Introduction
Hypertrophic cardiomyopathy (HCM) is an autosomal
dominant heart disease characterized by asymmetric hypertrophy of
the left ventricle, primarily caused by enlarged myocytes, in the
absence of other diseases (1,2). The
etiology of HCM is mainly associated with genetic factors,
endocrine disorders or autoimmune diseases (3). The estimated prevalence of HCM in the
general population is ~0.6%, and varies among children, adolescents
and adults (4). With advancements
in diagnostic techniques, the prevalence of HCM has shown an
increasing trend (5). Clinical
manifestations of HCM include chest tightness, angina, dyspnea and
syncope. These symptoms are progressive and may lead to serious
complications such as heart failure and sudden cardiac death
(6-8).
Despite substantial progress in the treatment of HCM using
personalized strategies (9,10), a
definitive cure for this condition remains unknown.
Autophagy (AT) is a biochemical process that
involves the degradation and recycling of damaged or discarded
intracellular components by lysosomes to protect cells against
external environmental conditions, such as hypoxia and oxidative
stress (11). Administration of AT
inducers, such as rapamycin, in animal models, inhibits mTOR and
promotes AT, effectively protecting cardiomyocytes and improving
cardiomyopathy phenotypes (12,13).
In addition, studies have demonstrated a close relationship between
AT and HCM, suggesting that targeting AT is a promising therapeutic
strategy for HCM (14-16).
However, the precise roles of AT-related genes (ARGs) in HCM remain
unclear, necessitating further investigation of the relationship
between ARGs and HCM.
In this study, key ARGs related to the development
of HCM were identified using bioinformatics analysis. The
association between these genes and HCM was determined through
functional annotation, pathway enrichment, protein-protein
interaction (PPI) and immune infiltration analyses. In addition,
potential drugs for the treatment of HCM were predicted, and the
results of bioinformatics analysis were validated through reverse
transcription-quantitative PCR (RT-qPCR). The findings provide a
valuable theoretical foundation for the development of novel
diagnostic and therapeutic strategies for HCM.
Materials and methods
Data collection and processing
The HCM dataset GSE180313(17) was obtained from the Gene Expression
Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). This dataset
contains heart tissues from 7 healthy individuals and 13 patients
with HCM. Preprocessed and merged data were integrated into a
unified dataset. The ‘limma’ package (18) in R software (v.3.6.3) (19) was used to identify differentially
expressed genes (DEGs) between the HCM and control groups, with the
screening criteria being set as a |log2FC| value of
<0.5 and a P-value of <0.05. The ‘ggplot2’ package (20) in R was used to generate heat maps
and volcano plots. Information regarding ARGs was obtained from the
Human Autophagy Database (http://www.autophagy.lu/index.html) and the Gene Set
Enrichment Analysis (GSEA) website (http://software.broadinstitute.org/gsea/index.jsp).
The extracted ARGs were processed and integrated into a gene set
known as ARGs. GSEA was used to investigate the overall association
between AT and HCM and identify potential biological processes
involving ARGs that were associated with the pathogenesis of HCM.
Genes in the GSE180313 dataset were scored using the AT dataset
from GSEA, resulting in the calculation of normalized enrichment
scores (NESs). P<0.05 was considered to indicate significant
enrichment. The workflow of the present study is shown in Fig. S1.
Identification and functional
enrichment analysis of differentially expressed ARGs (DEARGs)
DEARGs were obtained by intersecting the DEGs
identified in the GSE180313 dataset with the integrated ARG set.
The overlapping genes (DEARGs) were visualized on a Venn diagram.
Subsequently, the cluster profiler package (v4.8.3) (21) in R and DAVID (david.ncifcrf.gov/) were used to implement Gene
Ontology (GO; https://geneontology.org/) and Kyoto Encyclopedia of
Genes and Genomes (KEGG; https://www.genome.jp/kegg/) enrichment analyses of
DEARGs. P<0.05 was considered to indicate significant
enrichment.
PPI network and identification of hub
genes and key modules
DEARGs were imported into the Search Tool for the
Retrieval of Interacting Genes/Proteins (STRING) database (v11.09)
(https://string-db.org/) for PPI analysis with
default settings. The resulting PPI network was visualized using
Cytoscape software (v.3.7.2) (22). Subsequently, the MCODE plug-in (V
3.7.1) (20) was used to filter
and visualize key PPI networks, resulting in the identification of
key modules containing hub genes.
