Introduction
Colorectal cancer (CRC) is the third most common
cancer and the fourth leading cause of cancer-related mortality
worldwide (1,2). Approximately 1 million new cases and
600,000 deaths due to CRC are estimated to occur annually around
the world (3). CRC is one of the
most prevalent cancers in western populations (4) but has low incidence rates in Asia,
Africa and South America (5).
However, the frequency of CRC in China has rapidly increased such
that this carcinoma has emerged as the fifth most common cancer and
the fourth most common cause of cancer-related deaths in the
country (6). Surgical resection
followed by chemotherapy and/or radiotherapy is currently the
effective modality for CRC patients (7). Prognosis of CRC patients remains
unsatisfactory despite the remarkable developments in the diagnosis
and treatments of CRC (8). Local
recurrence and distant metastasis are the primary causes of the
unfavourable prognosis of CRC (9).
Therefore, understanding the molecular mechanisms underlying CRC
progression is essential to identify effective biomarkers and novel
therapeutic methods for CRC patients.
MicroRNAs (miRNAs) are an emerging group of
single-strand non-coding small RNAs (~22 nucleotides) first
discovered in the early 1990s in Caenorhabditis elegans
(10). miRNAs negatively regulate
gene expression by interacting with the 3-untranslated regions
(UTRs) of corresponding target messenger RNAs (mRNAs) in a
base-pairing manner, leading to translational repression or
degradation of target mRNAs (11).
Computational estimations suggest ~1,000 miRNAs in the human
genome, which regulate one-third of human protein-encoding genes
(12). Multiple bodies of evidence
have indicated that miRNAs are abnormally expressed in almost all
human neoplasms (13,14). Previous studies have demonstrated
that miRNA deregulation may be significantly correlated with
diagnosis, treatment and prognosis in human cancer (15–17).
miRNAs play important roles in tumourigenesis and tumour
development by regulating many diverse biological processes, such
as cell proliferation, cycle, apoptosis, invasion, migration,
metastasis, angiogenesis and epithelial-mesenchymal transition
(18–20). miRNAs can act as tumour suppressors
or oncogenes, depending on their transcript targets (21,22).
Therefore, miRNAs have been proposed to be potential indicators and
therapeutic targets in various types of cancers (23,24).
miR-411, located on chromosome 14q32, was previously
observed to be aberrantly expressed in multiple human cancers
(25–28). However, the expression pattern,
function and underlying molecular mechanism of miR-411 in CRC
remain unclear. Therefore, the present study was performed to
detect miR-411 expression, investigate the biological roles of
miR-411 and identify its mechanism of action in CRC cells.
Materials and methods
Tumour specimens
The present study was approved by the Human Ethics
Committee of the Cancer Hospital of China Medical University,
Liaoning Cancer Hospital and Institute. Written informed consent
was obtained from all patients. A total of 46 CRC tissues and
corresponding adjacent normal tissues were obtained from patients
who underwent surgerical resection at the Department of Colorectal
Surgery, Cancer Hospital of China Medical University, Liaoning
Cancer Hospital and Institute between May 2014 and February 2016.
None of these patients with CRC had been treated with chemotherapy,
radiotherapy or any adjuvant therapy before surgery. All tissue
specimens were immediately frozen in liquid nitrogen after
resection and stored at −80°C.
Cell lines and culture condition
CRC cell lines (SW480, SW620, HCT116, HT29, CaCo-2
and LoVo) and the 293T cell line were purchased from the Type
Culture Collection of Chinese Academy of Sciences (Shanghai,
China). Normal human colon epithelium cell line (FHC) was obtained
from the American Type Culture Collection (ATCC; Manassas, VA,
USA). All cells were maintained in Dulbeccos modified Eagles medium
(DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 10% fetal bovine serum (FBS; Invitrogen; Thermo
Fisher Scientific), 100 U/ml streptomycin and 100 U/ml penicillin
in a humidified incubator with an atmosphere of 5% CO2
at 37°C.
Oligonucleotide transfection
miR-411 mimics, miRNA mimic negative control
(miR-NC) and small-interfering RNA targeting PIK3R3 (PIK3R3 siRNA)
and its negative control (NC siRNA) were acquired from Suzhou
GenePharma Co., Ltd. (Shanghai, China). PIK3R3 overexpression
plasmid (pcDNA3.1-PIK3R3) and empty pcDNA3.1 plasmid were obtained
from Guangzhou RiboBio Co., Ltd. (Guangzhou, China). Cells were
seeded into 6-well plates at 5×105 cells/well. After 24
h, the cells were transfected with these oligonucleotides using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific) following
the manufacturers protocol.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissue specimens or
cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA),
according to the manufacturers instructions. Total RNA
concentration was determined using a NanoDrop ND-100
spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). For
miR-411 detection, the total RNA was reverse-transcribed to cDNA
using a TaqMan MicroRNA Reverse Transcription kit (Applied
Biosystems, Carlsbad, CA, USA). The relative expression of miR-411
was examined by TaqMan MicroRNA PCR kit (Applied Biosystems). The
miR-411 expression levels were normalised to those of U6 snRNA.
PIK3R3 mRNA was quantified by synthesising cDNA using PrimeScript
RT reagent kit (Takara Bio, Dalian, China). These cDNAs were used
to detect the expression of PIK3R3 mRNA by quantitative PCR using a
SYBR Premix Ex Taq™ kit (Takara Bio). The PIK3R3 mRNA expression
was normalised to those of GAPDH. The primers were designed as
follows: miR-411, 5-GGGGTAGTAGACCGTATAG-3 (forward) and
5-TGCGTGTCGTGGAGTC-3 (reverse); U6 snRNA, 5-CTC GCTTCGGCAGCACA-3
(forward) and 5-TGGTGTCGTG GAGTCG-3 (reverse); PIK3R3,
5-CTTGCTCTGTGGTGG CCGAT-3 (forward) and 5-GACGTTGAGGGAGTCGTT GT-3
(reverse); and GAPDH, 5-GAAGGTGAAGGTCGGA GTC-3 (forward) and
5-GAAGATGGTGATGGGATTTC-3 (reverse). Relative expression levels were
calculated using the 2−∆∆Ct method (29).
Cell Counting Kit-8 (CCK-8) assay
Cell proliferation was determined using the CCK-8
assay. Briefly, transfected cells were harvested at 24 h
post-transfection. A total of 3×103 cells suspended in
200 µl of DMEM with 10% FBS were seeded into each well of a 96-well
plate and cultured at 37°C for 0, 1, 2 or 3 days. At each
time-point, 10 µl of CCK-8 solution (Dojindo Molecular
Technologies, Kumamoto, Japan) was added to each well. After an
additional 2 h of incubation at 37°C, the optical density of each
well at a wavelength of 450 nm was measured with a microplate
reader (Model 550; Bio-Rad Laboratories, Shanghai, China). Each
assay was performed in quintuplicate and repeated at least
thrice.
