Introduction
Breast cancer is one of the leading causes of
cancer-associated mortality among females worldwide, which affects
about one in eight females in developed countries (1,2). Increased
incidence rates and mortality rates have been reported in most
developed and developing countries, respectively (3). In China, the health burden of cancer is
increasing. Every year, more than 1.6 million individuals are
diagnosed with breast cancer and 1.2 million patients succumb to
breast cancer. In numerous countries, including China, breast
cancer is now the most common type of cancer diagnosed in females
(4).
Numerous studies have confirmed that different
molecular subtypes of breast cancer differ in their
clinicopathological characteristics, differ in regards to the
benefit from various chemotherapy regimens and differ in regards to
patient prognosis (5–7). Breast cancer has been classified into
distinct subtypes, including luminal, human epidermal growth factor
receptor-2 (Her-2)-enriched and basal-like (8,9). In 2003,
Sorlie et al (10,11) divided the luminal type into two
subtypes of luminal A and luminal B.
Luminal subtype A is defined as estrogen receptor
(ER)- or progesterone receptor (PR)-positive, Her-2-negative and
Ki67 low expression as determined by immunohistochemistry. This
type is the most common subtype of breast cancer and is associated
with the best prognosis (12).
Luminal subtype B is a subtype of breast cancer with ER- or
PR-positive status, either positive or negative regarding Her-2 and
high expression of Ki67. Luminal subtype B is most commonly
identified in elderly patients with breast cancer, where ductal
carcinoma is common (13).
Her-2-positive breast cancer is Her-2-positive upon
immunohistochemistry (3+) or fluorescence in situ
hybridization, negative for ER and PR, and has high expression of
Ki67. The molecular characteristics of Her-2-positive breast cancer
are obvious amplification of the erb-b2 receptor tyrosine kinase 2
(ERBB2) gene, and upregulation of associated genes,
including growth factor receptor bound protein 7 (GRB7) and
TRAP100 (14,15). Basal-like is a molecular subtype that
has been studied more frequently. Its characteristics are lack of
ER and Her-2 expression and high expression of the biomarkers of
the basal epithelium, which is derived from the outer layer of the
ductal epithelium (16,17). Basal-like breast cancer and
triple-negative breast cancer (TNBC) are basically the same
subtypes. Basal-like is a subtype of breast cancer identified
according to the gene expression profile, while TNBC is identified
by immunohistochemistry and exhibits certain differences in its
characteristics (18,19).
TNBC refers to breast cancer with negative ER, PR
and proto-oncogene Her-2 upon immunohistochemical examination of
cancer tissues. This type of breast cancer accounts for 12-17% of
all pathological types of breast cancer, with a distinct biological
behavior and clinicopathological characteristics, and a worse
prognosis than other types (20).
According to research, the local recurrence of TNBC is not
significantly different from that of non-TNBC, but the incidence of
distant metastasis is higher. The incidence rates of lung
metastasis and liver metastasis were reported to be high, but there
was no difference regarding bone and brain metastasis (21–23). It is
important to identify biomarkers to predict the sensitivity to
chemotherapy and prognosis of TNBC patients.
Exosomes are described as vesicles of 40–100 nm in
diameter, which are secreted by certain cells (24). Exosomes may be secreted by tumor cells
during tumor progression and metastasis. They have been reported to
have vital roles in the progression and metastasis of the tumor by
communication via molecules including lipids, proteins, mRNAs and
microRNAs (miRNAs/miRs) (25–27). Exosomes may hold promise as potential
biomarkers, and separation and detection of circulating
tumor-associated exosomes may be used for the diagnosis of cancer
patients (28). Blocking of exocrine
secretions may inhibit the occurrence of tumors (29).
miRNAs are a type of non-coding, conserved and
single-stranded RNAs consisting of 18–24 nucleotides. Mature
single-stranded miRNA fragments are integrated with the argonaut
protein (Ago2) and target mRNA to form the RNA-mediated silencing
complex to regulate post-transcriptional gene expression by either
inhibiting the translation process or promoting the degradation
process of the target mRNA (30,31).
miRNAs regulate the stability of target mRNAs or inhibit
translation to control cell proliferation, differentiation,
invasion, migration and apoptosis, and have a key role in
tumorigenesis and progression (32).
In the present study, RNA sequencing was applied to detect
tumor-specific miRNAs in exosomes that may be associated with the
onset and progression of breast cancer.
Materials and methods
Subjects
All of the subjects included in the present study
presented themselves to the Cancer Center of Meizhou People's
Hospital (Meizhou, China) for medical examination between October
2015 and January 2016. The patients were aged between 30 and 69
years and their mean age was 49.83±9.763 years. The study was
performed on the basis of the Declaration of Helsinki, and was
supported by the Ethics Committee of Meizhou People's Hospital.
Sample collection and total RNA
extraction
Two peripheral blood samples were taken from each
subject at the same time; 4 ml was used for detection of tumor
markers, while 5.0 ml of peripheral venous blood was collected in
tubes with EDTA anti-coagulant for detection of miRNAs from
patients with breast cancer and healthy controls. Plasma was
separated and transferred to 1.5-ml RNA centrifuge tubes
(RNase-free) and then packaged and stored at −80°C for exosome
isolation and RNA extraction. Prior to sampling, the subjects did
not receive any pre-operative radiotherapy and/or chemotherapy and
were diagnosed with breast cancer by pathology.
Exosome isolation was performed using Exosome
Isolation Reagent (RiboBio Co., Ltd.) according to the
manufacturer's protocol. Total RNA was extracted from the exosomes
using TRIzol reagent (Invitrogen, Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. The quantity and purity
of total RNA were evaluated with a Nanodrop 2000 (Thermo Fisher
Scientific, Inc.), and the RNA Nano 6000 Assay Kit of the Agilent
Bioanalyzer 2100 system (Agilent Technologies) was used to analyze
RNA integrity.
Library construction and high
throughput sequencing
Library construction of small RNA was performed with
the Illumina TruSeq small RNA sample pre Kit (Illumina, Inc.), with
which joints were directly added to the specific structure of the
3′ and 5′end of the small RNA (5′end phosphate group, 3′end
hydroxyl group). Subsequently, the RNA was reverse-transcribed to
complementary (c)DNA. Following PCR amplification, the target DNA
fragments were separated by PAGE, and the cDNA libraries were
recovered and purified. PAGE was used to separate the target DNA
fragments after PCR and recover and purify the cDNA library.
After the libraries were constructed, they were
preliminarily quantified by Qubit2.0 and the libraries were diluted
to 1 ng/µl. An Agilent 2100 (Agilent Technologies, Inc.) was used
to detect the library size. The insert size was in line with the
expectations, and accurate quantification of the effective
concentration of the libraries (libraries with an effective
concentration of >2 nM) was performed using quantitative (q)PCR
in order to ensure the quality of the libraries. After the
qualification of the libraries, the different libraries were
sequenced on the Illumina HiSeq 2500 platform (Illumina, Inc.)
according to the protocols of RiboBio Co., Ltd.