Receiver operating characteristic
(ROC) analysis of hub genes
The ROC curves of hub genes were constructed using
data from the GSE180313 and GSE36961(23) datasets. The area under the curve
(AUC) was quantified for comparison, and only genes with AUC values
of >0.6 were considered to have statistically significant
diagnostic potential.
Immune infiltration analysis
To investigate the immune microenvironment of HCM
and key DEARGs, CIBERSORTx (https://cibersortx.stanford.edu/) was employed to
analyze the differences in immune infiltration between patients
with HCM and healthy controls. Additionally, this algorithm was
utilized to determine the proportions of various immune cell
types.
Prediction of therapeutic drugs
Key differentially expressed AT-associated genes and
compound interaction data from the Drug Signatures Database
(DSigDB; http://dsigdb.tanlab.org/) were
extracted using the Enrichr online tool (http://amp.pharm.mssm.edu/Enrichr). Drugs for the
treatment of HCM were predicted based on hub genes.
Cell culture and model
construction
The H9c2 immortalized rat cardiomyocyte-like cell
line (The Cell Bank of Type Culture Collection of The Chinese
Academy of Sciences) is commonly used in cardiac research in
vitro (24). H9c2 cells were
cultured in high-glucose DMEM (Hyclone; Cytiva) supplemented with
10% fresh fetal bovine serum (HyClone; Cytiva) and 1%
penicillin-streptomycin (Gibco; Thermo Fisher Scientific, Inc.)
under standard conditions (37˚C, 5% CO2, 95% humidity
and 21% oxygen). Cells from passages 3-10 were used for subsequent
experiments. Angiotensin II (AngII; GlpBio Technology, Inc.) was
used to induce hypertrophy in H9c2 cells. The cells were incubated
with AngII at different concentrations (50, 100, 200 and 400 nM) at
37˚C for 24 h, and the optimal concentration was determined based
on the mRNA and protein expression of atrial natriuretic peptide
(ANP) and brain natriuretic peptide (BNP). For further
experimentation, the cells were incubated with the determined
optimal concentration of AngII at 37˚C for 12, 24, 36 and 48 h.
RT-qPCR
Total RNA was extracted from H9c2 cells using TRIzol
reagent (Beijing Solarbio Science & Technology Co., Ltd.) and
reverse transcribed using the PrimeScript RT reagent kit (Monad
Biotech Co., Ltd.) according to the manufacturer's instructions.
qPCR was conducted using the SYBR Green qPCR Master Mix (cat. no.
B21203; Bimake.com) on a BIO-RAD CFX Connect Real-Time
PCR Detection System (Bio-Rad Laboratories, Inc.). The qPCR
protocol included an initial denaturation step at 95˚C for 10 min,
followed by 40 cycles of thermal cycling with denaturation at 95˚C
for 15 sec and annealing at 60˚C for 1 min. The primer sequences
used for qPCR are shown in Table
SI. ACTB served as the internal reference, and the relative
mRNA expression of target genes was calculated using the Cq
(2-ΔΔCq) method (25).
Western blotting
To extract total proteins, H9c2 cells were lysed in
RIPA buffer supplemented with protease inhibitors (Beijing Solarbio
Science & Technology Co., Ltd.) on ice. The extracted proteins
were quantified using a BCA assay kit (GlpBio Technology, Inc.) and
denatured by boiling for 5 min, and subsequently separated by on
12% gels by sodium dodecyl sulfate-polyacrylamide gel
electrophoresis, with 40 µg of protein loaded per lane. The
separated proteins were transferred to a PVDF membrane
(MilliporeSigma), which was incubated with 5% skimmed milk powder
at room temperature for 2 h on a shaker. Subsequently, the membrane
was incubated with primary antibodies against ANP (1:500; cat. no.
27426-1-AP; Proteintech Group, Inc.), BNP (1:500; cat. no. A2179;
ABclonal, Inc.), LC3 (1:1,000; cat.no. 381544; Chengdu
Zen-Bioscience Co., Ltd.), P62 (1:1,000; cat. no. 380612; Chengdu
Zen-Bioscience Co., Ltd.) and β-actin (1:1,000; cat. no.