Matrigel invasion assay
Transwell chambers (Corning Inc., Cambridge, MA,
USA) with 8-µm pore size filters covered with Matrigel (BD
Biosciences, Franklin Lakes, NJ, USA) were used to perform the
Matrigel invasion assay. Transfected cells were harvested at 48 h
post-transfection. Transfected cells (5×104) in FBS-free
DMEM medium were seeded in the upper chambers, and DMEM containing
10% FBS was supplemented into the lower chambers to serve as a
chemoattractant. After incubation at 37°C under 5% CO2
for 24 h, cells that did not invade through the pores were removed
carefully by a cotton swab. Invasive cells were fixed using 4%
paraformaldehyde, stained with 0.1% crystal violet solution, washed
in phosphate-buffered saline (PBS) and dried in air. Images were
captured, and cell number was counted in five randomly selected
fields under an inverted microscope (×200 magnifications; X71;
Olympus Corp., Tokyo, Japan).
Flow cytometry
Cell apoptosis was analysed using the Annexin V-FITC
apoptosis detection kit (Invitrogen). Transfected cells were
harvested at 48 h post-transfection, washed with PBS and fixed
using 80% ice-cold ethanol in PBS. Afterwards, the cells were
resuspended in 300 µl of 1X binding buffer and stained with 5 µl of
FITC-Annexin V and 5 µl of propidium iodide (PI) in the dark at
room temperature for 20 min. BD FACSCalibur™ flow cytometer was
used to detect the cell apoptosis.
Bioinformatic analysis and luciferase
reporter assay
The potential targets of miR-411 were predicted by
performing bioinformatic analysis using TargetScan (www.targetscan.org) and microRNA.org
(http://www.microrna.org/microrna/home.do).
pMIR-PIK3R3-3′-UTR wild-type (Wt) and
pMIR-PIK3R3-3′-UTR mutant (Mut) were synthesised and confirmed by
Shanghai Genepharma. 293T cells were plated in 24-well plates at a
density of 60–70% confluence. After incubation overnight, cells
were cotransfected with miR-411 mimics or miR-NC and
pMIR-PIK3R3-3′-UTR Wt or pMIR-PIK3R3-3′-UTR Mut, using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific) according
to the manufacturers protocol. After 24 h post-transfection,
luciferase activities were detected using Dual-Luciferase reporter
system (Promega, Madison, WI, USA) following the manufacturers
protocol. Firefly luciferase activity was normalised to
Renilla luciferase activity. Transfections were performed in
triplicate and repeated in three individual experiments.
Western blot analysis
Protein from tissue specimens or cells was extracted
using radioimmunoprecipitation assay cell lysis buffer (Beyotime
Institute of Biotechnology, Shanghai, China). Protein extracts were
quantified using Bicinchoninic Acid protein assay kit (Beyotime
Institute of Biotechnology). Equal amounts of protein were
separated by 10% sodium dodecyl sulphate-polyacrylamide gel
electrophoresis, transferred onto polyvinylidene difluoride
membranes (Millipore, Billerica, MA, USA), blocked with 5% fat-free
milk in Tris-buffered saline with Tween (TBST) buffer and incubated
with the specific primary antibodies against PIK3R3 antibody
(sc-376615; 1:1,000 dilution; Santa Cruz Biotechnology, Santa Cruz,
CA, USA), p-AKT antibody (sc-81433; 1:1,000 dilution; Santa Cruz
Biotechnology), AKT antibody (sc-81434; 1:1,000 dilution; Santa
Cruz Biotechnology), p-mTOR ser 2481 antibody (sc-293132; 1:1,000
dilution; Santa Cruz Biotechnology), mTOR antibody (sc-293089;
1:1,000 dilution; Santa Cruz Biotechnology) and GAPDH antibody
(sc-47724; 1:1,000 dilution; Santa Cruz Biotechnology) at 4°C
overnight. Subsequently, the membranes were washed thrice with TBST
for 10 min and probed with goat anti-mouse horseradish
peroxidase-conjugated secondary antibody (sc-2005; 1:5,000
dilution; Santa Cruz Biotechnology) at room temperature for 1 h.
Protein bands were visualised using ECL Protein Detection kit
(Pierce Biotechnology, Inc., Rockford, IL, USA) and analysed with
Quantity One software (Bio-Rad Laboratories, Inc., Hercules, CA,
USA). GAPDH was used as the loading control.
Statistical analysis
Data are presented as the mean ± standard deviation
and were compared with the Students t-test or one way ANOVA. All
statistical analyses were performed using the SPSS 13.0 software
(SPSS, Inc., Chicago, IL, USA). Spearmans correlation analysis was
adopted to investigate the correlation between miR-411 and PIK3R3
mRNA expression level. P<0.05 was considered statistically
significant.
Results
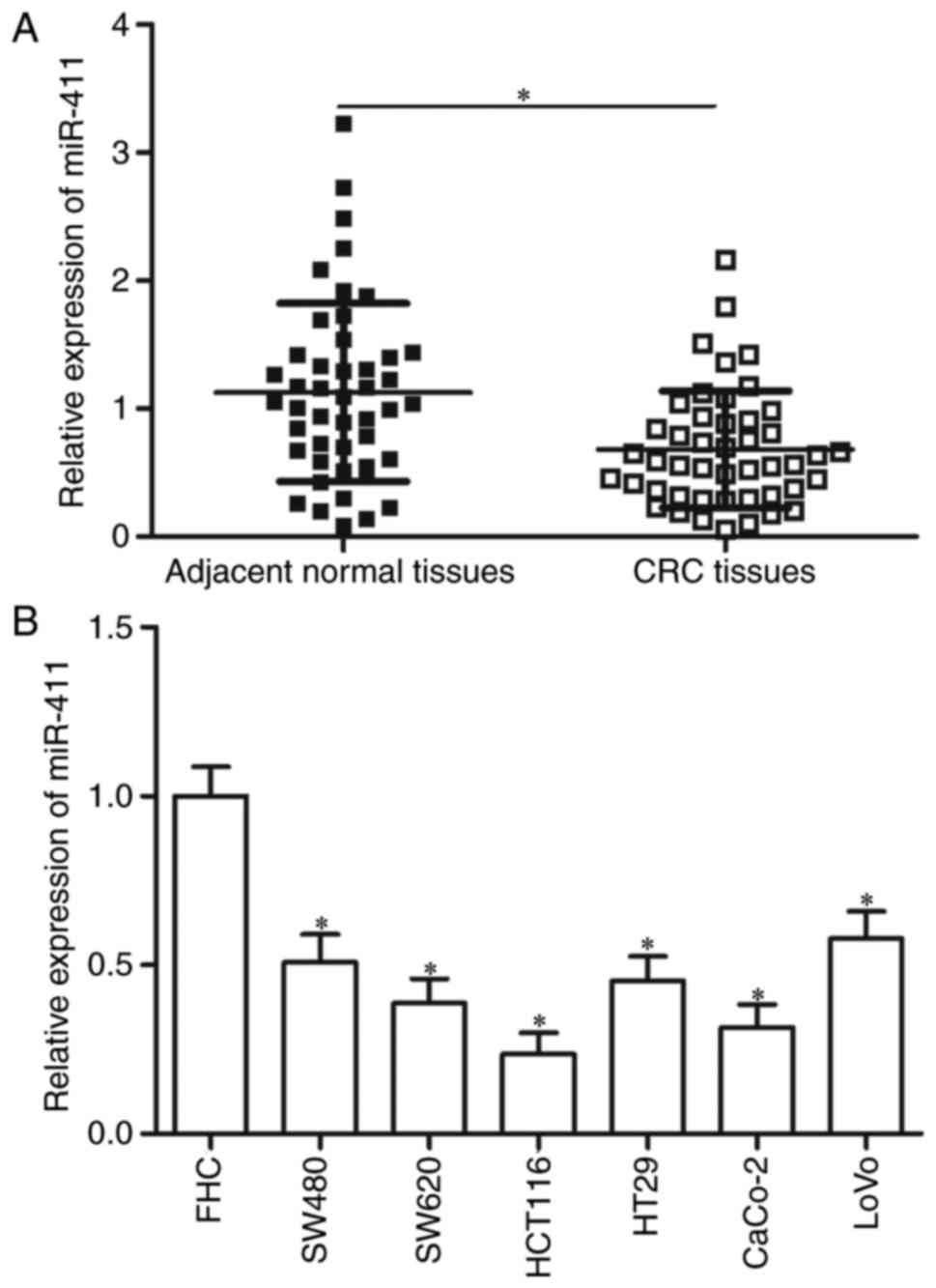
miR-411 is downregulated in human CRC
tissues and cell lines
RT-qPCR was performed on 46 paired CRC tissues and
corresponding adjacent normal tissues to investigate the status of
miR-411 in CRC. Compared with the corresponding adjacent normal
tissues, the expression of miR-411 was significantly downregulated
in the CRC tissues (Fig. 1A;
P<0.05). miR-411 expression levels in six CRC cell lines (SW480,
SW620, HCT116, HT29, CaCo-2 and LoVo) and normal FHC cells were
also analysed by RT-qPCR. Fig. 1B
shows that all tested CRC cells showed significantly lower
expression levels of miR-411 compared with the normal FHC
(P<0.05). These data suggest that miR-411 plays important roles
in CRC formation and progression.
