin into dynamic
extracellular networks and interacts with multiple binding proteins
with different biological functions, has been associated with human
cancer proliferation, migration and invasion (50), and thus is correlated with cancer
development (51,52). Furthermore, TLN1, which encodes a
cytoskeletal protein important in the assembly of actin filaments,
is considered as a diagnostic and prognostic marker in human HCC
(53,54). FBLN1, another component of the
fibrillary extracellular matrix, has also been associated with
cancer (55). As these cell
microenvironment-associated proteins have been associated with
cancer migration and metastasis, afforded by their biological
functions, alterations in the expression of these proteins in
exosomes may impact on the target cell micro environment, and
ultimately cause a change in the motile ability of HCC cells, as
observed in the present study. Indeed, the results of the present
study were consistent with previous observations, indicating that
exosomes may not only affect cells but also the surrounding
environment (56).
Also noteworthy was the identification of two
established oncogenes (MET and RAN) in the differentially expressed
subnetwork. The subnetwork structure indicated that these oncogenes
were indirectly associated with the extracellular microenvironment
and membrane structure, since they are highly interactive with each
other (57). This result suggests
that oncogenes may affect HCC cell status and migration by changing
the extracellular environment, thus leading to a change in the
motile ability of cells. MET, which is frequently identified as a
key gene in the process of cancer cell migration, serves a role in
epithelial-to-mesenchymal transition and contributes to the tumor
microenvironment (58). Meanwhile,
RAN is associated with the formation and organization of the
microtubule network, and previous results have demonstrated that
microtubule perturbation may regulate remodeling of the tumor
microenvironment to alter cell migratory potential (57). The tumor cell secretome has also been
implicated in this mechanism (59).
The identification of MET and RAN oncogenes within HCC exosomes
indicates that these genes may not only influence the origin cells,
but also have an impact on adjacent cells through exosome
transportation. Thus, the identification of these genes provides
insight into the complex mechanisms of exosome-associated processes
and pathways, particularly regarding cancer development and
migration. Further studies of these genes may provide more novel
perspectives on the association between exosomes and cancer.
In conclusion, the results from the present study
indicated that exosomes may alter the metastatic potential of
cancer cells. To the best of our knowledge, the present study was
the first to conduct protein profiling of exosomes from different
HCC cell origins, which identified protein candidates associated
with metastasis and recurrence. Collectively, these data may
provide the foundation for further studies into the regulatory role
of exosomes in cell-cell communication in HCC and other
cancers.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant nos. 31201008 and
31400634), the Specialized Science and Technology Key Project of
Fujian Province (grant no. 2013YZ0002-3), the Science and
Technology Infrastructure Construction Program of Fujian Province
(grant no. 2014Y2005), the Scientific Foundation of Fuzhou City
(grant nos. 2015-S-143-1 and 2015-S-143-21), the Natural Science
Foundation of Fujian Province (grant no. 2015-J-05173) and the
Scientific Research Project of the Health and Family Planning
Commission of Fujian province (grant no. 2015-1-96).
References
|
1
|
Thery C, Zitvogel L and Amigorena S:
Exosomes: Composition, biogenesis and function. Nat Rev Immunol.
2:569–579. 2002.PubMed/NCBI
|
|
2
|
Stoorvogel W, Kleijmeer MJ, Geuze HJ and
Raposo G: The biogenesis and functions of exosomes. Traffic.
3:321–330. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Valenti R, Huber V, Iero M, Filipazzi P,
Parmiani G and Rivoltini L: Tumor-released microvesicles as
vehicles of immunosuppression. Cancer Res. 67:2912–2915. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Skog J, Wurdinger T, van Rijn S, Meijer
DH, Gainche L, Sena-Esteves M, Curry WT Jr, Carter BS, Krichevsky
AM and Breakefield XO: Glioblastoma microvesicles transport RNA and
proteins that promote tumour growth and provide diagnostic
biomarkers. Nat Cell Biol. 10:1470–1476. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Valadi H, Ekström K, Bossios A, Sjöstrand
M, Lee JJ and Lötvall JO: Exosome-mediated transfer of mRNAs and
microRNAs is a novel mechanism of genetic exchange between cells.
Nat Cell Biol. 9:654–659. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Atay S, Banskota S, Crow J, Sethi G, Rink
L and Godwin AK: Oncogenic KIT-containing exosomes increase
gastrointestinal stromal tumor cell invasion. Proc Natl Acad Sci
USA. 111:pp. 711–716. 2014; View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Luga V, Zhang L, Viloria-Petit AM,
Ogunjimi AA, Inanlou MR, Chiu E, Buchanan M, Hosein AN, Basik M and
Wrana JL: Exosomes mediate stromal mobilization of autocrine
Wnt-PCP signaling in breast cancer cell migration. Cell.