Identification of differentially
expressed miRNAs
The adapter sequences and low-quality sequences were
removed prior to data analysis. The remaining reads were referred
to as ‘clean reads’ and acquired for the transcriptome assembly and
quantification. Next, the index of the reference genome was built
using Bowtie v2.0.6 (http://bowtie-bio.sourceforge.net) and paired-end
clean reads were mapped to the reference genome using TopHat v2.0.9
(http://ccb.jhu.edu/software/tophat/index.shtml). The
transcriptome assemblies were then generated using Cufflinks v2.1.1
(https://cole-trapnell-lab.github.io/cufflinks) with
the default parameters. The miRNA sequencing reads of each sample
were normalized to fragments per kilobase of transcript per million
mapped reads values using Cuffdiff v2.1.1 (http://cole-trapnell-lab.github.io/cufflinks).
The P-value and fold change were calculated for each gene.
P<0.05 and |log2(foldchange)|>1 were set as the
threshold for significantly differential expression.
Gene Ontology (GO) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) enrichment analysis
GO analysis was performed to elucidate the potential
biological process terms of the differentially expressed genes in
GO annotations. Furthermore, pathway analysis was utilized to
screen out the significant pathways of the differentially expressed
genes according to the KEGG database. Fisher's exact test was
applied to evaluate whether the GO terms or the KEGG pathways were
enriched by the differentially expressed genes, and P<0.05 was
set as the threshold of significant difference.
Reverse transcription (RT)-qPCR
Differentially expressed miRNAs were randomly
selected and their expression levels were detected by RT-qPCR. To
validate the sequencing results regarding differentially expressed
miRNAs, six upregulated miRNAs [Homo sapiens
(hsa)-miR-148a-5p, hsa-miR-200a-5p, hsa-miR-210a-3p,
hsa-miR-378a-3p, hsa-miR-483-5p and hsa-miR-7110-5p] and two
downregulated miRNAs (hsa-miR-92b-3p and hsa-miR-150-5p) were
selected. The qPCR forward primer sequences were as follows:
5′-AAGTTCTGAGACACTCC-3′, 5′-CATCTTACCGGACAGT-3′,
5′-TGTGACAGCGGCTAA-3′, 5′-CTGGACTTGGAGTCAGAAG-3′,
5′-AGACGGGAGGAAAGAA-3′, 5′-TGTGGGGAGAGAGAG-3′,
5′-TATTGCACTCGTCCCG-3′ and 5′-CAACCCTTGTACCAGTG-3′, respectively.
Total RNA from exosomes was extracted using an RNeasy Kit (TianGene
Co., Ltd.) and reverse-transcribed to cDNA using the following
conditions: 42°C for 1 h, 70°C for 10 min and a hold at 4°C. The
cDNA was amplified using miRNA assay primers and the SYBR Green Mix
(RiboBio Co., Ltd.) on the Roche Lightcycler 480 (Roche Molecular
Systems, Inc.). In addition, a common reverse primer was used in
the present study, with the sequence: 5′-CAGTGCGTGTCGTGGAGT-3′.
Each reaction was performed under the following conditions:
initialization for 10 min at 95°C, followed by 40 cycles of
amplification, with 2 sec at 95°C for denaturation, 20 sec at 60°C
for annealing, and 10 sec at 70°C for elongation. The expression of
exosomal miRNAs was quantified using the 2−ΔΔCq
(33) method and all miRNA expression
values were normalized against the U6 internal control.
Comparison of exosomal miRNA profiles
between patients with and without recurrence of breast cancer
The 27 subjects were followed up for 2 years. The
differential expression of miRNAs in patients with and without
recurrence of breast cancer was analyzed. Blood samples were
collected from all of these patients at the time-point of
diagnosis. Based on the follow-up results, the differential
expression of miRNAs at the initial diagnosis was analyzed, aiming
to identify miRNAs at the time-point of initial diagnosis that are
able to predict the presence of metastasis.
Statistical analysis
SPSS statistical software version 19.0 (IBM Corp.)
was used for data analysis. Values are expressed as the mean ±
standard deviation. Chi-square was used to compare the frequencies.
Multigroup comparisons of the means were carried out by one-way
analysis of variance (ANOVA) test with post hoc contrasts test were
used to determine the statistical significance of differences
between groups. P<0.05 was considered to indicate statistical
significance. Receiver operating characteristic (ROC) curve
analysis was performed and the area under the ROC curve (AUC) was
calculated to determine the diagnostic value of certain
differentially expressed miRNAs randomly selected for
differentiating between breast cancer patients and healthy
controls.
Results
Clinical characteristics
A total of 27 breast cancer patients and 3 healthy
controls participated in the present study. The clinical
characteristics of the 30 subjects of this study, including age,
clinical stage, immunohistochemistry, tumor markers and blood lipid
levels, are presented in Table
SI.
Statistics of the sequencing data
A total of 395,775,015 raw reads were obtained, with
an average of 13,192,500 raw reads for each sample. After data
filtering and normalization, 372,788,760 clean reads were obtained,
with an average 12,426,292 clean reads for each sample. The Q30
mean value was 94.19%, with a range of 91.13–97.83% for each
sample. This indicated that the quality of these 30 libraries was
suitable for the subsequent analysis. Detailed information on the
quality of the sequencing data is presented in Table SII.
Differentially expressed miRNAs in
exosomes
miRNAs were analyzed with strict quality control of
the data, and a total of 1,835 miRNAs were identified. In order to
systematically study the level of miRNA expression associated with
breast cancer, 27 patients with breast cancer and 3 healthy
controls were analyzed in the present study. Hierarchical
clustering of the miRNAs in breast cancer patients and healthy
controls is provided in Fig. 1.
