Introduction
At present, colorectal cancer (CRC) is considered to
be a major health issue due to its marked morbidity and mortality
in humans (1). Of course, studies
concerning the underlying mechanisms involved in colorectal
carcinogenesis and drug development are unceasing. Genetic
alterations have provided a molecular mechanism for colon
carcinogenesis and potential targets for anticancer durgs; however,
the results obtained in preclinical studies remain unsatisfactory
(2,3). Recently, epigenetic alterations, which
are defined as heritable changes in gene expression resulting from
mechanisms instead of changes in underlying DNA sequences, are
believed to contribute to the formation and development of CRC. The
best understood epigenetic mechanisms include DNA methylation,
histone modifications and microRNAs (miRNAs, miRs) (4). DNA methylation events in mammalian
cells are mainly caused by two major types of enzymatic activities,
namely, maintenance methylation by DNA methyltransferase (DNMT)1
and de novo methylation by DNMT3A and DNMT3B. miRNAs can
regulate gene expression by degradation of target mRNAs; however,
whether these miRNAs are in turn regulated by target genes remains
elusive.
The initiation of colon cancer cannot be completely
explained from the viewpoint of genetic and family history. Robust
in vitro and in vivo evidence indicates that
microenvironmental factors are involved in the onset of CRC
(5). The extremely complex tumor
microenvironment (TME) consists of numerous elements including
extracellular matrix (ECM) components, such as laminin and
collagen, vascular endothelial growth factor (VEGF), transforming
growth factor (TGF), glucose and varying concentrations of oxygen
(6), as well as various types of
cells, such as tumor, endothelial and epithelial cells,
fibroblasts, and immune and mesenchymal stem cells (7). Cancer development and progression
including initiation, proliferation, invasion, metastasis and
therapeutic resistance are driven by increasingly accumulated
genetic and epigenetic alterations in cancer cells themselves as
well as constructional changes in their microenvironment (8). Stromal cells and the ECM of the
microenvironment can serve as a significant regulator of cancer
initiation, growth and progression (9,10).
Fifteen years ago, Hanahan and Weinberg concluded that cancer cells
do not exist in isolation and that tumors are complex
collaborations between multiple cell types (11), implying that an optimal
microenvironment is required for cancer progression. TGF-β is an
important component of the microenvironment and is secreted by
stromal cells and tumor cells serving as a crucial player in tumor
initiation (12). TGF-β has a dual
function. It is a suppressor in early tumorigenesis but a promoter
at the later stage since its continued elevation or perturbation of
its signaling promotes malignant transformation. However, at the
microenvironment level, the TGF-β pathway facilitates the
generation of an optimal microenvironment in favor of tumor growth
and metastasis throughout all the steps of carcinogenesis (13). In light of the above findings, in
the present study, we designed a model of malignant transformation
induced by transforming growth factor-β1 (TGF-β1) to analyze the
relationship between DNA methyltransferases (DNMTs) and target
miRNAs and the intervening effect of evodiamine and berberine on
this interaction.
Materials and methods
Experimental animals and reagents
Neonatal rats, 7 days of age, were purchased from
the Experimental Animal Center of Guangdong Province. Dulbeccos
modified Eagles medium (DMEM) (Gibco, Grand Island, NY, USA),
berberine chloride (Sigma, St. Louis, MO, USA), evodiamine (Wako,
Tokyo, Japan), 5-Aza-dC (Sigma-Aldrich, St. Louis MO, USA), D-Hanks
solution, fetal bovine serum (FBS), penicillin-streptomycin (all
from Gibco), TGF-β1 (PeproTech, Inc., Rocky Hill, NJ, USA), bovine
pituitary extract (Sigma) were used. MMLV reverse transcriptase,
RNA enzyme inhibitor (Epicentre, Madison, WI, USA), 2X PCR Master
Mix (SuperArray, Frederick, MD, USA), 10X RT buffer solution (200
mM KCl, 5 mM DTT, 250 mM Tris-HCl, pH 8.3, 40 mM MgCl2;
Epicentre), and 2.5 mM dNTP mix (2.5 mM dATP, 2.5 mM dCTP, 2.5 mM
dGTP and 2.5 mM dTTP; HyTest Ltd., Turku, Finland) were used.
Medium preparation
Basic medium preparation included 10 ml FBS, bovine
pituitary extract 5 ng/ml, penicillin-streptomycin 100 U/ml. A
TGF-β1 stock solution (1,000 ng/ml) was prepared by dissolving 1 µg
TGF-β1 in 1 ml sterile double distilled water and maintained at
−20°C. The medium containing TGF-β1 was namely the diluted TGF-β1
stock solution with basic medium (10 ng/ml). A berberine chloride
stock solution (10,500 µM) was obtained by dissolving 21.4 mg
berberine in 5 ml double distilled water and sterilized with
0.22-µm filter membrane, and diluted to 105 µM with basic medium
when used. Evodiamine (2.3 mg) was dissolved in dimethyl sulfoxide
(DMSO) and processed with ultrasonic vibrating to prepare a 1,500
µM stock, and diluted with basic medium to a working concentration
(15 µM). The medium containing 5-Aza-dC (2.5 µM) was from the
diluent of the 250 µM stock with basic medium.