151:1542–1556. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dimitroulis D, Damaskos C, Valsami S,
Davakis S, Garmpis N, Spartalis E, Athanasiou A, Moris D,
Sakellariou S, Kykalos S, et al: From diagnosis to treatment of
hepatocellular carcinoma: An epidemic problem for both developed
and developing world. World J Gastroenterol. 23:5282–5294. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tang ZY, Ye SL, Liu YK, Qin LX, Sun HC, Ye
QH, Wang L, Zhou J, Qiu SJ, Li Y, et al: A decade's studies on
metastasis of hepatocellular carcinoma. J Cancer Res Clin Oncol.
130:187–196. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li Y, Tang ZY and Hou JX: Hepatocellular
carcinoma: Insight from animal models. Nat Rev Gastroenterol
Hepatol. 9:32–43. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
He M, Qin H, Poon TC, Sze SC, Ding X, Co
NN, Ngai SM, Chan TF and Wong N: Hepatocellular carcinoma-derived
exosomes promote motility of immortalized hepatocyte through
transfer of oncogenic proteins and RNAs. Carcinogenesis.
36:1008–1018. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Andreola G, Rivoltini L, Castelli C, Huber
V, Perego P, Deho P, Squarcina P, Accornero P, Lozupone F, Lugini
L, et al: Induction of lymphocyte apoptosis by tumor cell secretion
of FasL-bearing microvesicles. J Exp Med. 195:1303–1316. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Valenti R, Huber V, Filipazzi P, Pilla L,
Sovena G, Villa A, Corbelli A, Fais S, Parmiani G and Rivoltini L:
Human tumor-released microvesicles promote the differentiation of
myeloid cells with transforming growth factor-beta-mediated
suppressive activity on T lymphocytes. Cancer Res. 66:9290–9298.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kim JW, Wieckowski E, Taylor DD, Reichert
TE, Watkins S and Whiteside TL: Fas ligand-positive membranous
vesicles isolated from sera of patients with oral cancer induce
apoptosis of activated T lymphocytes. Clin Cancer Res.
11:1010–1020. 2005.PubMed/NCBI
|
|
15
|
Liu C, Yu S, Zinn K, Wang J, Zhang L, Jia
Y, Kappes JC, Barnes S, Kimberly RP, Grizzle WE and Zhang HG:
Murine mammary carcinoma exosomes promote tumor growth by
suppression of NK cell function. J Immunol. 176:1375–1385. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Clayton A, Mitchell JP, Court J, Linnane
S, Mason MD and Tabi Z: Human tumor-derived exosomes down-modulate
NKG2D expression. J Immunol. 180:7249–7258. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wu SD, Ma YS, Fang Y, Liu LL, Fu D and
Shen XZ: Role of the microenvironment in hepatocellular carcinoma
development and progression. Cancer Treat Rev. 38:218–225. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Vidal-Vanaclocha F: The liver
prometastatic reaction of cancer patients: Implications for
microenvironment-dependent colon cancer gene regulation. Cancer
Microenviron. 4:163–180. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hong C, Funk S and Whiteside T: Isolation
of biologically active exosomes from plasma of patients with
cancer. Methods Mol Biol. 1633:257–265. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Keshava Prasad TS, Goel R, Kandasamy K,
Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R,
Shafreen B, Venugopal A, et al: Human Protein Reference Database −
2009 update. Nucleic Acids Res. 37:D767–D77. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Khurana E, Fu Y, Chen J and Gerstein M:
Interpretation of genomic variants using a unified biological
network approach. PLoS Comput Biol. 9:e10028862013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Razick S, Magklaras G and Donaldson IM:
iRefIndex: A consolidated protein interaction database with
provenance. BMC Bioinformatics. 9:4052008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Raposo G, Nijman HW, Stoorvogel W,
Liejendekker R, Harding CV, Melief CJ and Geuze HJ: B lymphocytes
secrete antigen-presenting vesicles. J Exp Med. 183:1161–1172.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Caby MP, Lankar D, Vincendeau-Scherrer C,
Raposo G and Bonnerot C: Exosomal-like vesicles are present in
human blood plasma. Int Immunol. 17:879–887. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ding SJ, Li Y, Shao XX, Zhou H, Zeng R,
Tang ZY and Xia QC: Proteome analysis of hepatocellular carcinoma
cell strains, MHCC97-H and MHCC97-L, with different metastasis
potentials. Proteomics. 4:982–994. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Matsuo H, Chevallier J, Mayran N, Le Blanc
I, Ferguson C, Fauré J, Blanc NS, Matile S, Dubochet J, Sadoul R,
et al: Role of LBPA and Alix in multivesicular liposome formation
and endosome organization. Science. 303:531–534. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Leiserson MD, Vandin F, Wu HT, Dobson JR,
Eldridge JV, Thomas JL, Papoutsaki A, Kim Y, Niu B, McLellan M, et
al: Pan-cancer network analysis identifies combinations of rare
somatic mutations across pathways and protein complexes. Nat Genet.