Low expression of miR-411 correlates
with adverse clinicopathological characteristics of CRC
patients
miR-411 expression and clinical characteristics of
patients with CRC were associated to explore the clinical value of
miR-411 in CRC. The CRC patients were subsequently divided into
either the miR-411 low-expression group (n=23) or the miR-411
high-expression group (n=23). The median expression level of
miR-411 in all samples was regarded as the cut-off. As shown in
Table I, the low expression level
of miR-411 was significantly correlated with lymph node metastasis
(P=0.018), distant metastasis (P=0.007) and TNM stage (P=0.008).
However, no correlation was observed between miR-411 expression and
other clinicopathological characteristics, such as sex (P=0.555),
age (P=0.548), tumour size (P=0.345) and differentiation (P=0.552).
These results suggest that miR-411 is a possible prognostic
biomarker for CRC patients.
 | Table I.Correlation between the miR-411
expression and clinicopathological features of CRC cases. |
Table I.
Correlation between the miR-411
expression and clinicopathological features of CRC cases.
|
|
| miR-411
expression |
|
|---|
|
|
|
|
|
|---|
| Features | Cases | Low | High | P-value |
|---|
| Sex |
|
|
| 0.555 |
|
Male | 24 | 13 | 11 |
|
|
Female | 22 | 10 | 12 |
|
| Age (years) |
|
|
| 0.548 |
|
<60 | 28 | 15 | 13 |
|
|
≥60 | 18 | 8 | 10 |
|
| Tumour size
(cm) |
|
|
| 0.345 |
|
<5 | 15 | 9 | 6 |
|
| ≥5 | 31 | 14 | 17 |
|
|
Differentiation |
|
|
| 0.552 |
| Well
and moderate | 26 | 12 | 14 |
|
|
Poor | 20 | 11 | 9 |
|
| Lymph node
metastasis |
|
|
| 0.018a |
|
Absence | 24 | 8 | 16 |
|
|
Present | 22 | 15 | 7 |
|
| Distant
metastasis |
|
|
| 0.007a |
|
Absence | 27 | 9 | 18 |
|
|
Present | 19 | 14 | 5 |
|
| TNM stage |
|
|
| 0.008a |
|
I–II | 22 | 6 | 15 |
|
|
III–IV | 24 | 17 | 8 |
|
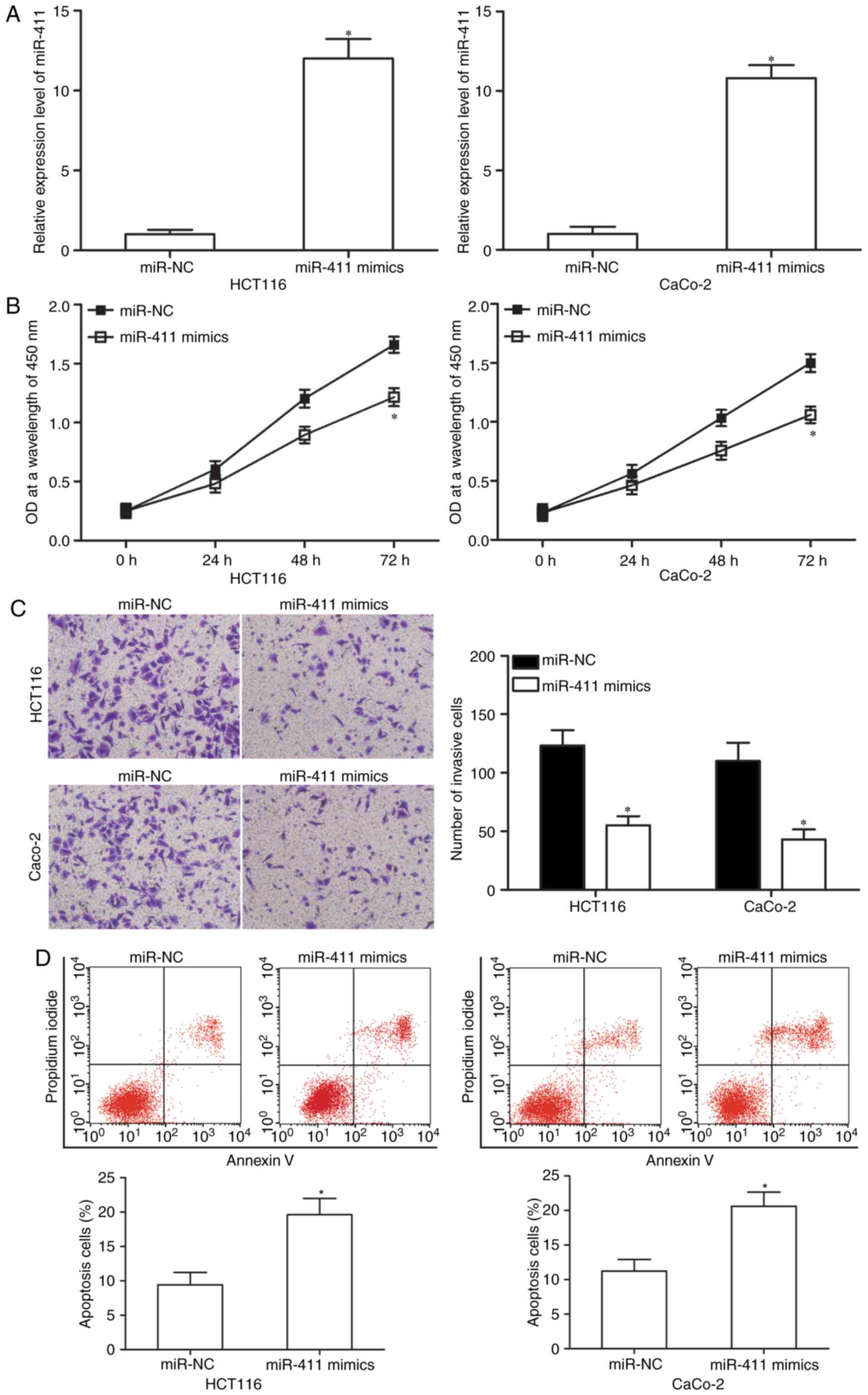
miR-411 inhibits cell proliferation
and invasion but promotes apoptosis of CRC
We selected HCT-116 and CaCo-2 cells, both with
relatively lower endogenous miR-411, to transfect miR-411 mimics to
investigate the functional roles of miR-411 in CRC cells. After
transfection, RT-qPCR analysis demonstrated that miR-411 was
markedly upregulated in the HCT-116 and CaCo-2 cells after
transfection with miR-411 mimics (50 nM; Fig. 2A, P<0.05). Subsequently, the
effect of miR-411 overexpression on cell proliferation was
assessed. CCK-8 assay indicated that the ectopic expression of
miR-411 inhibited HCT-116 and CaCo-2 cell proliferation (Fig. 2B; P<0.05). Matrigel invasion
assay was performed to examine the invasion abilities of HCT-116
and CaCo-2 cells after transfection with miR-411 mimics or miR-NC.