Tissue culture and treatment
Since our previous study found that colon tissue
from neonatal rats of 7 days of age display robust vitality and
were easily cultured (data not provided), we used the tissues from
these rats as our in vitro model in the present study. The
rats were sacrificed by cervical spine dislocation and were
immediately dissected in a sterile console to obtain the colon
tissues. The tissues were immediately washed in cold D-Hanks
cleaning solution, followed by longitudinal dissection and repeated
washings with D-Hanks to eliminate mesentery, blood clots, fatty
tissues and debris in the intestine. Afterwards, the cleaned colon
tissues were cut into 1–2 mm3 tissue blocks, and then
plated into a 6-well plate. To note, a 0.5-cm interval between each
block was considered to be suitable. After redundant fluid around
the tissue blocks was drained, the plates were cultured in an
incubator with 5% CO2 at 37°C until the redundant fluid
around the blocks became dry. The plates were then overturned, the
medium was added and the culturing was continued under the previous
conditions. In the present study, the tissues were divided into 3
groups: TGF-β1, control and treatment groups, which were further
divided into the berberine, evodiamine and 5-Aza-dC groups. The
control group was cultured with basic medium for 3 days, and the
TGF-β1 group was cultured with medium containing TGF-β1 of 10 ng/ml
for 3 days, while the treatment groups were first cultured with
medium of 10 ng/ml TGF-β1 for 2 days, and then berberine chloride,
evodiamine and 5-Aza-dC were respectively added and were again
cultured for 24 h (Fig. 1). All
tissue samples were collected after they were cultured for 24, 48
and 72 h, and were then frozen at −80°C for RT-PCR detection.
Hematoxylin and eosin (H&E)
staining for observation of the histological structures in the
colon tissues
The colon tissues were cultured and fixed with 4%
paraformaldehyde, and then paraffin-embedded samples were cut into
4-µm sections. After a 10- and a 3-min treatment with
dimethylbenzene and ethanol, respectively, the slices were stained
with hematoxylin for 10 min and subsequently 0.5% eosin for 3 min,
and then again treated with ethanol. The slices were sealed with
neutral gum and observed for obtaining images using an inverted
phase contrast microscope.
Immunohistochemistry (IHC)
Samples fixed with 4% paraformaldehyde for 24–48 h
were dehydrated, permeabilized and embedded with paraffin.
Paraffin-embedded slides (4 µm) were deparaffinized by
dimethylbenzene for 10 min and were incubated with 5% FBS at room
temperature for 10 min. Antigen retrieval was performed separately
using moderate heat-induced antigen retrieval two times in a
microwave oven. IHC was performed using rabbit monoclonal
antibodies against α-SMA, Crumbs3 and E-cadherin. Primary
antibodies were diluted in Antibody Diluent (Beyotime, Wuhan,
China) to the indicated optimal dilutions of 1:100 for Crumbs3;
1:50 for E-cadherin; and 1:1,000 for α-SMA. The slides were stained
with DAB staining used as the chromogen for the optimal time, and
then hematoxylin for 1 min. Optical density (OD) values of the
positive immune product for each group were obtained by
pathological analysis software IPP 6.0.
Detection of DNMT and miRNA expression
by real-time PCR
To extract the total RNA of each sample, the
collected samples were fully ground into a powder-like material in
liquid nitrogen, and a certain amount of powder was added to 1 ml
of TRIzol, and RNA concentration was performed using isopropanol.
In addition, the RNA quality and quantity in the samples were
confirmed through ultraviolet absorption measurement and denatured
agarose gel electrophoresis. To minimize variation, all RNA samples
from a single experiment were reverse-transcribed simultaneously
according to the instructions of the manufacturer. The RT reaction
mix contained a total of 800 ng RNA, 2 µl of l0X RT buffer, 2 µl
dNTP (2.5 mM each), 0.2 µl MMLV reverse transcriptase (200 U/µl),
0.3 µl RT-specific primer (1 µM) and 0.3 µl of RNase inhibitor (40
µ/µl) in 20 µl RNase-free water. The following bi-directional
primers of DNMTs and miRNAs were designed using primer design
software Primer 5.0 (Bio-Rad, Hercules, CA, USA) and were used for
RT-PCR (Table I). Thermal cycling
was performed using 40 cycles at 95°C for 10 min, 95°C for 10 sec
and 60°C for 60 sec using CFX96 (Bio-Rad). Each PCR of the RNA
samples from 3 independent experiments was repeated 3 times, and
the average median threshold cycle values were used for analysis
owing to the random error of each sample. Control genes GAPDH and
U6 were respectively used for normalization comparison of DNMTs and
miRNAs in the samples, and the relative expression level was
obtained by comparative CT method in which the fold change of the
gene was calculated by 2−ΔΔCt.
 | Table I.Primer sequences of the DNMTs and
microRNAs for RT-PCR. |
Table I.
Primer sequences of the DNMTs and
microRNAs for RT-PCR.
| Gene | Bi-directional
primer sequences 5′-3′ |
|---|
| DNMTs |
| GAPDH
(internal control) | Forward |
5′-ggtcggtgtgaacggatttgg-3′ |
|
| Reverse |
5′-gtagaccatgtagttgaggtc-3′ |
|
DNMT1 | Forward |
5′-atggcttaacagaaaaggagtg-3′ |
|
| Reverse |
5′-gccaggtagccttcctcagacaa-3′ |
|
DNMT3A | Forward |
5′-ggcactcgctgggtcatgtgg-3′ |
|
| Reverse |
5′-gctggccacctggaggacttc-3′ |
|
DNMT3B | Forward |
5′-agtgggtatgaggactgtatcat-3′ |
|
| Reverse |
5′-agactggctctgtgcagattg-3′ |
| microRNAs |
| U6
(internal control) | Forward |
5′-gcttcggcagcacatatactaaaat-3′ |
|
| Reverse |
5′-cgcttcacgaatttgcgtgtcat-3′ |
|
microRNA-29a | Forward |
5′-tgactgatttcttttggtgttc-3′ |
|
| Reverse |
5′-gtcgtatccagtgcagggtccgaggtattcgcactggatacgacctgaa-3′ |
|
microRNA-152 | Forward |
5′-aggttctgtgatacactccg-3′ |
|
| Reverse |
5′-gtcgtatccagtgcagggtccgaggtattcgcactggatacgacagtcg-3′ |
|
microRNA-429 | Forward |
5′-tgtaatactgtctggtaatgc-3′ |
|
| Reverse |
5′-gtcgtatccagtgcagggtccgaggtattcgcactggatacgacacggc-3′ |
Statistical analyses
All data were processed by statistical software SPSS
19.0 (SPSS Inc. Chicago, IL, USA). The RT-PCR results are presented
as mean ± SEM. One-way ANOVA analysis was adapted to accomplish the
comparison among 3 groups. Level of significance α was 0.05, and a
probability value P<0.05 was considered to indicate a
statistically significant result.