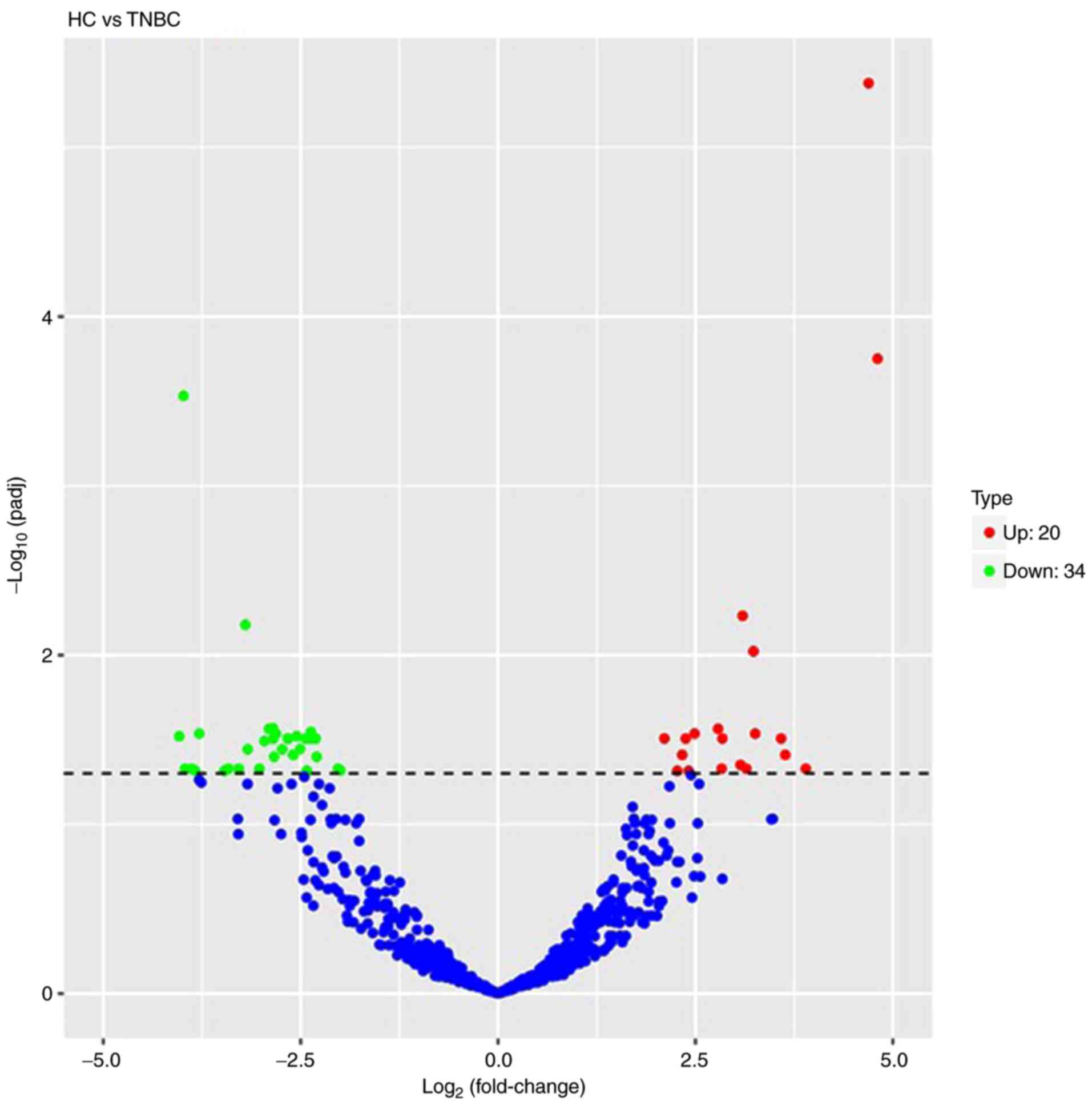
Volcano plots were used to analyze the overall
distribution of differentially expressed miRNAs. The fold change
and corrected significance level were used to screen differentially
expressed miRNAs. No significant differences in miRNAs were
identified among the basal-like, HER-2+, luminal A,
luminal B and HC groups. Compared to the miRNA expression profiles
of the healthy controls, a total of 54 differentially expressed
miRNAs were discriminated in TNBC patients, including 20
upregulated miRNAs and 34 downregulated miRNAs (P<0.05). A
volcano plot is displayed in Fig. 2
and a list of miRNAs identified to be differentially expressed
between TNBC patients and HCs is provided in Table SIII. The blue and red in the figure
indicate that reduced and increased expression, respectively. The
frequency of base usage among miRNA sequences is indicated in
Fig. 3.
GO and KEGG pathway analyses
To study the functions and associated pathways of
the miRNAs identified in the present study, GO and pathway analyses
were performed. The hierarchical classification of genes in the
categories cellular component, molecular function of biological
process revealed gene regulatory networks.
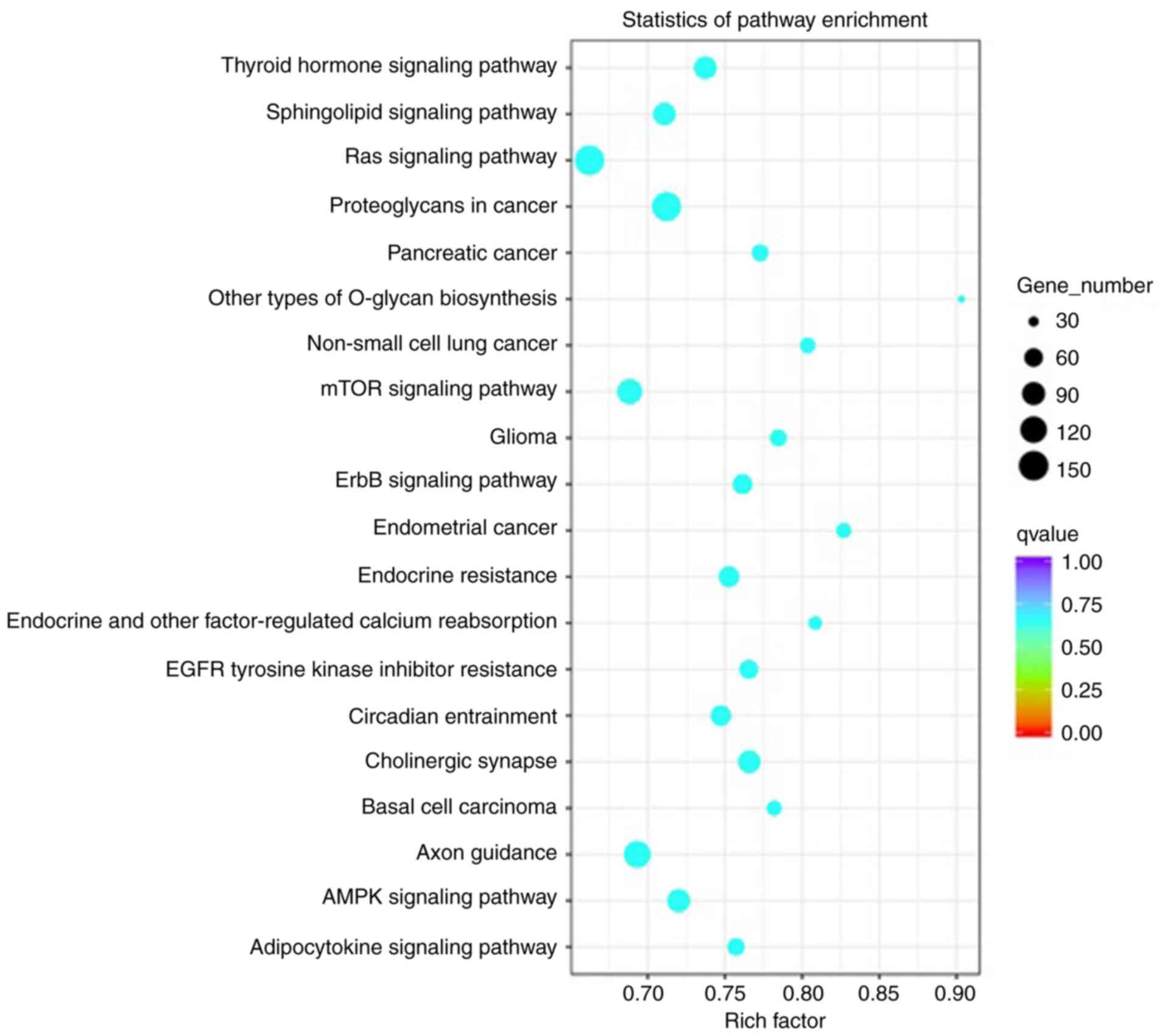
TNBC-associated miRNAs were selected and the
biological process pathways enriched by these miRNAs were studied.
Analysis indicated statistically significant accumulation of
pathways linked to proliferation, signal transduction and
migration, and the associated functions are presented in Fig. 4. These pathways included the ‘Ras
signaling pathway’, ‘mTOR signaling pathway’, ‘Axon guidance’,
‘proteoglycans in cancer’, ‘sphingolipid signaling pathway’,
‘adenosine monophosphate kinase (AMPK) signaling pathway’ and
‘thyroid hormone signaling pathway’.