The results revealed that restored expression of miR-411 decreased
cell invasion abilities in HCT-116 and CaCo-2 cells (Fig. 2C; P<0.05). Then, we analysed the
effect of miR-411 on cell apoptosis using flow cytometry. Fig. 2D shows that upregulation of miR-411
increased the apoptosis rate in the HCT-116 and CaCo-2 cells
(P<0.05). Thus, these findings suggest that miR-411 plays a
suppressive role in CRC progression.
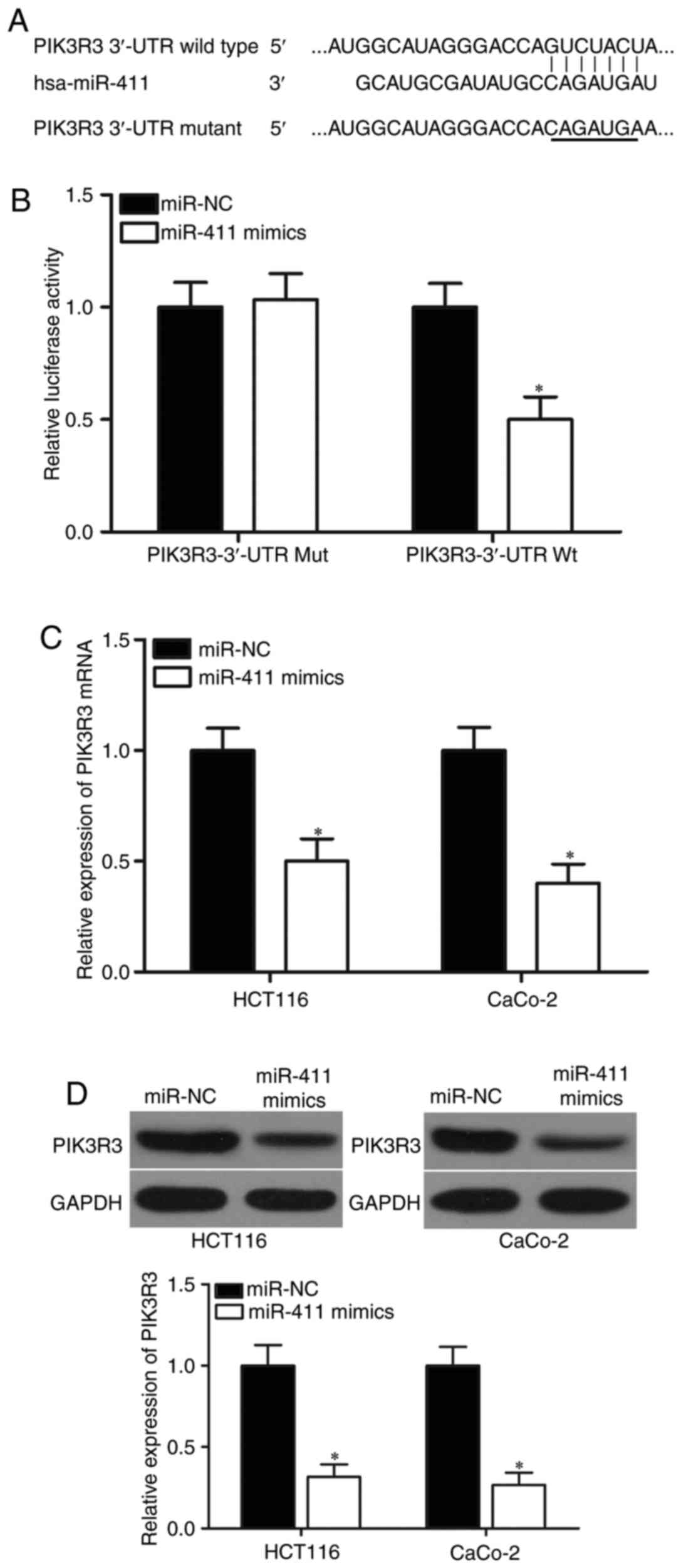
PIK3R3 is a direct target of miR-411
in CRC
Potential targets of miR-411 were predicted using
bioinformatic analysis to determine the molecular mechanisms
responsible for the tumour-suppressing roles of miR-411. Among
hundreds of potential candidates, PIK3R3 was selected for further
target identification (Fig. 3A)
because of its important roles in CRC tumourigenesis and tumour
development (30). In addition,
PIK3R3 was also validated as a direct target of other miRNAs in CRC
(31). To verify this prediction,
luciferase reporter assay was conducted in 293T cells cotransfected
with miR-411 mimics or miR-NC and pMIR-PIK3R3-3′-UTR Wt or
pMIR-PIK3R3-3′-UTR Mut. The results showed that miR-411
significantly repressed the relative luciferase activities with the
wild-type PIK3R3 3′-UTR (Fig. 3B;
P<0.05) but did not alter the activity of the mutant PIK3R3
3′-UTR. To further confirm the potential role of miR-411 in the
regulation of PIK3R3, we detected PIK3R3 expression in HCT-116 and
CaCo-2 cells following transfection with miR-411 mimics or miR-NC.
RT-qPCR and western blot analysis revealed that miR-411
overexpression caused a significant downregulation in PIK3R3
expression at the mRNA (Fig. 3C;
P<0.05) and protein (Fig. 3D;
P<0.05) levels in the HCT-116 and CaCo-2 cells. These results
showed that PIK3R3 is a direct target of miR-411 in CRC.