Results
Morphological alterations during
tissue culture
As shown in Fig. 2,
out-migration of cells in the tissues was found in the process of
culture, and the cells revealed a paved stone-like proliferation
from the tissue edge towards the outside. Out-migrated cells at the
edge of the tissue were increasingly dense following TGF-β1
treatment than that noted in cells treated with basic medium
revealing sporadically distribution of cells around the tissue,
suggesting that TGF-β markedly promoted cell proliferation. In
contrast, the numbers of cells at the edge of the tissues treated
respectively with berberine, evodiamine and 5-Aza-dC were decreased
to a varied extent, namely, the cytostatic effect of evodiamine was
highly obvious, which was followed by berberine, and that of
5-Aza-dC was the weakest, indicating that these drugs inhibited the
cell growth induced by TGF-β1 through various mechanisms which are
likely to be associated with epigenetics.
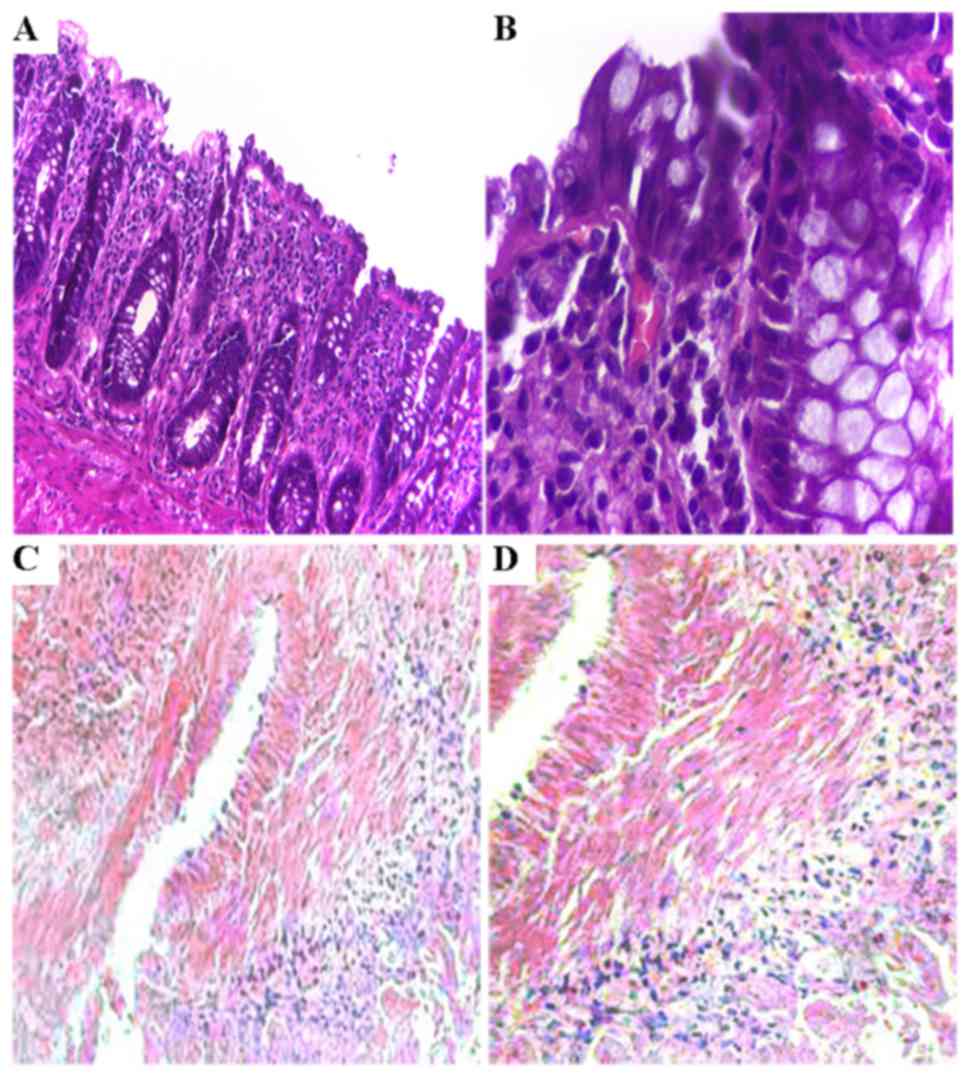
TGF-β1 stimulates histological changes
in the colon tissues as determined by H&E staining
After a 48-h culture, various sizes and larger gaps
between cells were observed in tissues treated with TGF-β1,
compared to that noted in the control revealing a paved stone-like
epithelium tightly arranged. Our H&E staining revealed that
epithelial structures were tightly arranged in the mucosa involving
more chromatin in the nucleus (detected by deep H&E staining,
as shown in Fig. 3A and B) in the
control tissues. However, the epithelial cells were loosely
arranged in the tissues exposed to TGF-β1 presenting changes of
phenotype including many fibroblast-like cells as demonstrated by
high expression of α-SMA (Fig. 3C and
D), a marker of fibroblasts.