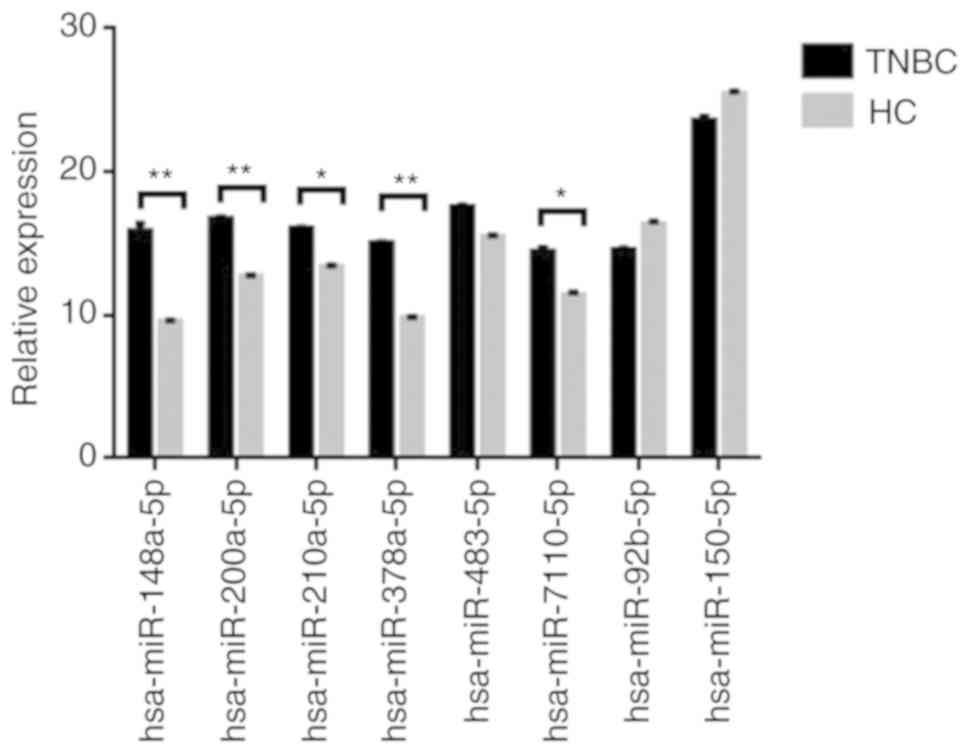
RT-qPCR validation of miRNA
expression
To validate the sequencing data for certain miRNAs,
the expression levels of six upregulated miRNAs (hsa-miR-148a-5p,
hsa-miR-200a-5p, hsa-miR-210a-3p, hsa-miR-378a-3p, hsa-miR-483-5p
and hsa-miR-7110-5p) and two downregulated microRNAs
(hsa-miR-92b-3p and hsa-miR-150-5p) in the exosome of TNBC patients
(n=20) and healthy controls (n=20) were verified by RT-qPCR.
Details concerning the patient collective used for verification of
the data are provided in Table SI.
U6 small nuclear RNA was used as the internal control. As presented
in Fig. 5, there were significant
differences in the expression of eight miRNAs between TNBC patients
and healthy controls. The results of RT-qPCR were consistent with
those of the RNA sequencing.
Comparison of exosomal miRNA profiles
between patients with and without recurrence of breast cancer and
ROC analysis
Certain miRNAs were differentially expressed in
exosomes of patients with vs. without recurrence. Hierarchical
clustering of differentially expressed miRNAs between patients with
and without recurrence in breast cancer is provided in Fig. 6. hsa-miR-150-5p, hsa-miR-576-3p and
hsa-miR-4665-5p were considered as potential biomarkers in the
patients with recurrence.
Certain differentially expressed miRNAs in the
exosome were randomly selected for evaluating their ability to
distinguish patients with recurrence of breast cancer from those
without recurrence, and their diagnostic value was determined by
ROC curve analysis. miRNAs with a P<0.05 and an AUC>0.6 were
selected as markers with the ability to distinguishing between
patients with and without recurrence in breast cancer. More
specifically, the ROC curves yielded the following AUCs: miR-150-5p
(AUC=0.705, upregulated), miR-576-3p (AUC=0.691, upregulated),
miR-4665-5p (AUC=0.681, upregulated). These miRNAs were identified
to be able to discriminate breast cancer patients with recurrence
from without recurrence. The ROC curves are provided in Fig. 7.
Discussion
The development of human cancers is a multi-step
process in which normal cells acquire traits that eventually lead
them to become cancer cells. The interaction between tumor cells
and the surrounding microenvironment is key in overcoming this
obstacle and promoting tumor progression and metastasis. Exosomes
are membrane-derived vesicles that are regarded as important
mediators for intercellular communication in recent years. They
carry lipids, proteins, mRNAs and miRNAs and may be transferred to
recipient cells by fusion of exosomes and target cell membranes. In
the context of cancer cells, this process requires the transfer of
pro-cancer cell content to surrounding cells within the tumor
microenvironment or into the circulation to function at a distance
to allow the cancer to progress. In this process, exosomal small
RNAs are transferred to recipient cells, where they regulate the
expression of target genes.
miRNAs are endogenous, non-coding RNAs that regulate
gene expression mainly at the post-transcriptional level, and their
expression depends on the physiological and pathological state of
the body. To date, characteristic miRNA expression patterns have
been identified in malignant tumors, which are different from those
in normal tissues. Detection of these miRNAs may be an important
means of early diagnosis, targeted therapy and prognostication of
patients with malignant tumors. However, miRNAs are intracellular
molecules that reside inside the cells, and it is difficult to
assess the expression levels of miRNAs from the cells in deep
tissues. miRNAs in exosomes are protected from exposure to
nucleases and shear stress, and these characteristics make exosomal
miRNAs potential markers for disease detection. It is well known
that exosome-encapsulated miRNAs serve as ideal biomarkers for
disease detection at an early stage.
In breast cancer patients, certain miRNAs may be
abnormally expressed, thereby promoting the proliferation, survival
or metastasis of tumor cells. For instance, miR-2l, miR-10b, and
miR-155 are frequently overexpressed in breast cancer cells
(34), and miR-30b, miR-126, miR-17P,
miR-335 and miR-205 are frequently downregulated in breast cancer
cells (35,36). miRNAs that are abnormally expressed in
breast cancer have multiple roles, but they may be divided into
cancer-promoting or tumor-suppressor miRNAs. They exert their
cancer-promoting or tumor-suppressing effects by affecting the
corresponding signaling pathways and transcription factors. For
instance, miR-21 exerts its promoting effect on the anti-apoptotic
protein Bcl-2, as well as programmed cell death 4 and p53 (37), and its concentration in serum and
breast cancer tissues is positively associated with metastasis and
the poor prognosis of breast cancer patients. By contrast, miR-155,
let-7e, miR-10a, miR-144 and miR-193 exert anti-apoptotic effects
by acting on caspase-3 (38).
Overexpression of miR-155 may also activate the JAK/STAT3 pathway
and promote tumorigenesis and development. miR-10b promotes
downregulation of E-cadherin expression (39). Others, including miR-27a, miR-96 and
miR-182, may have a cancer-promoting role by inhibiting forkhead
box O1 (FOXO1) (40).