47:106–114. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Buschow SI, van Balkom BW, Aalberts M,
Heck AJ, Wauben M and Stoorvogel W: MHC class II-associated
proteins in B-cell exosomes and potential functional implications
for exosome biogenesis. Immunol Cell Biol. 88:851–856. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Santhana Kumar K, Tripolitsioti D, Ma M,
Grählert J, Egli KB, Fiaschetti G, Shalaby T, Grotzer MA and
Baumgartner M: The Ser/Thr kinase MAP4K4 drives c-Met-induced
motility and invasiveness in a cell-based model of SHH
medulloblastoma. Springerplus. 4:192015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang F, Yang Z, Cao M, Xu Y, Li J, Chen
X, Gao Z, Xin J, Zhou S, Zhou Z, et al: MiR-203 suppresses tumor
growth and invasion and down-regulates MiR-21 expression through
repressing Ran in esophageal cancer. Cancer Lett. 342:121–129.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Nakamura K, Sawada K, Kinose Y, Yoshimura
A, Toda A, Nakatsuka E, Hashimoto K, Mabuchi S, Morishige KI,
Kurachi H, et al: Exosomes promote ovarian cancer cell invasion
through transfer of CD44 to peritoneal mesothelial cells. Mol
Cancer Res. 15:78–92. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lemoinne S, Thabut D, Housset C, Moreau R,
Valla D, Boulanger CM and Rautou PE: The emerging roles of
microvesicles in liver diseases. Nat Rev Gastroenterol Hepatol.
11:350–361. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Choi D, Lee T, Spinelli C,
Chennakrishnaiah S, D'Asti E and Rak J: Extracellular vesicle
communication pathways as regulatory targets of oncogenic
transformation. Semin Cell Dev Biol. 67:11–22. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Théry C, Ostrowski M and Segura E:
Membrane vesicles as conveyors of immune responses. Nat Rev
Immunol. 9:581–593. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu Y, Cui X, Hu B, Lu C, Huang X, Cai J,
He S, Lv L, Cong X, Liu G, et al: Upregulated expression of CAP1 is
associated with tumor migration and metastasis in hepatocellular
carcinoma. Pathol Res Pract. 210:169–175. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Baum B, Li W and Perrimon N: A
cyclase-associated protein regulates actin and cell polarity during
Drosophila oogenesis and in yeast. Curr Biol. 10:964–973. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yamazaki K, Takamura M, Masugi Y, Mori T,
Du W, Hibi T, Hiraoka N, Ohta T, Ohki M, Hirohashi S and Sakamoto
M: Adenylate cyclase-associated protein 1 overexpressed in
pancreatic cancers is involved in cancer cell motility. Lab Invest.
89:425–432. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Mincheva-Nilsson L and Baranov V: Cancer
exosomes and NKG2D receptor-ligand interactions: Impairing
NKG2D-mediated cytotoxicity and anti-tumour immune surveillance.
Semin Cancer Biol. 28:24–30. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Choi DS, Park JO, Jang SC, Yoon YJ, Jung
JW, Choi DY, Kim JW, Kang JS, Park J, Hwang D, et al: Proteomic
analysis of microvesicles derived from human colorectal cancer
ascites. Proteomics. 11:2745–2751. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Meckes DG Jr, Gunawardena HP, Dekroon RM,
Heaton PR, Edwards RH, Ozgur S, Griffith JD, Damania B and
Raab-Traub N: Modulation of B-cell exosome proteins by gamma
herpesvirus infection. Proc Natl Acad Sci USA. 110:pp. E2925–E2933.
2013; View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Al Kaabi A, Traupe T, Stutz M, Buchs N and
Heller M: Cause or effect of arteriogenesis: Compositional
alterations of microparticles from CAD patients undergoing external
counterpulsation therapy. PLoS One. 7:e468222012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Tauro BJ, Greening DW, Mathias RA, Ji H,
Mathivanan S, Scott AM and Simpson RJ: Comparison of
ultracentrifugation, density gradient separation, and
immunoaffinity capture methods for isolating human colon cancer
cell line LIM1863-derived exosomes. Methods. 56:293–304. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liang B, Peng P, Chen S, Li L, Zhang M,
Cao D, Yang J, Li H, Gui T, Li X and Shen K: Characterization and
proteomic analysis of ovarian cancer-derived exosomes. J
Proteomics. 80:171–182. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ramakrishnaiah V, Thumann C, Fofana I,
Habersetzer F, Pan Q, de Ruiter PE, Willemsen R, Demmers JA, Stalin
Raj V, Jenster G, et al: Exosome-mediated transmission of hepatitis
C virus between human hepatoma Huh7.5 cells. Proc Natl Acad Sci
USA. 110:pp. 13109–13113. 2013; View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Setti M, Osti D, Richichi C, Ortensi B,
Del Bene M, Fornasari L, Beznoussenko G, Mironov A, Rappa G, Cuomo
A, et al: Extracellular vesicle-mediated transfer of CLIC1 protein
is a novel mechanism for the regulation of glioblastoma growth.