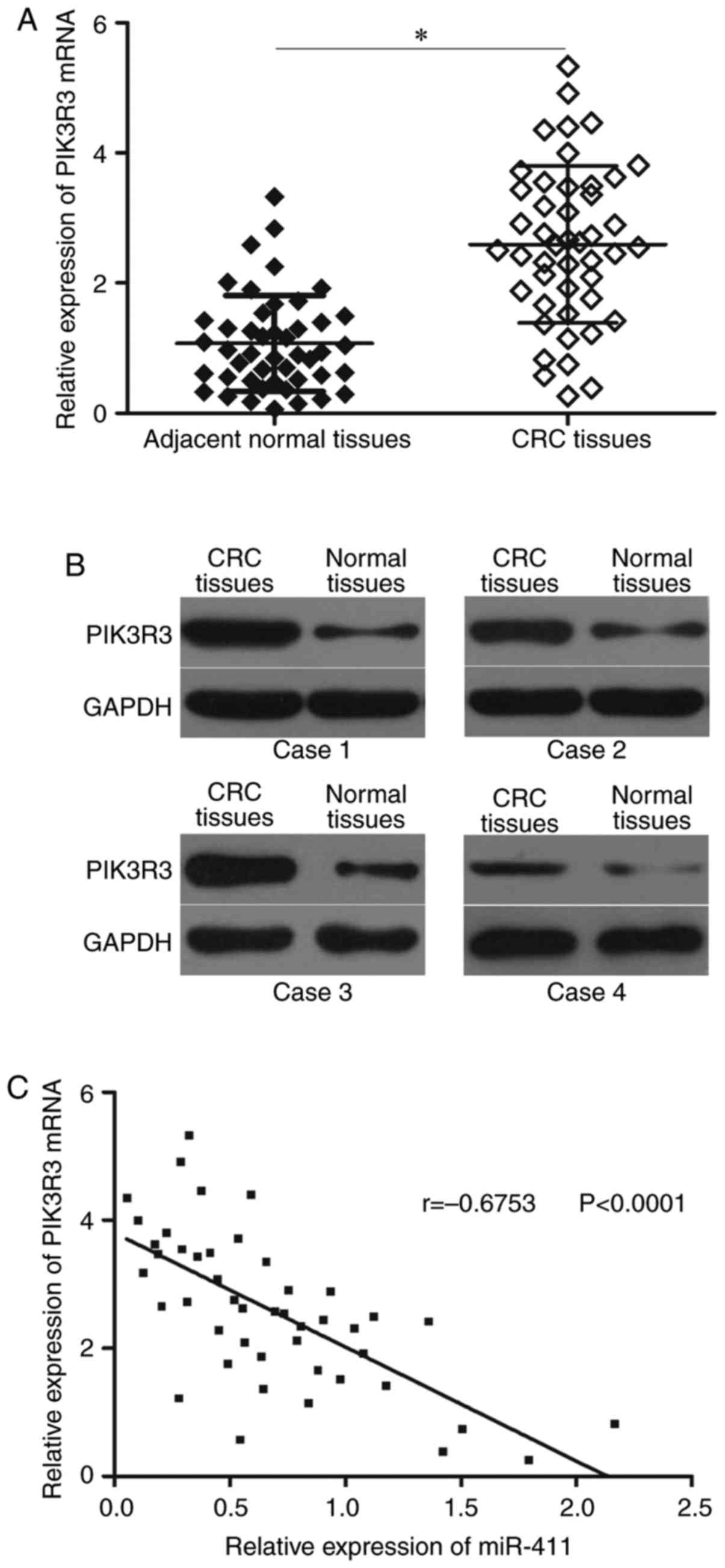
miR-411 level is negatively correlated
with PIK3R3 level in CRC tissues
We measured PIK3R3 expression in 46 paired CRC
tissues and corresponding adjacent normal tissues to further
examine the relationship between miR-411 and PIK3R3. Fig. 4A shows that PIK3R3 mRNA was
upregulated in CRC tissues compared with that in adjacent normal
tissues (P<0.05). Western blot results showed that PIK3R3
protein was highly expressed in CRC tissues (Fig. 4B; P<0.05). Furthermore, an
inverse association between miR-411 and PIK3R3 mRNA level in CRC
tissues was observed using Spearmans correlation analysis (Fig. 4C, r=−0.6753; P<0.0001).
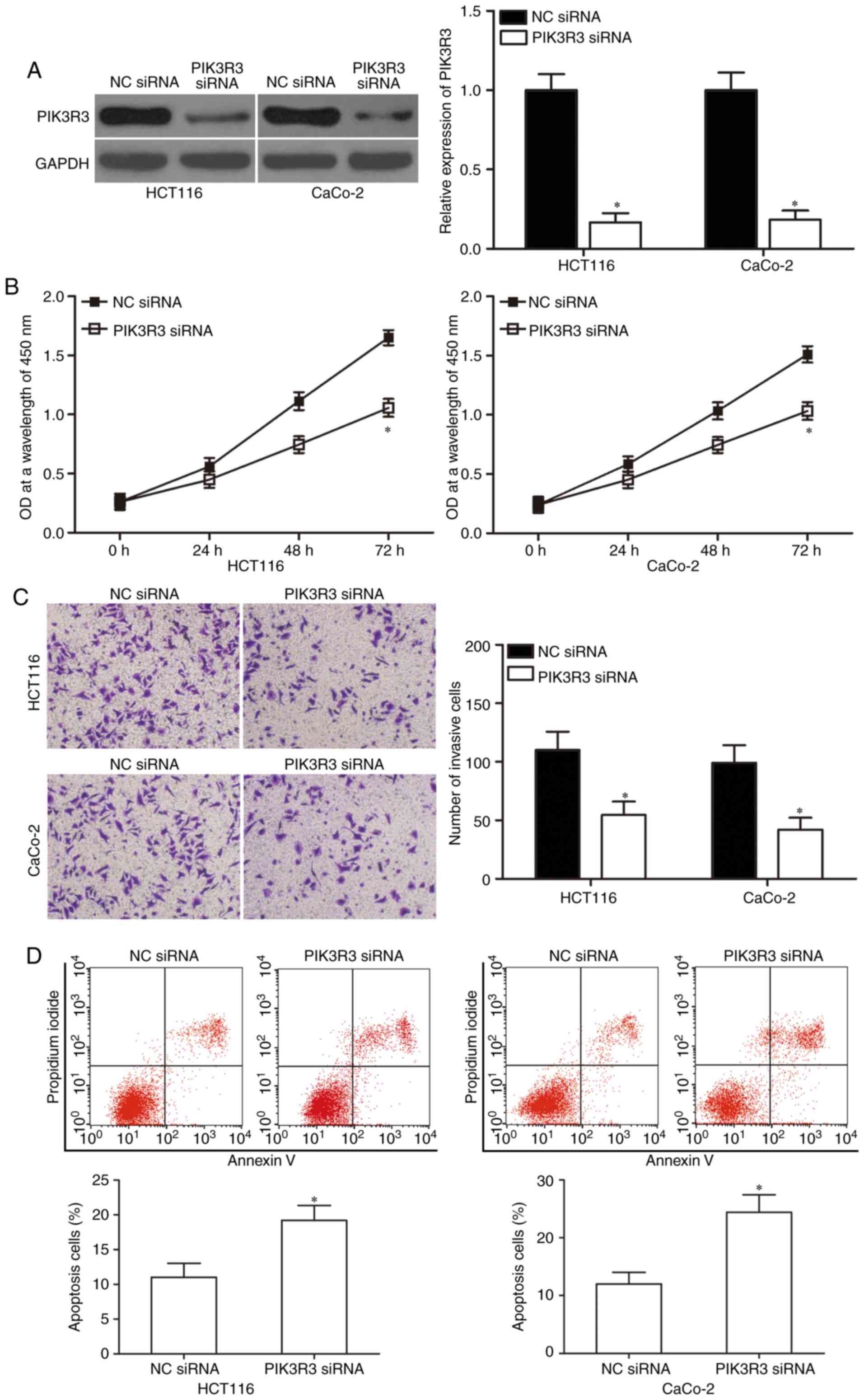
PIK3R3 knockdown inhibits
proliferation and invasion but increases apoptosis in CRC
cells
We used PIK3R3-specific siRNA to genetically
knockdown endogenous PIK3R3 expression in HCT-116 and CaCo-2 cells
to verify whether PIK3R3 knockdown would simulate the
miR-411-mediated effects. Western blot analysis was used to
determine the transfection efficiency and confirmed that PIK3R3 was
significantly downregulated in the PIK3R3 siRNA-transfected HCT-116
and CaCo-2 cells (Fig. 5A;
P<0.05). Next, CCK-8 assay, Matrigel invasion assay and flow
cytometric analysis demonstrated that down-regulation of PIK3R3
suppressed cell proliferation (Fig.