66009-1-Ig; Proteintech Group, Inc.) at 4˚C overnight. The
following day, the membrane was incubated with horseradish
peroxidase-conjugated mouse and rabbit secondary antibodies
(1:5,000; cat. nos. 511103 and 511203, respectively; Chengdu
Zen-Bioscience, Co., Ltd.) at room temperature for 2 h. Protein
bands were visualized using the Ultra High Sensitivity ECL kit
(catalog no. GK10008; GlpBio Technology, Inc.) and captured using
the FluorChem FC3 System (ProteinSimple). ImageJ software
(v1.8.0.345; National Institutes of Health) was used to
semi-quantify the optical density of protein bands.
Detection of autolysosome
acidification
To assess the level of AT in cells, LysoTracker Red
was used to label intracellular lysosomes, as autophagosomes can
bind to lysosomes. Briefly, H9c2 cells were incubated with 50 nM
LysoTracker Red working solution (Beyotime Institute of
Biotechnology) at 37˚C for 30 min in the dark and subsequently
examined using a fluorescence microscope.
Immunofluorescence analysis
LC3 expression in H9c2 cells was detected through
immunofluorescence staining. Briefly, the cells were washed twice
with PBS, fixed with 4% paraformaldehyde (biosharp life sciences)
at room temperature for 10 min, blocked with 2% BSA (cat. no.
CAS#9048-46-8; Shanghai Yuanye Bio-Technology Co., Ltd.) at room
temperature for 30 min and incubated with anti-LC3 antibody (1:200;
cat. no. 381544; Chengdu Zen-Bioscience Co., Ltd.) at 4˚C
overnight. The following day, the cells were incubated with a
fluorescently labeled secondary antibody (1:200; cat. no. A32732;
Thermo Fisher Scientific, Inc.) at room temperature for 1 h.
Thereafter, nuclei were stained with DAPI at room temperature for 5
min and the cells were examined using a fluorescence
microscope.
Statistical analysis
Statistical analysis was performed using the
GraphPad Prism 8 software (Dotmatics). The unpaired two-tailed
Student's t-test was used to compare the differences between two
groups. One-way analysis of variance and Dunnett's multiple
comparison test were employed to assess the differences among
multiple groups. Data are expressed as the mean ± standard
deviation of 3 repeats. P<0.05 was considered to indicate a
statistically significant difference.
Results
Study protocol
The overall protocol of the present study is shown
in Fig. S1. The patient data used