Oncotarget. 6:31413–31427. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Landskron G, De la Fuente M, Thuwajit P,
Thuwajit C and Hermoso M: Chronic inflammation and cytokines in the
tumor microenvironment. J Immunol Res. 2014:1491852014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Park JE, Tan HS, Datta A, Lai RC, Zhang H,
Meng W, Lim SK and Sze SK: Hypoxic tumor cell modulates its
microenvironment to enhance angiogenic and metastatic potential by
secretion of proteins and exosomes. Mol Cell Proteomics.
9:1085–1099. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ahles T, Li Y, McDonald BC, Schwartz GN,
Kaufman PA, Tsongalis GJ, Moore JH and Saykin AJ: Longitudinal
assessment of cognitive changes associated with adjuvant treatment
for breast cancer: The impact of APOE and smoking. Psychooncology.
23:1382–1390. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chandler P, Song Y, Lin J, Zhang S, Sesso
HD, Mora S, Giovannucci EL, Rexrode KE, Moorthy MV, Li C, et al:
Lipid biomarkers and long-term risk of cancer in the Women's Health
Study. Am J Clin Nutr. 103:1397–1407. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhang K, Zhu T, Gao D, Zhang Y, Zhao Q,
Liu S, Su T, Bernier M and Zhao R: Filamin A expression correlates
with proliferation and invasive properties of human metastatic
melanoma tumors: Implications for survival in patients. J Cancer
Res Clin Oncol. 140:1913–1926. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Tian ZQ, Shi JW, Wang XR, Li Z and Wang
GY: New cancer suppressor gene for colorectal adenocarcinoma:
Filamin A. World J Gastroenterol. 21:2199–2205. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wieczorek K and Niewiarowska J: Filamin A
as a mediator of alterations in cancer cells. Postepy Biochem.
60:77–83. 2014.(In Polish). PubMed/NCBI
|
|
53
|
Zhang JL, Qian YB, Zhu LX and Xiong QR:
Talin1, a valuable marker for diagnosis and prognostic assessment
of human hepatocelluar carcinomas. Asian Pac J Cancer Prev.
12:3265–3269. 2011.PubMed/NCBI
|
|
54
|
Youns MM, Abdel Wahab AH, Hassan ZA and
Attia MS: Serum talin-1 is a potential novel biomarker for
diagnosis of hepatocellular carcinoma in Egyptian patients. Asian
Pac J Cancer Prev. 14:3819–3823. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Xiao W, Wang J, Li H, Xia D, Yu G, Yao W,
Yang Y, Xiao H, Lang B, Ma X, et al: Fibulin-1 is epigenetically
down-regulated and related with bladder cancer recurrence. BMC
Cancer. 14:6772014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Hosseini-Beheshti E, Choi W, Weiswald LB,
Kharmate G, Ghaffari M, Roshan-Moniri M, Hassona MD, Chan L, Chin
MY, Tai IT, et al: Exosomes confer pro-survival signals to alter
the phenotype of prostate cells in their surrounding environment.
Oncotarget. 7:14639–14658. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Yuen H, Chan K, Grills C, Murray JT,
Platt-Higgins A, Eldin OS, O'Byrne K, Janne P, Fennell DA, Johnston
PG, et al: Ran is a potential therapeutic target for cancer cells
with molecular changes associated with activation of the
PI3K/Akt/mTORC1 and Ras/MEK/ERK pathways. Clin Cancer Res.
18:380–391. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Banyard J and Bielenberg D: The role of
EMT and MET in cancer dissemination. Connect Tissue Res.
56:403–413. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Berbari NF, Sharma N, Malarkey EB,
Pieczynski JN, Boddu R, Gaertig J, Guay-Woodford L and Yoder BK:
Microtubule modifications and stability are altered by cilia
perturbation and in cystic kidney disease. Cytoskeleton (Hoboken).
70:24–31. 2013. View Article : Google Scholar : PubMed/NCBI
|




