Expression of Crumbs3, E-cadherin and
α-SMA by IHC and RT-PCR
TGF-β1, a pleiotropic cytokine, is primarily
released from tumor cells and stroma in the tumor microenvironment
(TME) and promotes the creation of a pro-TME and functions in
epithelial-mesenchymal transition (14) showing loss of epithelial markers
such as E-cadherin, destruction of polarity and acquirement of
mesenchymal markers such as vimentin and α-SMA (15,16).
Therefore, E-cadherin, Crumbs3 and α-SMA were used as indicators in
the present study. In the control, IHC analysis revealed that
Crumbs3 expression was mainly found in the apex of the epithelial
membrane (Fig. 4E), E-cadherin
expression was mainly noted in the epithelial layer of the mucosa
and was positively associated with differentiation (Fig. 4F). α-SMA expression with a monolayer
was mainly along the crypt axis and slight expression was also
found in the smooth muscle layer (Fig.
4G). By contrast, 48 h after exposure to TGF-β1, expression of
Crumbs3 and E-cadherin in the tissues was mainly observed in the
cytoplasm (Fig. 4A and B), while
expression of α-SMA was prominently found in activated
myofibroblasts and mesenchyme (Fig.
4C). Furthermore, in addition to the altered location, in
regards to quantification, the OD values of Crumbs3 (0.1005±0.0488)
and E-cadherin (0.1967±0.0541) were significantly lower in the
tissues treated with TGF-β1 than the OD values in the control
(0.2706±0.0484 and 0.2643±0.0192, respectively; P<0.05), while
the α-SMA level (0.2957±0.0249) in the TGF-β1 group was
significantly higher than that noted in the control (0.1867±0.0633;
P<0.01). However, after a 24-h treatment with evodiamine and
berberine, the tissues revealed higher growth viability, such as
the existence of paved stone-like epithelial cells around the colon
tissues. However, it was not determined whether evodiamine and
berberine mediated the phenotypic changes induced by TGF-β1. Hence,
we detected the expression of phenotypic markers using qRT-PCR. As
revealed in Fig. 4, the results
showed that expression of Crumbs3 in the TGF-β1 group was
consistently lower than that of the control during culture
(P<0.05), particularly at 54 h. Twenty-four hours after
stimulation with berberine, expression of Crumbs3 was markedly
different from that of the TGF-β1 group and control (P<0.05).
Unfortunately, its expression was lower in the evodiamine group
than that noted in the TGF-β1 group (P>0.05), which suggests
that the biological effects of TGF-β were reversed by evodiamine
and berberine, particularly the latter. In addition, although there
was no statistical significance, slightly upregulated E-cadherin
was induced by evodiamine and berberine. Although there was no
evident effect of evodiamine and berberine on α-SMA expression
following a 6-h treatment, its significant downregulation was noted
after a 24-h stimulation (P<0.05), and its expression was close
to a normal level compared to the control (P>0.05).
Expression of DNMTs and miRNAs as
detected by RT-PCR pre-treatment and post-treatment
Sample RNA quality control
The integrity of RNA was assessed by electrophoresis
on a denaturing agarose gel. Intact total RNA running on a
denaturing gel had sharp 28S and 18S rRNA bands (Fig. 5). The total RNAs of these samples
had not been degraded and had adequate quality for the follow-up
study.
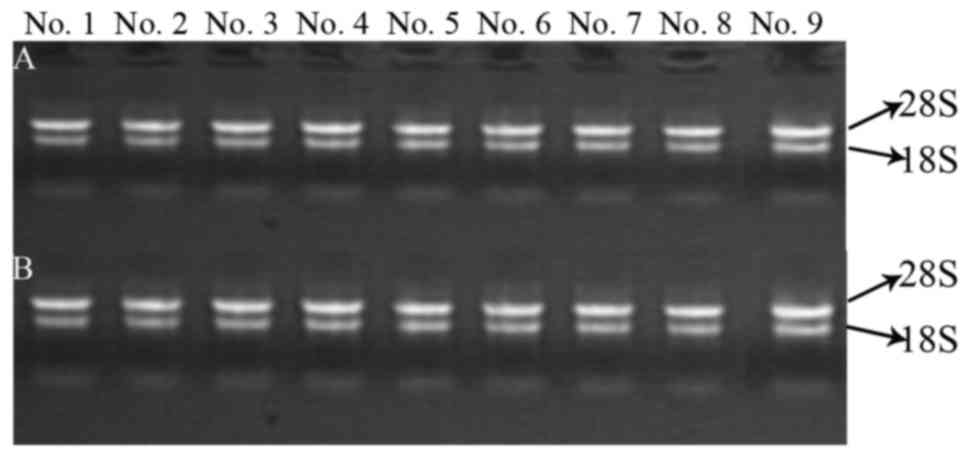 | Figure 5.Electrophoretogram for RNA in the
samples. (A and B) RNA quality of extracted DNMTs and microRNAs,
respectively. No. 1, control group for 24 h; No. 2, TGF-β1 group
for 24 h; No. 3, control group for 48 h; No. 4, TGF-β1 group for 48
h; No. 5, control group for 72 h; No. 6, TGF-β1 group for 72 h; No.
7, berberine treatment for another 24 h; No. 8, evodiamine
treatment for another 24 h; and No. 9, 5-Aza-dC treatment for
another 24 h. |
TGF-β1 upregulates the expression of DNMTs in the
colon tissue
Two epigenetic regulatory factors, DNMTs and miRNAs,
have pivotal roles in carcinogenesis and epigenetic modification
(17,18). To demonstrate whether upregulation
or downregulation of DNMTs and target miRNAs occurs in a CRC
canceration model induced by TGF-β1, we performed quantitative
RT-PCR analyses. Our results revealed that expression levels of
DNMT1, DNMT3A and DNMT3B were 30.7% (P<0.001), 57.49%
(P<0.001) and 60.26% (P=0.0004), respectively, as compared with
the control. After stimulation with TGF-β1 for 24 h, the relative
expression of DNMT1 in the control and TGF-β1 treatment group was
0.49044±0.02890 and 0.15064±0.0137, respectively, and for DNMT3A
the values were 0.53803±0.01410 and 0.30929±0.02669, respectively,
and for DNMT3B, 3.94082±0.37565 and 1.71198±0.22057, respectively.