Previous studies on the association between miRNAs
and breast cancer were summarized and compared with the present
results. By targeting GLUT1, miR-22 may inhibit the proliferation
and invasion of breast cancer cells, and the expression of miR-22
and GLUT1 in breast cancer tissue samples exhibits a negative
correlation (41). NF-E2-related
factor 2 (Nrf2) is an important transcription factor that activates
the expression of cellular detoxifying enzymes. miR-200a regulates
the Keap1/Nrf2 pathway in mammary epithelium and re-activates the
Nrf2-dependent anti-oxidant pathway in breast cancer (42,43).
Estradiol-G-protein-coupled estrogen receptor 1 induces the level
of HOTAIR through the suppression of miR-148a. miR-148a levels were
reported to be negatively correlated with HOTAIR levels in breast
cancer patients (44–46). Studies have reported an association
between miR-210 and neo-adjuvant chemotherapy in HER2-positive
breast cancer (47,48). Furthermore, the expression level of
miR-378 was found to be highly associated with breast cancer.
Overexpression of miR-378 in formalin-fixed and paraffin-embedded
tissues of breast cancer patients may serve as an ideal diagnostic
biomarker for breast cancer (49).
miR-378a-3p was reported to modulate the sensitivity of breast
cancer MCF-7 cells to tamoxifen through targeting Golgi transport
1A (50). Various studies suggest
that treatment with miR-296 is effective in ER-negative breast
cancers. A combination of let-7 with miR-296 may be an efficient
molecular medicine for any type of breast cancer (51). The miR-320a/metadherin pathway is a
putative therapeutic target in breast cancer and dysregulation of
miR320a may be involved in invasive breast cancer progression
(52,53). Another study suggested that modulating
the levels of miR-34a, a tumor-suppressor miRNA, may be a novel and
useful approach for breast cancer therapy (54). The present study also indicated
differential expression of the above mentioned miRNAs in breast
cancer.
Among the miRNAs identified to be differentially
expressed in the present study, certain miRNAs have been previously
reported in breast cancer. Chabre et al (55) indicated that circulating miR-483-5p is
a promising non-invasive biomarker for the clinical outcome of
adrenocortical carcinoma. miR-486-5p was also reported to be
downregulated in patients with lymph node metastases (56), lung adenocarcinoma (57) and breast cancer (58). miR-490-3p is downregulated in
triple-negative breast cancer (TNBC) and has a suppressive role in
cancer cell proliferation, invasion and tumorigenesis (59), and the present results were consistent
with this finding. Lower expression levels of miR-378a-3p were
reported to be associated with poor prognosis for tamoxifen-treated
patients with breast cancer (50).
Decreased expression of miR-122-5p was reported in patients with
all stages and grades of breast cancer (60). Several studies found that miR-210
expression is associated with breast cancer patient survival
(61). miR-320a was found to inhibit
the proliferation, invasion (62) and
metastasis (53) of breast cancer.
miR-34a-5p was suggested as a potential biomarker of
chemotherapy-associated cardiac dysfunction in breast cancer
patients (63). Circulating miRNAs
are potential biomarkers for distinguishing inflammatory breast
cancer (IBC) from non-IBC, and may also be a candidate for early
detection of breast cancer (64).
High levels of miR-374b-5p were found to be associated with a
favorable outcome for TNBC (65). The
expression levels of miR-155-5p were reported to be upregulated in
TNBC (66–68). The present results were consistent
with these studies. The clinical outcome for patients with breast
cancer presenting with positive miR-361-5p expression was
significantly better than that of patients with negative miR-361-5p
expression (69). Another study found
that miR-361-5p is downregulated in TNBC tissues and that
overexpression of miR-361-5p inhibits the proliferation, migration
and invasion of TNBC cells (70). A
further study indicated that miR-429 is upregulated in TNBC
(71). miR-92a-3p may be part of
target pathways for selective intervention to suppress
hormone-regulated metastasis in breast cancer (72).
In the present study, TNBC-specific miRNAs were
identified and subjected to GO functional enrichment analysis in
the category biological process and KEGG pathway analysis,
revealing that these miRNAs were significantly accumulated in
pathways associated with proliferation, signal transduction and
migration. Certain pathways were found to be associated with
cancer, mainly including the ‘Ras signaling pathway’ (73,74), ‘mTOR
signaling pathway’ (75,76), ‘axon guidance’ (77), ‘proteoglycans in cancer’ (78,79),
‘sphingolipid signaling pathway’ (80), ‘AMPK signaling pathway’ (81,82),
‘thyroid hormone signaling pathway’ (83), ‘ErbB signaling pathway’ (84) and ‘epidermal growth factor receptor
tyrosine kinase inhibitor resistance’ (85,86).
ROC curve analysis was performed to assess the
sensitivity and specificity of miRNAs as diagnostic biomarkers. The
exosome levels of hsa-miR-150-5p, hsa-miR-576-3p and
hsa-miR-4665-5p were higher in breast cancer with recurrence
compared to those in patients without recurrence. Therefore, the
results indicated that these exosomal miRNAs may serve as novel
biomarkers with a predictive value for breast cancer recurrence. It
is important to explore the association between miRNAs and the
diagnosis, treatment and prognosis of breast cancer. However,
current understanding of the regulatory mechanisms of miRNAs is
still superficial and their interaction with other regulatory
mechanisms requires further analysis. The present study may provide
insight into the important roles of certain miRNAs in the
occurrence and development of breast cancer.
As a diagnostic marker, exosomes are stable,
capturable, bioactive and real-time. However, there are currently
limitations regarding the application of exosomes as clinical
markers, including specificity, source clarity, enrichment
efficiency, detection methods and convenience of extraction. In
addition to miRNAs, exosomal mRNA and protein will become a novel
research ‘hotspot’. Regarding precise applications as treatment
targets and biomarkers, the mechanisms of exosomes in complex
tumor-associated processes require further elucidation.
In conclusion, in the present study, miRNA
expression profiles between TNBC and healthy controls were produced
and analyzed by bioinformatics, revealing that certain miRNAs may
have important roles in various biological and pathological
processes of TNBC. These results may provide useful biological
information for the early diagnosis of TNBC patients. In addition,
certain exosomal miRNAs may serve as novel biomarkers, as they were
determined to be of predictive value for the recurrence of breast
cancer compared with non-recurrence. Yet, further studies are
required to reveal the functional significance of abnormally
expressed miRNAs in breast cancer, which is one of our next major
research goals.
Supplementary Material
Supporting Data
Acknowledgements
The author would like to thank their other
colleagues of the Center for Precision Medicine, Meizhou People's
Hospital who were not listed in the authorship for their helpful
comments on the manuscript.
Funding
This study was supported by Medical and Health
Research Project of Meizhou City, Guangdong Province, China (grant
no. 2016-B-33 to QW) and the Science and Technology Planning
Program of Meizhou City, Guangdong Province, China (grant no.
2015B037 to HZ).
Availability of data and materials
The data used to support the findings of this study
are available from the corresponding author upon request.
Authors' contributions
HW and QW conceived and designed the experiments.