5B; P<0.05) and invasion (Fig.
5C; P<0.05) but increased apoptosis (Fig. 5D; P<0.05) in the HCT-116 and
CaCo-2 cells. Thus, our data confirmed that PIK3R3 downregulation
had similar tumour-suppressive effects as miR-411 overexpression in
CRC. These results further suggest that PIK3R3 is a direct
downstream target of miR-411 in CRC.
Overexpression of PIK3R3 abrogates the
tumour-suppressing effects of miR-411 in CRC cells
We confirmed that PIK3R3 is a direct target of
miR-411. Thus, rescue experiments were employed to disclose whether
PIK3R3 overexpression could abolish the tumour-suppressing effects
induced by miR-411 in CRC cells. HCT-116 and CaCo-2 cells were
transfected with miR-411 mimics with or without PIK3R3
overexpression plasmid pcDNA3.1-PIK3R3. After transfection, western
blot results showed that the reduced PIK3R3 protein expression
caused by miR-411 mimics was markedly restored by transfection of
the pcDNA3.1-PIK3R3 in the HCT-116 and CaCo-2 cells (Fig. 6A; P<0.05). In addition, the
effects of miR-411 on CRC cell proliferation (Fig. 6B; P<0.05), invasion (Fig. 6C; P<0.05) and apoptosis (Fig. 6D; P<0.05) were significantly
reversed by PIK3R3 overexpression. These results indicated that
miR-411 served as a tumour suppressor in CRC, at least in part, by
directly regulating PIK3R3.
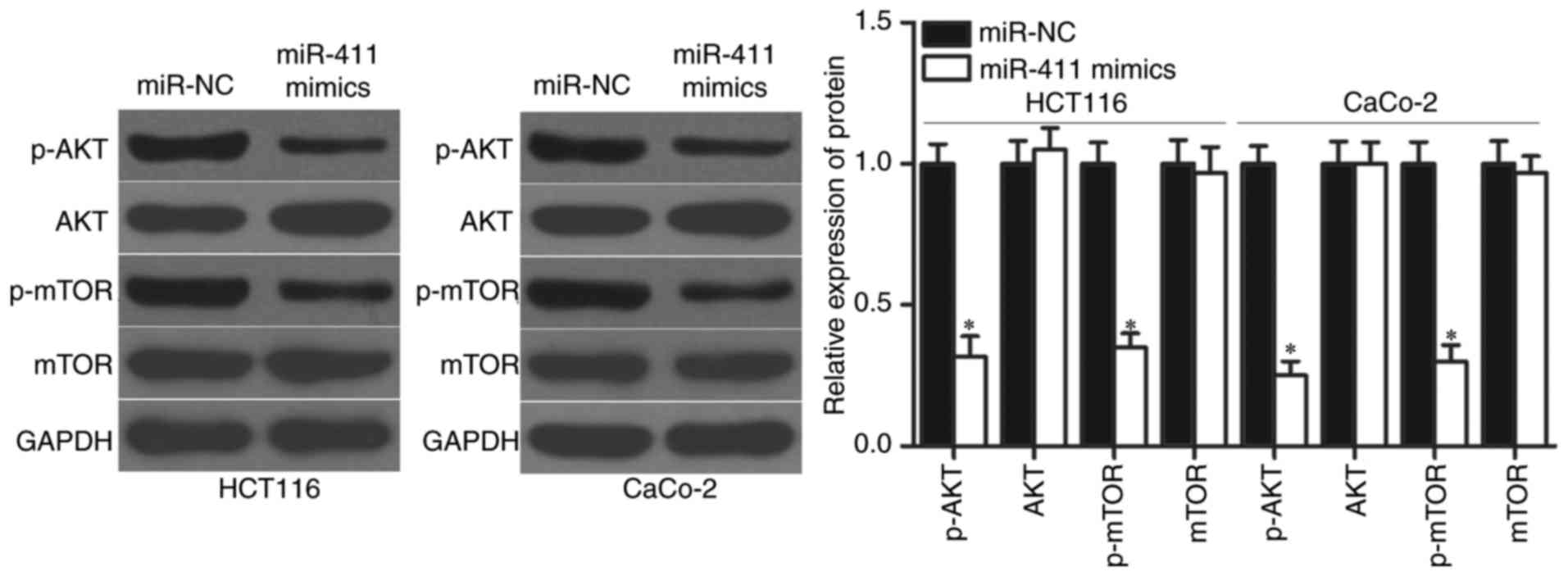
miR-411 regulates the AKT/mTOR
signalling pathways in CRC
PIK3R3 plays crucial roles in tumourigenesis and
tumour development by regulating the AKT/mTOR signalling pathway
(32,33). We detected p-AKT, AKT, p-mTOR and
mTOR expression in HCT-116 and CaCo-2 cells transfected with
miR-411 mimics or miR-NC to determine whether AKT/mTOR signalling
pathway was involved in the regulatory effects of miR-411 on CRC.
The results showed that upregulation of miR-411 significantly
reduced the p-AKT and p-mTOR expression in HCT-116 and CaCo-2
cells, whereas transfection with miR-411 mimics did not affect the
total AKT and mTOR protein levels (Fig.
7; P<0.05). Thus, miR-411 suppressed the activation of the
AKT/mTOR signalling pathway by targeting PIK3R3 to inhibit CRC
progression.
Discussion
Aberrant expression of miRNAs plays important roles
in the development and progression of various cancers by modulating
oncogenic and tumour-suppressor pathways (22,34).