After treatment with TGF-β1 for 48 h, the expression levels of
DNMT1, DNMT3A and DNMT3B were increased, but these levels were
still lower than that of the control (Fig. 6). At this time, the relative
expression of DNMT1 in the control and TGF-β1 treatment group was
0.46046±0.04390 and 0.40820±0.02852, respectively, and for DNMT3A
these values were 0.52721±0.04919 and 0.3390±0.00727, respectively,
and for DNMT3B, 4.0348±0.05953 and 1.04989±0.19734, respectively.
5-Aza-dC, an inhibitor of DNMTs, has been widely used in the
clinic. After treatment with 5-Aza-dC, expression levels of the
DNMTs were not detected by RT-PCR, suggesting that the activity of
the DNMTs was fully inhibited, which lays a foundation for studying
the regulation of target miRNAs by DNMTs.
Downregulation of microRNAs targeting DNMTs is
induced by TGF-β1 and reversed by DNMT inhibitor
miRNAs, the most widely studied class of non-coding
RNAs (ncRNAs), mediate post-transcriptional gene silencing by
inhibiting the translation of target mRNAs (19,20).
Recent studies concerning tumor miRNA expression profiles have
displayed that the downregulation of miRNAs frequently exists in
cancer tissues compared to their expression levels in normal tissue
(21). Thus, in order to determine
the expression profiles of miRNAs in colon carcinogenesis and
whether these miRNAs mediate the expression of each DNMT gene,
using the miRNA target prediction database (http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/,
http://www.microrna.org/microrna/home.do, http://mir-db.org/miRDB/), we detected the expression
of the target miRNAs via qRT-PCR and used a DNMT inhibitor,
5-Aza-dC. Our results revealed (Fig.
6) that miRNA-29a, miRNA-152 and miRNA-429 were significantly
downregulated after a 24- and 48-h treatment with TGF-β1, as
compared with the control. However, these levels at 48 h were lower
than those at 24 h, which is consistent with a study of Lu et
al (21). The relative
expression of miRNA-152 (a target of DNMT1) in the control and
TGF-β1 treatment group following a 24- and 48-h treatment was
22.28703±0.40663, 1.78214±0.18116 and 22.44430±0.03726,
0.74761±0.31879, respectively, and the relative expression of
miRNA-429 (a target of DNMT3A) was 0.83347±0.06146, 0.54788±0.02543
and 0.82344±0.00379, 0.22779±0.00867, respectively, and the
relative expression of miRNA-29a (a target of DNMT3A and DNMT3B)
was 6.11961±0.31352, 2.90970±0.07024 and 4.55873±0.50071,
0.88628±0.02706, respectively. When DNMT1, DNMT3A and DNMT3B were
inhibited by 5-Aza-dC, notably, these target miRNAs were
upregulated. These levels were still lower in the treatment group
than that in the control. These results indicate that target miRNAs
may serve as tumor suppressors during colon carcinogenesis and they
can also be regulated by their corresponding DNMTs.
Berberine and evodiamine mediate the expression
of DNMTs and target miRNAs
The results of the present study found that both
berberine and evodiamine had an effect on the expression levels of
DNMTs and target miRNAs (Fig. 6).
Seventy-two hours after stimulation with TGF-β1, qRT-PCR revealed
that expression of DNMT1 in the berberine and evodiamine groups was
0.29639±0.07215 and 0.27462±0.02630, respectively, while its
expression in the TGF-β1 group was 0.10827±0.02736, as compared
with DNMT1 expression in the control (0.47688±0.06386); for DNMT3A,
72 h after cultured with TGF-β1, its expression in the two groups
increased (0.89135±0.08597 and 0.62777±0.07734, respectively),
while its expression in the TGF-β1 group increased as well
(0.48348±0.05992), but was lower than that of the two former groups
and the control (0.57838±0.09560). Consistent with the results for
DNMT3A, 72 h later, expression of DNMT3B in the berberine,
evodiamine, TGF-β1 and control group was 3.29743±1.08786,
3.13845±0.36048, 1.44989±0.19734 and 4.33481±0.31815,
respectively.
Since these DNMTs were affected by berberine and
evodiamine, to determine whether the miRNAs were also influenced by
the two drugs, we tested the expression levels of the target miRNAs
by qRT-PCR. After a 24 h treatment with the drugs, expression of
miRNA-152 (target of DNMT1) was higher in the berberine and
evodiamine groups than that in the TGF-β1 group, but lower than the
control (14.51982±3.25912, 6.26986±1.41350, 1.53881±0.29361 and
22.65288±1.95338, respectively). However, the expression profile of
miRNA-429 (target of DNMT3A) was not similar to that of DNMT1; its
expression in the evodiamine group was upregulated
(1.49523±0.02687), but downregulated in the berberine and TGF-β1
group (0.20130±0.00671 and 0.09948±0.00293, respectively). In the
presence of berberine and evodiamine, expression of miRNA-29a
(target of DNMT3A and DNMT3B) in the berberine and TGF-β1 group was
markedly upregulated (54.02511±3.59926 and 2.53252±0.05201,
respectively), particularly its expression in the evodiamine group
was significantly more than that of the control (222.25954±5.46865
vs. 5.19267±2.00633). These results suggest that berberine and
evodiamine influence the regulation of miRNAs targeting DNMTs.