HZ, LL, QZ, QH and ZY recruited subjects and collected and analyzed
the clinical data. HW and ZY conducted the laboratory testing. ZY
and HZ helped to analyze the data. HW and QW prepared the
manuscript. HW reviewed the manuscript. All authors read and
approved the manuscript and agree to be accountable for all aspects
of the research in ensuring that the accuracy or integrity of any
part of the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
This study was conducted on the basis of the
Declaration of Helsinki, and was supported by the Ethics Committee
of the Meizhou People's Hospital (Meizhou, Guangdong, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
TNBC
|
triple-negative breast cancer
|
|
RT-qPCR
|
reverse transcription quantitative
PCR
|
|
GO
|
Gene Ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
FC
|
fold change
|
|
ANOVA
|
one-way analysis of variance
|
|
ROC
|
receiver operating characteristic
|
References
|
1
|
Siegel RL, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
DeSantis C, Ma J, Bryan L and Jemal A:
Breast cancer statistics, 2013. CA Cancer J Clin. 64:52–62. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Unger-Saldaña K: Challenges to the early
diagnosis and treatment of breast cancer in developing countries.
World J Clin Onco. 5:465–477. 2014. View Article : Google Scholar
|
|
4
|
Fan L, Strasser-Weippl K, Li JJ, St Louis
J, Finkelstein DM, Yu KD, Chen WQ, Shao ZM and Goss PE: Breast
cancer in China. Lancet Oncol. 15:e279–e289. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wiechmann L, Sampson M, Stempel M, Jacks
LM, Patil SM, King T and Morrow M: Presenting features of breast
cancer differ by molecular subtype. Ann Surg Oncol. 16:2705–2710.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Parise CA, Bauer KR, Brown MM and Caggiano
V: Breast cancer subtypes as defined by the estrogen receptor (ER),
progesterone receptor (PR), and the human epidermal growth factor
receptor 2 (HER2) among women with invasive breast cancer in
California, 2004. Breast J. 15:593–602. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hugh J, Hanson J, Cheang MC, Nielsen TO,
Perou CM, Dumontet C, Reed J, Krajewska M, Treilleux I, Rupin M, et
al: Breast cancer subtypes and response to docetaxel in
node-positive breast cancer: Use of an immunohistochemical
definition in the BCIRG 001 trial. J Clin Oncol. 27:1168–1178.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Park S, Koo JS, Min SK, Park HS, Lee JS,
Lee JS, Kim SI and Park BW: Characteristics and outcomes according
to molecular subtypes of breast cancer as classified by a panel of
four biomarkers using immunohistochemistry. Breast. 21:50–57. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Meyers MO, Klauber-Demore N, Ollila DW,
Amos KD, Moore DT, Drobish AA, Burrows EM, Dees EC and Carey LA:
Impact of breast cancer molecular subtypes on locoregional
recurrence in patients treated with neoadjuvant chemotherapy for
locally advanced breast cancer. Ann Surg Oncol. 18:2851–2857. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sørlie T, Perou CM, Tibshirani R, Aas T,
Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey
SS, et al: Gene expression patterns of breast carcinomas
distinguish tumor subclasses with clinical implications. Proc Natl
Acad Sci USA. 98:10869–10874. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sorlie T, Tibshirani R, Parker J, Hastie
T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, et
al: Repeated observation of breast tumor subtypes in independent
gene expression data sets. Proc Natl Acad Sci USA. 100:8418–8423.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Badve S, Turbin D, Thorat MA, Morimiya A,
Nielsen TO, Perou CM, Dunn S, Huntsman DG and Nakshatri H: FOXA1
expression in breast cancer-correlation with luminal subtype A and
survival. Clin Cancer Res. 13:4415–4421. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tang LC, Jin X, Yang HY, He M, Chang H,
Shao ZM and Di GH: Luminal B subtype: A key factor for the worse
prognosis of young breast cancer patients in China. BMC Cancer.
15:2012015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Böcker W, Moll R, Poremba C, Holland R,
Van Diest PJ, Dervan P, Bürger H, Wai D, Ina Diallo R, Brandt B, et
al: Common adult stem cells in the human breast give rise to
glandular and myoepithelial cell lineages: A new cell biological
concept. Lab Invest. 82:737–746. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Birnbaum D, Bertucci F, Ginestier C,
Tagett R, Jacquemier J and Charafe-Jauffret E: Basal and luminal
breast cancers: Basic or luminous. Int J Oncol. 25:249–258.
2004.PubMed/NCBI
|
|
16
|
Turner NC, Reisfilho JS, Russell AM,
Springall RJ, Ryder K, Steele D, Savage K, Gillett CE, Schmitt FC,
Ashworth A and Tutt AN: BRCA1 dysfunction in sporadic basal-like
breast cancer. Oncogene. 26:2126–2132. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Millikan RC, Newman B, Tse CK, Moorman PG,
Conway K, Dressler LG, Smith LV, Labbok MH, Geradts J, Bensen JT,
et al: Epidemiology of basal-like breast cancer. Breast Cancer Res
Treat. 109:123–139. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Thike AA, Cheok PY, Jaralazaro AR, Tan B,
Tan P and Tan PH: Triple-negative breast cancer:
Clinicopathological characteristics and relationship with
basal-like breast cancer. Mod Pathol. 23:123–133. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rakha E, Ellis I and Reis-Filho J: Are
triple-negative and basal-like breast cancer synonymous? Clin
Cancer Res. 14:618–619. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chacón RD and Costanzo MV: Triple-negative
breast cancer. Breast Cancer Res. 12 (Suppl 2):S32010. View Article : Google Scholar
|
|
21
|
Podo F, Buydens LM, Degani H, Hilhorst R,
Klipp E, Gribbestad IS, Van Huffel S, van Laarhoven HW, Luts J,
Monleon D, et al: Triple-negative breast cancer: Present challenges
and new perspectives. Mol Oncol. 4:209–229. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Oakman C, Moretti E, Galardi F, Biagioni
C, Santarpia L, Biganzoli L and Di Leo A: Adjuvant systemic
treatment for individual patients with triple negative breast
cancer. Breast. 20 (Suppl 3):S135–S141. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
André F and Zielinski CC: Optimal
strategies for the treatment of metastatic triple-negative breast
cancer with currently approved agents. Ann Oncol. 23 (Suppl
6):vi46–vi51. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chevillet JR, Kang Q, Ruf IK, Briggs HA,
Vojtech LN, Hughes SM, Cheng HH, Arroyo JD, Meredith EK,
Gallichotte EN, et al: Quantitative and stoichiometric analysis of
the microRNA content of exosomes. Proc Natl Acad Sci USA.
111:148882014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wolfers J, Lozier A, Raposo G, Regnault A,
Théry C, Masurier C, Flament C, Pouzieux S, Faure F, Tursz T, et
al: Tumor-derived exosomes are a source of shared tumor rejection
antigens for CTL cross-priming. Nat Med. 7:297–303. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Clayton A, Mitchell JP, Court J, Mason MD
and Tabi Z: Human tumor-derived exosomes selectively impair
lymphocyte responses to interleukin-2. Cancer Res. 67:7458–7466.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Clayton A, Mitchell JP, Court J, Linnane
S, Mason MD and Tabi Z: Human tumor-derived exosomes down-modulate
NKG2D expression. J Immunol. 180:72492008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hannafon BN and Ding WQ: Intercellular
communication by exosome-derived microRNAs in cancer. Int J Mol
Sci. 14:14240–14269. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chaput N, Taïeb J, Schartz NE, André F,
Angevin E and Zitvogel L: Exosome-based immunotherapy. Cancer
Immunol Immun. 53:234–239. 2004. View Article : Google Scholar
|
|
30
|
Chendrimada TP, Gregory RI, Kumaraswamy E,
Norman J, Cooch N, Nishikura K and Shiekhattar R: TRBP recruits the
Dicer complex to Ago2 for microRNA processing and gene silencing.