Therefore, exploring the functions of miRNAs that specifically
contribute to CRC tumourigenesis and tumour development would
greatly aid in obtaining more information on CRC and provide new
targets for its diagnosis and treatment. In the present study,
miR-411 was significantly downregulated in the CRC tissues and cell
lines. In addition, the low expression of miR-411 was significantly
correlated with lymph node metastasis, distant metastasis and TNM
stage of CRC. Furthermore, restored expression of miR-411
suppressed cell proliferation and invasion but promoted apoptosis
in CRC. Moreover, PIK3R3 was identified as a novel direct target of
miR-411 in CRC, and miR-411 could diminish the AKT/mTOR signalling
pathway in CRC. These results suggested that miR-411 may be used to
design novel prognostic biomarker and therapeutic strategies for
CRC patients.
miR-411 is aberrantly expressed in several types of
cancers. For example, miR-411 was observed to be highly expressed
in hepatocellular carcinoma (25).
miR-411 was also found to be upregulated in lung cancer (26). However, contradictory reports have
demonstrated the downregulated miR-411 expression in breast cancer
tissues and cell lines (27,28).
Downregulated miR-411 expression was correlated with lymph node
metastasis and histological grade of breast cancer (27). These contradictory studies indicated
that miR-411 expression may be subject to tissue-specific
regulatory processes in various types of human cancer and that
miR-411 could serve as a useful prognosis marker in human
cancers.
miR-411 has recently been shown to be an oncogenic
miRNA in multiple types of cancer. For instance, Xia et al
(25)reported that miR-411 play an
oncogenic role in hepatocellular carcinoma by promoting cell
proliferation and anchorage-independent growth. Zhao et al
(26) found that miR-411
overexpression increased cell proliferation in lung cancer by
regulating cell cycle regulators. However, Sun et al
(35) found that miR-411 served as
a tumour suppressor miRNA in rhabdomyosarcoma by inhibiting cell
growth both in vitro and in vivo. Guo et al
revealed that resumed expression of miR-411 repressed cell growth
and metastasis in breast cancer (27,28).
These conflicting findings indicated that miR-411 acted as an
oncogene in certain types of cancer and as a tumour suppressor in
others. These findings also suggested that miR-411 may be a novel
therapeutic target for the development of antineoplastic
agents.
Targets of miR-411 should be identified to elucidate
the functions of miR-411 in CRC occurrence and progression. Novel
therapeutic targets may be determined for the treatment of CRC
patients. Several direct targets of miR-411 have been validated.
These targets include IL-18 (36)
in malignant pleural mesothelioma, ITCH (25) in hepatocellular carcinoma, SPRY4 in
rhabdomyosarcoma (35), FOXO1
(26) in lung cancer, SP1 (27) and GRB2 (28) in breast cancer. In the present
study, bioinformatic analysis predicted that PIK3R3 is a potential
target of miR-411. Luciferase reporter assay confirmed that the
3′-UTR of PIK3R3 may be directly targeted by miR-411. The results
from RT-qPCR and western blot analysis revealed that miR-411
negatively regulated PIK3R3 expression in CRC, suggesting that
miR-411 regulated PIK3R3 expression in transcriptional level. This
mainly due to that miRNAs induce mRNA degradation when the binding
complementarity is perfect, However, miRNAs regulate gene
expression at post-transcriptional level when the binding at the
3UTR of target genes is only partially complementary (37). PIK3R3 was upregulated in CRC tissues
and inversely correlated with miR-411 expression levels.
Downregulation of PIK3R3 had similar tumour-suppressive effects as
miR-411 overexpression in CRC. Moreover, upregulation of miR-411
could rescue the tumour-suppressing effects of miR-411
overexpression on CRC cells. Thus, PIK3R3 is a novel direct and
functional downstream target of miR-411 in CRC.
PIK3R3 is a member of the phosphatidylinositol
3-kinase (PI3K) family and was found to be abnormally upregulated
in various types of human cancers, such as glioma (38), ovarian (39), gastric (40), hepatocellular carcinoma (32), lung (41) and breast cancer (42). The oncogenic roles of PIK3R3 on
cancer initiation and progression has been described in several
types of cancers, such as Ewing sarcoma (43), gastric (40), ovarian (39) and breast cancer (42). In CRC, PIK3R3 was upregulated in
clinical specimens and cell lines. In addition, upregulated PIK3R3
expression has been positively correlated with CRC metastasis.
Functional experiments revealed that PIK3R3 overexpression
increased tumour migration, invasion and epithelial-to-mesenchymal
transition in vitro and promoted metastasis in vivo
(30). The present study revealed
that miR-411 inhibited CRC progression by directly targeting PIK3R3
and indirectly regulating AKT/mTOR signalling pathway. Thus,
miR-411/PIK3R3/AKT/mTOR pathway may serve as a potential target to
treat CRC patients.
In conclusion, the present study provided first
evidence that miR-411 exhibited tumour suppressive roles against
CRC by directly targeting PIK3R3. These results suggested that
miR-411 could be a novel therapeutic target for patients with CRC.
In this study, we did not explore the effect of miR-411 on the
normal FHC cells.
In the following experiments, we will examine the
effects of miR-411 underexpression on FHC cells. The small sample
size is a limitation of the study, and we will collect more CRC
tissues and the relationship between the pathologies in the top and
bottom percentiles and miR-411 expression. Despite we found that
miR-411 increased apoptosis of CRC cells, the effect of miR-411
overexpression on the expression levels apoptotic markers did not
analyzed. In our further research we should examine the effects of
miR-411 on apoptotic markers in CRC cells. In this study,
luciferase reporter assays were performed in 293T cells. The
luciferase reporter assay will be repeated with the CRC cell lines
used in the following experiments.
Acknowledgements
The present study was supported by grants from the
National Natural Science Fund from the National Natural Science
Foundation of China (grant no. 81672427) and The Project of
Liaoning Clinical Research Center for Colorectal Cancer (grant no.
2015225005).
References
|
1
|
Haggar FA and Boushey RP: Colorectal
cancer epidemiology: Incidence, mortality, survival, and risk
factors. Clin Colon Rectal Surg. 22:191–197. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Center MM, Jemal A, Smith RA and Ward E:
Worldwide variations in colorectal cancer. CA Cancer J Clin.
59:366–378. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sung JJ, Lau JY, Goh KL and Leung WK; Asia
Pacific Working Group on Colorectal Cancer, : Increasing incidence
of colorectal cancer in Asia: Implications for screening. Lancet
Oncol. 6:871–876. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li S, Wang J, Lu Y and Fan D: Screening
and early diagnosis of colorectal cancer in China: A 12 year
retrospect (1994–2006). J Cancer Res Clin Oncol. 133:679–686. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sanz-Garcia E, Grasselli J, Argiles G,
Elez ME and Tabernero J: Current and advancing treatments for
metastatic colorectal cancer. Expert Opin Biol Ther. 16:93–110.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Meyerhardt JA and Mayer RJ: Systemic
therapy for colorectal cancer. N Engl J Med. 352:476–487. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Qiu Y, Liu Q, Chen G, Wang W, Peng K, Xiao
W and Yang H: Outcome of rectal cancer surgery in obese and
nonobese patients: A meta-analysis. World J Surg Oncol. 14:232016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
OHara SP, Mott JL, Splinter PL, Gores GJ
and LaRusso NF: MicroRNAs: Key modulators of posttranscriptional
gene expression. Gastroenterology. 136:17–25. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu J: Control of protein synthesis and
mRNA degradation by microRNAs. Curr Opin Cell Biol. 20:214–221.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
He L, Thomson JM, Hemann MT,
Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe
SW, Hannon GJ, et al: A microRNA polycistron as a potential human
oncogene. Nature. 435:828–833. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang J, Lin H, Wang XY, Zhang DQ, Chen
JX, Zhuang Y and Zheng XL: Predictive value of microRNA-143 in
evaluating the prognosis of patients with hepatocellular carcinoma.