Discussion
Accumulating evidence suggests that the development
of colorectal cancer (CRC) involves a multistep process entailing
the accumulation of genetic or epigenetic aberrations, which result
in the simultaneous failure of protective mechanisms and the
activation of tumorigenic pathways (22). Important findings of the present
study were that expression levels of DNMT1, DNMT3A and DNMT3B, as
well as their target microRNAs (miRNA-152, miRNA-429 and miRNA-29a)
were markedly downregulated in the colon cancer tissues of the
neonatal mice, as compared with the normal control, and that the
target miRNAs were also regulated by DNMT1, DNMT3A and DNMT3B,
respectively, and that both berberine and evodiamine had an effect
on the expression of DNMTs and target miRNAs. These results
indicate that both DNMTs and miRNAs are critical regulators of the
initiation, growth and development of colon cancer in neonatal
rats, and that berberine and evodiamine inhibited the onset and
growth of colon cancer through epigenetics by influencing the
expression of DNMTs and miRNAs. It is now known that epigenetic
events involving multiple interactions with DNMTs, small non-coding
RNAs and tumor-suppressor genes likely result in colon
carcinogenesis. In the present study, we mainly focused on the
inter-regulation between DNMTs and target miRNAs during colon
canceration induced by TGF-β1. As a major epigenetic modification,
DNA methylation plays a critical role in the regulation of
chromatin structure and gene expression and is involved in a
variety of biological processes, as well as their levels and
patterns are regulated by DNMT1, DNMT3A and DNMT3B (23). Epigenetic mechanisms can be promoted
as a response to various stimuli and stressors subjected to the
organism. Epigenetic modifications can be used as extraordinary
candidates for new targets for drug development (24,25).
DNA methylation frequently occurs in the cytosine of CpG motifs
methylated in the genome, but some regions called CpG islands in
which the concentration of CpG motifs is abundant are usually
demethylated. As such, the hypermethylation of such elements
frequently leads to silencing or a downregulated expression of
target genes. In contrast, their hypomethylation can contribute to
the upregulation of gene expression. Numerous studies have found
that global hypomethylation is associated with chromosomal
instability and hypermethylation of key tumor-suppressor genes
(TSGs) is used as a pivotal player in carcinogenesis (26,27).
In contrast, chromosomal instability playing a key role in CRC
involves changes in the chromosome copy number and structure or the
loss of the wild-type copy of TSGs, such as APC and TP53 (28). Unfortunately, these genes are
frequently silenced in the process of colorectal canceration due to
the hypermethylation of the promoter. Studies have revealed that
overexpression of either DNMT3A or DNMT3B is related to
carcinogenesis in a cancer type-dependent manner (29,30),
which suggests that both DNMT3A and DNMT3B play a pivotal role in
canceration.
Together with the data mentioned above, our present
results revealed that expression levels of the three DNMTs were
downregulated in the TGF-β1 group as compared with the normal
control after a 24-h treatment with TGF-β1, while these genes were
upregulated at 48 h after stimulation with TGF-β1. This finding can
be explained by the fact that various oncogenes may be activated or
this is only a response to TGF-β1 in the early phase of
carcinogenesis. However, in the persistent presence of TGF-β1, the
expression levels of DNMT1 and DNMT3A were significantly increased
indicating that hypermethylation occurred in promoter CpG islands,
resulting in epigenetic silencing of TSGs leading to the generation
and proliferation of neoplastic cells.
miRNAs, a cluster of small and non-coding RNAs of
18–23 nucleotides in length, regulate the expression of target
genes post-transcriptionally and determine the cell fate by
controlling the translation of target mRNAs in different tissues
and cell types. Therefore, miRNAs take part in a variety of key
biological processes involving cell growth, development, apoptosis,
differentiation and oncogenesis by means of the regulation of gene
expression (31). miRNA-429 mainly
targets DNMT3A while targets of miRNA-29a include DNMT3A and
DNMT3B. Downregulation of miRNA-29a whose level is associated with
prognosis (32) was found in
various tumors such as liver (33)
and lung (34). DNMT1 can be
targeted by miR-152 (35) whose
downregulation was also observed in many types of cancers, such as
ovarian (36), stomach and breast
cancer (37,38). miRNAs whose positive and negative
regulation has been described in colorectal carcinogenesis are
deemed as potential molecular biomarkers for human malignancies
(39,40). Our results in the present study
revealed that expression levels of miR-152, miR-429 and miR-29a
were continuously downregulated after treatment with TGF-β1,
indicating that these miRNAs may play a role as tumor suppressors
and are inactivated during colon carcinogenesis. Taken together,
the expression profiles of the DNMTs determined that expression
levels of the DNMTs were negatively regulated by their target
miRNAs. The underlying mechanisms that contributed to the
regulation of the DNMTs by target miRNAs have been addressed. A
recent study demonstrated that miRNAs downregulate DNMT3B
expression by directly targeting the 3′-UTR of DNMT3B mRNA
(18). Our data indicated that
miR-152, miR-429 and miR-29a directly bind to the DNMT1, DNMT3A and
DNMT3B transcript, respectively, and post-transcriptionally mediate
expression of those genes. In addition, after treatment with
5-Aza-dC, expression of these target miRNAs was markedly increased
but did not reach a normal level, revealing that the expression of
miRNAs was also regulated by their corresponding DNMTs. In this
context, there is crucial crosstalk between DNMTs and target miRNAs
through a double-negative feedback loop.
Berberin, an isoquinoline alkaloid (41), is extracted from the Chinese herbal
medicine Coptis chinensis, and has been found to have
multiple antitumor roles which are valuable in clinical treatment.