Nature. 436:740–744. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hagiwara K, Kosaka N, Yoshioka Y,
Takahashi RU, Takeshita F and Ochiya T: Stilbene derivatives
promote Ago2-dependent tumour-suppressive microRNA activity. Sci
Rep. 2:3142012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang J, Jiang C, Shi X, Yu H, Lin H and
Peng Y: Diagnostic value of circulating miR-155, miR-21, and
miR-10b as promising biomarkers in human breast cancer. Int J Clin
Exp Med. 9:10258–10265. 2016.
|
|
35
|
Shan HC and Toyokuni S: Malignant
mesothelioma as an oxidative stress-induced cancer: An update. Free
Radical Bio Med. 86:166–178. 2015. View Article : Google Scholar
|
|
36
|
Wang W and Luo YP: MicroRNAs in breast
cancer: Oncogene and tumor suppressors with clinical potential. J
Zhejiang Univer B. 16:18–31. 2015. View Article : Google Scholar
|
|
37
|
Li X, Xin S, He Z, Che X, Wang J, Xiao X,
Chen J and Song X: MicroRNA-21 (miR-21) post-transcriptionally
downregulates tumor suppressor PDCD4 and promotes cell
transformation, proliferation, and metastasis in renal cell
carcinoma. Cell Physiol Biochem. 33:1631–1642. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Corcoran C, Friel AM, Duffy MJ, Crown J
and O'Driscoll L: Intracellular and extracellular microRNAs in
breast cancer. Clin Chem. 57:18–32. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yong L, Jing Z, Zhang PY, Zhang Y, Sun SY,
Yu SY and Xi QS: MicroRNA-10b targets E-cadherin and modulates
breast cancer metastasis. Med Sci Monit. 18:BR299–BR308.
2012.PubMed/NCBI
|
|
40
|
Guttilla IK and White BA: Coordinate
regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast
cancer cells. J Biol Chem. 284:23204–23216. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen B, Tang H, Liu X, Liu P, Yang L, Xie
X, Ye F, Song C, Xie X and Wei W: miR-22 as a prognostic factor
targets glucose transporter protein type 1 in breast cancer. Cancer
Lett. 356:410–417. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Eades G, Yang M, Yao Y, Zhang Y and Zhou
Q: miR-200a regulates Nrf2 activation by targeting Keap1 mRNA in
breast cancer cells. J Biol Chem. 286:40725–40733. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liu M, Hu C, Xu Q, Chen L, Ma K, Xu N and
Zhu H: Methylseleninic acid activates Keap1/Nrf2 pathway via
up-regulating miR-200a in human oesophageal squamous cell carcinoma
cells. Bioscience Rep. 35:e002562015. View Article : Google Scholar
|
|
44
|
Tao S, He H, Chen Q and Yue W: GPER
mediated estradiol reduces miR-148a to promote HLA-G expression in
breast cancer. Biochem Biophys Res Commun. 451:74–78. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Tao S, He H and Chen Q: Estradiol induces
HOTAIR levels via GPER-mediated miR-148a inhibition in breast
cancer. J Transl Med. 13:1312015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Xu X, Zhang Y, Jasper J, Lykken E,
Alexander PB, Markowitz GJ, McDonnell DP, Li QJ and Wang XF:
miR-148a functions to suppress metastasis and serves as a
prognostic indicator in triple-negative breast cancer. Oncotarget.
7:20381–20394. 2016.PubMed/NCBI
|
|
47
|
Müller V, Gade S, Steinbach B, Loibl S,
von Minckwitz G, Untch M, Schwedler K, Lübbe K, Schem C, Fasching
PA, et al: Changes in serum levels of miR-21, miR-210, and miR-373
in HER2-positive breast cancer patients undergoing neoadjuvant
therapy: A translational research project within the Geparquinto
trial. Breast Cancer Res Treat. 147:61–68. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hong L, Yang J, Han Y, Lu Q, Cao J and
Syed L: High expression of miR-210 predicts poor survival in
patients with breast cancer: A meta-analysis. Gene. 507:135–138.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yin JY, Deng ZQ, Liu FQ, Qian J, Lin J,
Tang Q, Wen XM, Zhou JD, Zhang YY and Zhu XW: Association between
mir-24 and mir-378 in formalin-fixed paraffin-embedded tissues of
breast cancer. Int J Clin Exp Patho. 7:4261–4267. 2014.
|
|
50
|
Ikeda K, Horieinoue K, Ueno T, Suzuki T,
Sato W, Shigekawa T, Osaki A, Saeki T, Berezikov E, Mano H and
Inoue S: miR-378a-3p modulates tamoxifen sensitivity in breast
cancer MCF-7 cells through targeting GOLT1A. Sci Rep. 5:131702015.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Savi F, Forno I, Faversani A, Luciani A,
Caldiera S, Gatti S, Foa P, Ricca D, Bulfamante G, Vaira V and
Bosari S: miR-296/Scribble axis is deregulated in human breast
cancer and miR-296 restoration reduces tumour growth in vivo. Clin
Sci (Lond). 127:233–242. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yang H, Yu J, Wang L, Ding D, Zhang L, Chu
C, Chen Q, Xu Z, Zou Q and Liu X: miR-320a is an independent
prognostic biomarker for invasive breast cancer. Oncol Lett.
8:1043–1050. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Yu J, Wang JG, Zhang L, Yang HP, Wang L,
Ding D, Chen Q, Yang WL, Ren KH, Zhou DM, et al: MicroRNA-320a
inhibits breast cancer metastasis by targeting metadherin.
Oncotarget. 7:38612–38625. 2016.PubMed/NCBI
|
|
54
|
Li L, Yuan L, Luo J, Gao J, Guo J and Xie
X: miR-34a inhibits proliferation and migration of breast cancer
through down-regulation of Bcl-2 and SIRT1. Clin Exp Med.
13:109–117. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Chabre O, Libé R, Assie G, Barreau O,
Bertherat J, Bertagna X, Feige JJ and Cherradi N: Serum miR-483-5p
and miR-195 are predictive of recurrence risk in adrenocortical
cancer patients. Endocr Relat Cancer. 20:579–594. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Rask L, Balslev E, Søkilde R, Høgdall E,
Flyger H, Eriksen J and Litman T: Differential expression of
miR-139, miR-486 and miR-21 in breast cancer patients
sub-classified according to lymph node status. Cell Oncol.
37:215–227. 2014. View Article : Google Scholar
|
|
57
|
Song Q, Xu Y, Yang C, Chen Z, Jia C, Chen
J, Zhang Y, Lai P, Fan X, Zhou X, et al: miR-483-5p promotes
invasion and metastasis of lung adenocarcinoma by targeting RhoGDI1
and ALCAM. Cancer Res. 74:3031–3042. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhang M, Liu D, Li W, Wu X, Gao CE and Li
X: Identification of featured biomarkers in breast cancer with
microRNA microarray. Arch Gynecol Obstet. 294:1047–1053. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Jia Z, Liu Y, Gao Q, Han Y, Zhang G, Xu S,
Cheng K and Zou W: miR-490-3p inhibits the growth and invasiveness
in triple-negative breast cancer by repressing the expression of
TNKS2. Gene. 593:41–47. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Uen YH, Wang JW, Wang CC, Jhang Y, Chung
JY, Tseng T, Sheu M and Lee S: Mining of potential microRNAs with
clinical correlation-regulation of syndecan-1 expression by
miR-122-5p altered mobility of breast cancer cells and possible
correlation with liver injury. Oncotarget. 9:28165–28175. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Block I, Burton M, Sørensen KP, Andersen
L, Larsen MJ, Bak M, Cold S, Thomassen M, Tan Q and Kruse TA:
Association of miR-548c-5p, miR-7-5p, miR-210-3p, miR-128-3p with
recurrence in systemically untreated breast cancer. Oncotarget.