Cancer Biomark. 19:257–262. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Karatas OF, Oner M, Abay A and Diyapoglu
A: MicroRNAs in human tongue squamous cell carcinoma: From
pathogenesis to therapeutic implications. Oral Oncol. 67:124–130.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Teoh SL and Das S: The role of MicroRNAs
in diagnosis, prognosis, metastasis and resistant cases in breast
cancer. Curr Pharm Des. 23:1845–1859. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bucay N, Bhagirath D, Sekhon K, Yang T,
Fukuhara S, Majid S, Shahryari V, Tabatabai Z, Greene KL, Hashimoto
Y, et al: A novel microRNA regulator of prostate cancer
epithelial-mesenchymal transition. Cell Death Differ. 24:1263–1274.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cui L, Li Y, Lv X, Li J, Wang X, Lei Z and
Li X: Expression of MicroRNA-301a and its functional roles in
malignant melanoma. Cell Physiol Biochem. 40:230–244. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lee HW, Lee EH, Ha SY, Lee CH, Chang HK,
Chang S, Kwon KY, Hwang IS, Roh MS and Seo JW: Altered expression
of microRNA miR-21, miR-155, and let-7a and their roles in
pulmonary neuroendocrine tumors. Pathol Int. 62:583–591. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hwang HW and Mendell JT: MicroRNAs in cell
proliferation, cell death, and tumorigenesis. Br J Cancer.
94:776–780. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:pp. 2257–2261.
2006; View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tyagi N, Arora S, Deshmukh SK, Singh S,
Marimuthu S and Singh AP: Exploiting nanotechnology for the
development of MicroRNA-based cancer therapeutics. J Biomed
Nanotechnol. 12:28–42. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Barger JF and Nana-Sinkam SP: MicroRNA as
tools and therapeutics in lung cancer. Respir Med. 109:803–812.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xia K, Zhang Y, Cao S, Wu Y, Guo W, Yuan W
and Zhang S: miR-411 regulated ITCH expression and promoted cell
proliferation in human hepatocellular carcinoma cells. Biomed
Pharmacother. 70:158–163. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhao Z, Qin L and Li S: miR-411
contributes the cell proliferation of lung cancer by targeting
FOXO1. Tumour Biol. 37:5551–5560. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Guo L, Yuan J, Xie N, Wu H, Chen W, Song S
and Wang X: miRNA-411 acts as a potential tumor suppressor miRNA
via the downregulation of specificity protein 1 in breast cancer.
Mol Med Rep. 14:2975–2982. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang Y, Xu G, Liu G, Ye Y, Zhang C, Fan
C, Wang H, Cai H, Xiao R, Huang Z, et al: miR-411-5p inhibits
proliferation and metastasis of breast cancer cell via targeting
GRB2. Biochem Biophys Res Commun. 476:607–613. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang G, Yang X, Li C, Cao X, Luo X and Hu
J: PIK3R3 induces epithelial-to-mesenchymal transition and promotes
metastasis in colorectal cancer. Mol Cancer Ther. 13:1837–1847.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li B, Xie Z and Li B: miR-152 functions as
a tumor suppressor in colorectal cancer by targeting PIK3R3. Tumour
Biol. 37:10075–10084. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cao G, Dong W, Meng X, Liu H, Liao H and
Liu S: MiR-511 inhibits growth and metastasis of human
hepatocellular carcinoma cells by targeting PIK3R3. Tumour Biol.
36:4453–4459. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu K, Li X, Cao Y, Ge Y, Wang J and Shi
B: MiR-132 inhibits cell proliferation, invasion and migration of
hepatocellular carcinoma by targeting PIK3R3. Int J Oncol.
47:1585–1593. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hwang HW and Mendell JT: MicroRNAs in cell
proliferation, cell death, and tumorigenesis. Br J Cancer. 96
Suppl:R40–R44. 2007.PubMed/NCBI
|
|
35
|
Sun M, Huang F, Yu D, Zhang Y, Xu H, Zhang
L, Li L, Dong L, Guo L and Wang S: Autoregulatory loop between
TGF-β1/miR-411-5p/SPRY4 and MAPK pathway in rhabdomyosarcoma
modulates proliferation and differentiation. Cell Death Dis.
6:e18592015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yamamoto K, Seike M, Takeuchi S, Soeno C,
Miyanaga A, Noro R, Minegishi Y, Kubota K and Gemma A: MiR-379/411
cluster regulates IL-18 and contributes to drug resistance in
malignant pleural mesothelioma. Oncol Rep. 32:2365–2372. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Oliveto S, Mancino M, Manfrini N and Biffo
S: Role of microRNAs in translation regulation and cancer. World J
Biol Chem. 8:45–56. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhu Y, Zhao H, Rao M and Xu S:
MicroRNA-365 inhibits proliferation, migration and invasion of
glioma by targeting PIK3R3. Oncol Rep. 37:2185–2192. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang L, Huang J, Yang N, Greshock J,
Liang S, Hasegawa K, Giannakakis A, Poulos N, OBrien-Jenkins A,
Katsaros D, et al: Integrative genomic analysis of
phosphatidylinositol 3-kinase family identifies PIK3R3 as a
potential therapeutic target in epithelial ovarian cancer. Clin
Cancer Res. 13:5314–5321. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhou J, Chen GB, Tang YC, Sinha RA, Wu Y,
Yap CS, Wang G, Hu J, Xia X, Tan P, et al: Genetic and
bioinformatic analyses of the expression and function of PI3K
regulatory subunit PIK3R3 in an Asian patient gastric cancer
library. BMC Med Genomics. 5:342012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xu L, Wen Z, Zhou Y, Liu Z, Li Q, Fei G,
Luo J and Ren T: MicroRNA-7-regulated TLR9 signaling-enhanced
growth and metastatic potential of human lung cancer cells by
altering the phosphoinositide-3-kinase, regulatory subunit 3/Akt
pathway. Mol Biol Cell. 24:42–55. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Klahan S, Wu MS, Hsi E, Huang CC, Hou MF
and Chang WC: Computational analysis of mRNA expression profiles
identifies the ITG family and PIK3R3 as crucial genes for
regulating triple negative breast cancer cell migration. BioMed Res
Int. 2014:5365912014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Niemeyer BF, Parrish JK, Spoelstra NS,
Joyal T, Richer JK and Jedlicka P: Variable expression of PIK3R3
and PTEN in Ewing Sarcoma impacts oncogenic phenotypes. PLoS One.
10:e01168952015. View Article : Google Scholar : PubMed/NCBI
|