Recent results have demonstrated that berberin inhibited the
proliferation of colon cancer cells through downregulation of
epidermal growth factor receptor (EGFR) expression (42), suppressed colon cancer cell
migration by targeting AMP-activated protein kinase (AMPK)
signaling (43), and induced cancer
cell death via cell cycle arrest, inducing apoptosis and inhibiting
inflammation (44). Evodiamine, a
natural chemical isolated from Evodia rutaecarpa, shows
numerous biological effects including antitumor, antinociceptive
and vasorelaxant properties (45,46).
Recent results demonstrated that the viability of CRC cells can be
inhibited by evodiamine via induction of apoptosis (47).
Although pharmacological research of berberine and
evodiamine has been carried out, studies concerning their effect on
epigenetic alterations is sparse. Our present results revealed that
berberine and evodiamine had a prominent effect on expression of
DNMTs and target miRNAs during early colon canceration as compared
with TGF-β group. In particular, their effect on the expression of
miR-29a was more obvious, implying that berberine and evodiamine
mediate the expression of DNMTs and miRNAs by a mechanism involving
the targeting of multiple factors which warrant further
elucidation. Our results suggest that berberine and evodiamine are
potential therapeutic agents for the early treatment and prevention
of CRC.
Acknowledgements
The present study was funded by the National Natural
Science Foundation of China (no. 81173257).
References
|
1
|
Tenesa A and Dunlop MG: New insights into
the aetiology of colorectal cancer from genome-wide association
studies. Nat Rev Genet. 10:353–358. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Friess H, Langrehr JM, Oettle H, Raedle J,
Niedergethmann M, Dittrich C, Hossfeld DK, Stöger H, Neyns B,
Herzog P, et al: A randomized multi-center phase II trial of the
angiogenesis inhibitor cilengitide (EMD 121974) and gemcitabine
compared with gemcitabine alone in advanced unresectable pancreatic
cancer. BMC Cancer. 6:2852006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Philip PA, Benedetti J, Corless CL, Wong
R, O'Reilly EM, Flynn PJ, Rowland KM, Atkins JN, Mirtsching BC,
Rivkin SE, et al: Phase III study comparing gemcitabine plus
cetuximab versus gemcitabine in patients with advanced pancreatic
adenocarcinoma: Southwest Oncology Group-directed intergroup trial
S0205. J Clin Oncol. 28:3605–3610. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
van Kampen JG, Marijnissen-van Zanten MA,
Simmer F, van der Graaf WT, Ligtenberg MJ and Nagtegaal ID:
Epigenetic targeting in pancreatic cancer. Cancer Treat Rev.
40:656–664. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Garagnani P, Pirazzini C and Franceschi C:
Colorectal cancer microenvironment: Among nutrition, gut
microbiota, inflammation and epigenetics. Curr Pharm Des.
19:765–778. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Postovit LM, Seftor EA, Seftor RE and
Hendrix MJ: Influence of the microenvironment on melanoma cell fate
determination and phenotype. Cancer Res. 66:7833–7836. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Adjei IM and Blanka S: Modulation of the
tumor microenvironment for cancer treatment: A biomaterials
approach. J Funct Biomater. 6:81–103. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shi X and Wang X: The role of MTDH/AEG-1
in the progression of cancer. Int J Clin Exp Med. 8:4795–4807.
2015.PubMed/NCBI
|
|
9
|
Ma H, Xu H and Qin J: Biomimetic tumor
microenvironment on a microfluidic platform. Biomicrofluidics.
7:115012013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Schmaus A, Bauer J and Sleeman JP: Sugars
in the microenvironment: The sticky problem of HA turnover in
tumors. Cancer Metastasis Rev. 33:1059–1079. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zu X, Zhang Q, Cao R, Liu J, Zhong J, Wen
G and Cao D: Transforming growth factor-β signaling in tumor
initiation, progression and therapy in breast cancer: An update.
Cell Tissue Res. 347:73–84. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Neuzillet C, Tijeras-Raballand A, Cohen R,
Cros J, Faivre S, Raymond E and de Gramont A: Targeting the TGFβ
pathway for cancer therapy. Pharmacol Ther. 147:22–31. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ohshio Y, Teramoto K, Hashimoto M,
Kitamura S, Hanaoka J and Kontani K: Inhibition of transforming
growth factor-β release from tumor cells reduces their motility
associated with epithelial-mesenchymal transition. Oncol Rep.
30:1000–1006. 2013.PubMed/NCBI
|
|
15
|
Mao Y, Keller ET, Garfield DH, Shen K and
Wang J: Stromal cells in tumor microenvironment and breast cancer.
Cancer Metastasis Rev. 32:303–315. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Quante M, Varga J, Wang TC and Greten FR:
The gastrointestinal tumor microenvironment. Gastroenterology.
145:63–78. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lee JY, Jeong W, Lim W, Lim CH, Bae SM,
Kim J, Bazer FW and Song G: Hypermethylation and
post-transcriptional regulation of DNA methyltransferases in the
ovarian carcinomas of the laying hen. PLoS One. 8:e616582013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xue G, Ren Z, Chen Y, Zhu J, Du Y, Pan D,
Li X and Hu B: A feedback regulation between miR-145 and DNA
methyltransferase 3b in prostate cancer cell and their responses to
irradiation. Cancer Lett. 361:121–127. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mendell JT: MicroRNAs: Critical regulators
of development, cellular physiology and malignancy. Cell Cycle.
4:1179–1184. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Vaiopoulos AG, Athanasoula KCh and
Papavassiliou AG: NF-κB in colorectal cancer. J Mol Med.
91:1029–1037. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hamidi T, Singh AK and Chen T: Genetic
alterations of DNA methylation machinery in human diseases.