9:9030–9042. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Wang B, Yang Z, Wang H, Cao Z, Zhao Y,
Gong C, Ma L, Wang X, Hu X and Chen S: MicroRNA-320a inhibits
proliferation and invasion of breast cancer cells by targeting
RAB11A. Am J Cancer Res. 5:2719–2729. 2015.PubMed/NCBI
|
|
63
|
Frères P, Bouznad N, Servais L, Josse C,
Wenric S, Poncin A, Thiry J, Moonen M, Oury C, Lancellotti P, et
al: Variations of circulating cardiac biomarkers during and after
anthracycline-containing chemotherapy in breast cancer patients.
BMC Cancer. 18:1022018. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Hamdi K, Goerlitz D, Stambouli N, Islam M,
Baroudi O, Neili B, Benayed F, Chivi S, Loffredo C, Jillson IA, et
al: miRNAs in Sera of Tunisian patients discriminate between
inflammatory breast cancer and non-inflammatory breast cancer.
SpringerPlus. 3:6362014. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Liu Y, Cai Q, Bao PP, Su Y, Cai H, Wu J,
Ye F, Guo X, Zheng W, Zheng Y and Shu XO: Tumor tissue microRNA
expression in association with triple-negative breast cancer
outcomes. Breast Cancer Res Treat. 152:183–191. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Fassan M, Baffa R, Palazzo JP, Lloyd J,
Crosariol M, Liu CG, Volinia S, Alder H, Rugge M, Croce CM and
Rosenberg A: MicroRNA expression profiling of male breast cancer.
Breast Cancer Res. 11:R582009. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Farazi TA, Horlings HM, Ten Hoeve JJ,
Mihailovic A, Halfwerk H, Morozov P, Brown M, Hafner M, Reyal F,
van Kouwenhove M, et al: MicroRNA sequence and expression analysis
in breast tumors by deep sequencing. Cancer Res. 71:4443–4453.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Ouyang M, Li Y, Ye S, Ma J, Lu L, Lv W,
Chang G, Li X, Li Q, Wang S and Wang W: MicroRNA profiling implies
new markers of chemoresistance of triple-negative breast cancer.
PLoS One. 9:e962282014. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Cao ZG, Huang YN, Yao L, Liu YR, Hu X, Hou
YF and Shao ZM: Positive expression of miR-361-5p indicates better
prognosis for breast cancer patients. J Thorac Dis. 8:1772–1779.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Han J, Yu J, Dai Y, Li J, Guo M, Song J
and Zhou X: Overexpression of miR-361-5p in triple-negative breast
cancer (TNBC) inhibits migration and invasion by targeting RQCD1
and inhibiting the EGFR/PI3K/Akt pathway. Bosn J Basic Med Sci.
19:52–59. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Chang YY, Kuo WH, Hung JH, Lee CY, Lee YH,
Chang YC, Lin WC, Shen CY, Huang CS, Hsieh FJ, et al: Deregulated
microRNAs in triple-negative breast cancer revealed by deep
sequencing. Mol Cancer. 14:362015. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
McFall T, Mcknight B, Rosati R, Kim S,
Huang Y, Viola-Villegas N and Ratnam M: Progesterone receptor A
promotes invasiveness and metastasis of luminal breast cancer by
suppressing regulation of critical microRNAs by estrogen. J Biol
Chem. 293:1163–1177. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Benvenuti S, Sartorebianchi A, Di
Nicolantonio F, Zanon C, Moroni M, Veronese S, Siena S and Bardelli
A: Oncogenic activation of the RAS/RAF signaling pathway impairs
the response of metastatic colorectal cancers to anti-epidermal
growth factor receptor antibody therapies. Cancer Res.
67:2643–2648. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Downward J: Targeting RAS signalling
pathways in cancer therapy. Nat Rev Cancer. 3:11–22. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Guertin DA and Sabatini DM: Defining the
role of mTOR in cancer. Cancer Cell. 12:9–22. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Zoncu R, Efeyan A and Sabatini DM: mTOR:
From growth signal integration to cancer, diabetes and ageing. Nat
Rev Mol Cell Biol. 12:21–35. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Biankin AV, Waddell N, Kassahn KS, Gingras
MC, Muthuswamy LB, Johns AL, Miller DK, Wilson PJ, Patch AM, Wu J,
et al: Pancreatic cancer genomes reveal aberrations in axon
guidance pathway genes. Nature. 491:399–405. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Kidd PM: The use of mushroom glucans and
proteoglycans in cancer treatment. Altern Med Rev. 5:4–27.
2000.PubMed/NCBI
|
|
79
|
Iozzo RV and Sanderson RD: Proteoglycans
in cancer biology, tumour microenvironment and angiogenesis. J Cell
Mol Med. 15:1013–1031. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Van Brocklyn JR: Sphingolipid signaling
pathways as potential therapeutic targets in gliomas. Mini Rev Med
Chem. 7:984–990. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Yuan HD, Quan HY, Zhang Y, Kim SH and
Chung SH: 20(S)-Ginsenoside Rg3-induced apoptosis in HT-29 colon
cancer cells is associated with AMPK signaling pathway. Mol Med
Rep. 3:825–831. 2010.PubMed/NCBI
|
|
82
|
Green AS, Chapuis N, Lacombe C, Mayeux P,
Bouscary D and Tamburini J: LKB1/AMPK/mTOR signaling pathway in
hematological malignancies: From metabolism to cancer cell biology.
Cell Cycle. 10:2115–2120. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Dentice M, Luongo C, Ambrosio R, Sibilio
A, Casillo A, Iaccarino A, Troncone G, Fenzi G, Larsen PR and
Salvatore D: β-catenin regulates deiodinase levels and thyroid
hormone signaling in colon cancer cells. Gastroenterol.
143:1037–1047. 2012. View Article : Google Scholar
|
|
84
|
King CR, Kraus MH and Aaronson SA:
Amplification of a novel v-erbB-related gene in a human mammary
carcinoma. Science. 229:974–976. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Normanno N, De Luca A, Maiello MR,
Campiglio M, Napolitano M, Mancino M, Carotenuto A, Viglietto G and
Menard S: The MEK/MAPK pathway is involved in the resistance of
breast cancer cells to the EGFR tyrosine kinase inhibitor
gefitinib. J Cell Physiol. 207:420–427. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Oxnard GR, Arcila ME, Sima CS, Riely GJ,
Chmielecki J, Kris MG, Pao W, Ladanyi M and Miller V: Acquired
resistance to EGFR tyrosine kinase inhibitors in EGFR mutant lung
cancer: Distinct natural history of patients with tumors harboring
the T790M mutation. Clin Cancer Res. 17:1616–1622. 2011. View Article : Google Scholar : PubMed/NCBI
|