Epigenomics. 7:247–265. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cea M, Cagnetta A, Gobbi M, Patrone F,
Richardson PG, Hideshima T and Anderson KC: New insights into the
treatment of multiple myeloma with histone deacetylase inhibitors.
Curr Pharm Des. 19:734–744. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Andreoli F, Barbosa AJ, Parenti MD and Del
Rio A: Modulation of epigenetic targets for anticancer therapy:
Clinicopathological relevance, structural data and drug discovery
perspectives. Curr Pharm Des. 19:578–613. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Esteller M: Epigenetics in cancer. N Engl
J Med. 358:1148–1159. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Laird PW: The power and the promise of DNA
methylation markers. Nat Rev Cancer. 3:253–266. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Markowitz SD and Bertagnolli MM: Molecular
origins of cancer: Molecular basis of colorectal cancer. N Engl J
Med. 361:2449–2460. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wu Q and Ni X: ROS-mediated DNA
methylation pattern alterations in carcinogenesis. Curr Drug
Targets. 16:13–19. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Luczak MW and Jagodziński PP: The role of
DNA methylation in cancer development. Folia Histochem Cytobiol.
44:143–154. 2006.PubMed/NCBI
|
|
31
|
Pastuszak-Lewandoska D, Kordiak J,
Migdalska-Sęk M, Czarnecka KH, Antczak A, Górski P, Nawrot E,
Kiszałkiewicz JM, Domańska D and Brzeziańska-Lasota E: Quantitative
analysis of mRNA expression levels and DNA methylation profiles of
three neighboring genes: FUS1NPRL2/G21 and RASSF1A in non-small
cell lung cancer patients. Respir Res. 16:762015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Garzon R, Fabbri M, Cimmino A, Calin GA
and Croce CM: MicroRNA expression and function in cancer. Trends
Mol Med. 12:580–587. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xiong Y, Fang JH, Yun JP, Yang J, Zhang Y,
Jia WH and Zhuang SM: Effects of microRNA-29 on apoptosis,
tumorigenicity, and prognosis of hepatocellular carcinoma.
Hepatology. 51:836–845. 2010.PubMed/NCBI
|
|
34
|
Fabbri M, Garzon R, Cimmino A, Liu Z,
Zanesi N, Callegari E, Liu S, Alder H, Costinean S,
Fernandez-Cymering C, et al: MicroRNA-29 family reverts aberrant
methylation in lung cancer by targeting DNA methyltransferases 3A
and 3B. Proc Natl Acad Sci USA. 104:15805–15810. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xu H, Cheung IY, Guo HF and Cheung NK:
MicroRNA miR-29 modulates expression of immunoinhibitory molecule
B7-H3: Potential implications for immune based therapy of human
solid tumors. Cancer Res. 69:6275–6281. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ji W, Yang L, Yuan J, Yang L, Zhang M, Qi
D, Duan X, Xuan A, Zhang W, Lu J, et al: MicroRNA-152 targets DNA
methyltransferase 1 in NiS-transformed cells via a feedback
mechanism. Carcinogenesis. 34:446–453. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Langhe R, Norris L, Saadeh FA,
Blackshields G, Varley R, Harrison A, Gleeson N, Spillane C, Martin
C, O'Donnell DM, et al: A novel serum microRNA panel to
discriminate benign from malignant ovarian disease. Cancer Lett.
356:628–636. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Pinto R, De Summa S, Danza K, Popescu O,
Paradiso A, Micale L, Merla G, Palumbo O, Carella M and Tommasi S:
MicroRNA expression profiling in male and female familial breast
cancer. Br J Cancer. 111:2361–2368. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Omrane I and Benammar-Elgaaied A: The
immune microenvironment of the colorectal tumor: Involvement of
immunity genes and microRNAs belonging to the TH17 pathway. Biochim
Biophys Acta. 1856:28–38. 2015.PubMed/NCBI
|
|
40
|
Slaby O, Svoboda M, Michalek J and Vyzula
R: MicroRNAs in colorectal cancer: Translation of molecular biology
into clinical application. Mol Cancer. 8:1022009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen C, Yu Z, Li Y, Fichna J and Storr M:
Effects of berberine in the gastrointestinal tract - a review of
actions and therapeutic implications. Am J Chin Med. 42:1053–1070.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang L, Cao H, Lu N, Liu L, Wang B, Hu T,
Israel DA, Peek RM Jr, Polk DB and Yan F: Berberine inhibits
proliferation and down-regulates epidermal growth factor receptor
through activation of Cbl in colon tumor cells. PLoS One.
8:e566662013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Park JJ, Seo SM, Kim EJ, Lee YJ, Ko YG, Ha
J and Lee M: Berberine inhibits human colon cancer cell migration
via AMP-activated protein kinase-mediated downregulation of
integrin β1 signaling. Biochem Biophys Res Commun. 426:461–467.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Murthy KN Chidambara, Jayaprakasha GK and
Patil BS: The natural alkaloid berberine targets multiple pathways
to induce cell death in cultured human colon cancer cells. Eur J
Pharmacol. 688:14–21. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kan SF, Yu CH, Pu HF, Hsu JM, Chen MJ and
Wang PS: Anti-proliferative effects of evodiamine on human prostate
cancer cell lines DU145 and PC3. J Cell Biochem. 101:44–56. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kobayashi Y: The nociceptive and
anti-nociceptive effects of evodiamine from fruits of Evodia
rutaecarpa in mice. Planta Med. 69:425–428. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chien CC, Wu MS, Shen SC, Ko CH, Chen CH,
Yang LL and Chen YC: Activation of JNK contributes to
evodiamine-induced apoptosis and G2/M arrest in human colorectal
carcinoma cells: A structure-activity study of evodiamine. PLoS
One. 9:e997292014. View Article : Google Scholar : PubMed/NCBI
|