in the present study were derived from the GSE180313 dataset in the
GEO database.
Identification of DEGs between the HCM
and control groups
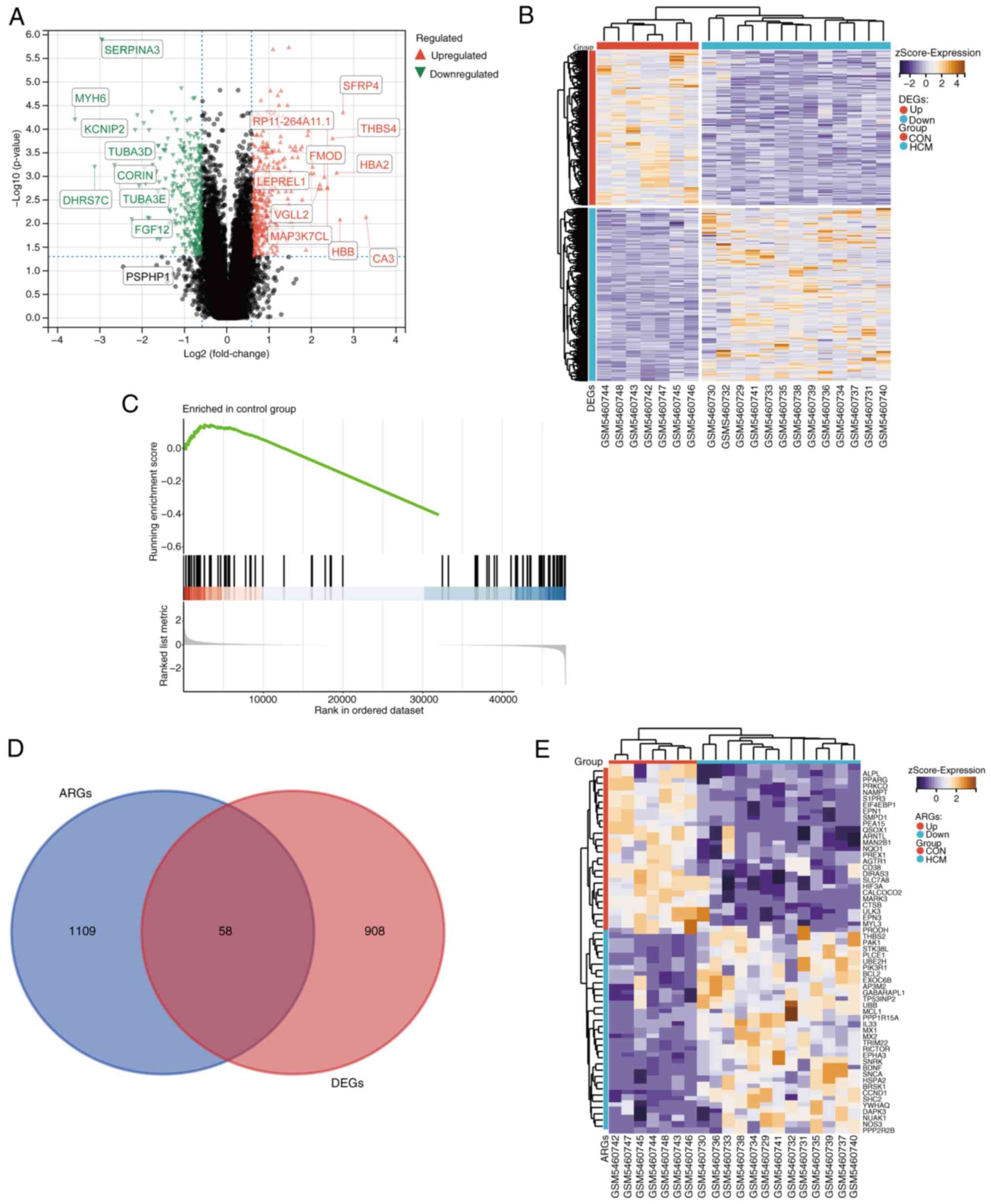
After pre-processing and normalization of the
GSE180313 dataset, a total of 966 DEGs were identified between the
HCM and control groups. Of these 966 DEGs, 510 genes were
upregulated and 456 genes were downregulated. A volcano plot and a
heat map were generated to visualize the DEGs (Fig. 1A and B).
Identification of DEARGs and
enrichment analysis
GSEA was used to compare ARGs between the HCM and
control groups. Fig. 1C
demonstrates the significant differences in ARG expression between
the two groups (|NES|=1.521; P<0.05). This indicates that ARGs
could be a crucial characteristic of HCM and that their
dysregulation supports a link between HCM and autophagy.
Furthermore, a total of 1,167 ARGs were identified after
pre-processing of the integrated gene set. These ARGs were
intersected with DEGs to obtain 58 DEARGs (Fig. 1D). A heat map was generated to
visualize the expression patterns of these DEARGs in the HCM and
control groups (Fig. 1E).
Functional and mechanistic analyses of
DEARGs
To investigate the functions and pathways of DEARGs,
GO and KEGG enrichment analyses were performed using DAVID. The
results demonstrated that DEARGs were significantly enriched in
biological processes such as ‘peptidyl-serine phosphorylation’,
‘cellular response to oxidative stress’, ‘regulation of autophagy’
and ‘response to UV’; molecular functions such as ‘protein serine
kinase activity’, ‘tau protein binding’ and ‘death domain binding’;
and cellular components such as the ‘mitochondrial outer membrane’
and ‘organelle outer membrane’ (Fig.
2A-C). KEGG analysis demonstrated that the DEARGs were notably
enriched in the ‘AGE-RAGE signaling pathway in diabetic
complications’, ‘PI3K-Akt signaling pathway’, ‘measles’, ‘hepatitis
C’, ‘AMPK signaling pathway’ and ‘EGFR tyrosine kinase inhibitor
resistance’ (Fig. 2D).
Interactions were identified between DEARGs and the aforementioned
functions and pathways through gene and pathway cross-talk mapping,
suggesting that multiple genes and pathways may be involved in the
regulation of DEARGs in HCM (Fig.
2E-H). Overall, these findings suggested that ARGs regulate the
progression of HCM through intricate interplay among multiple gene
functions and pathways.
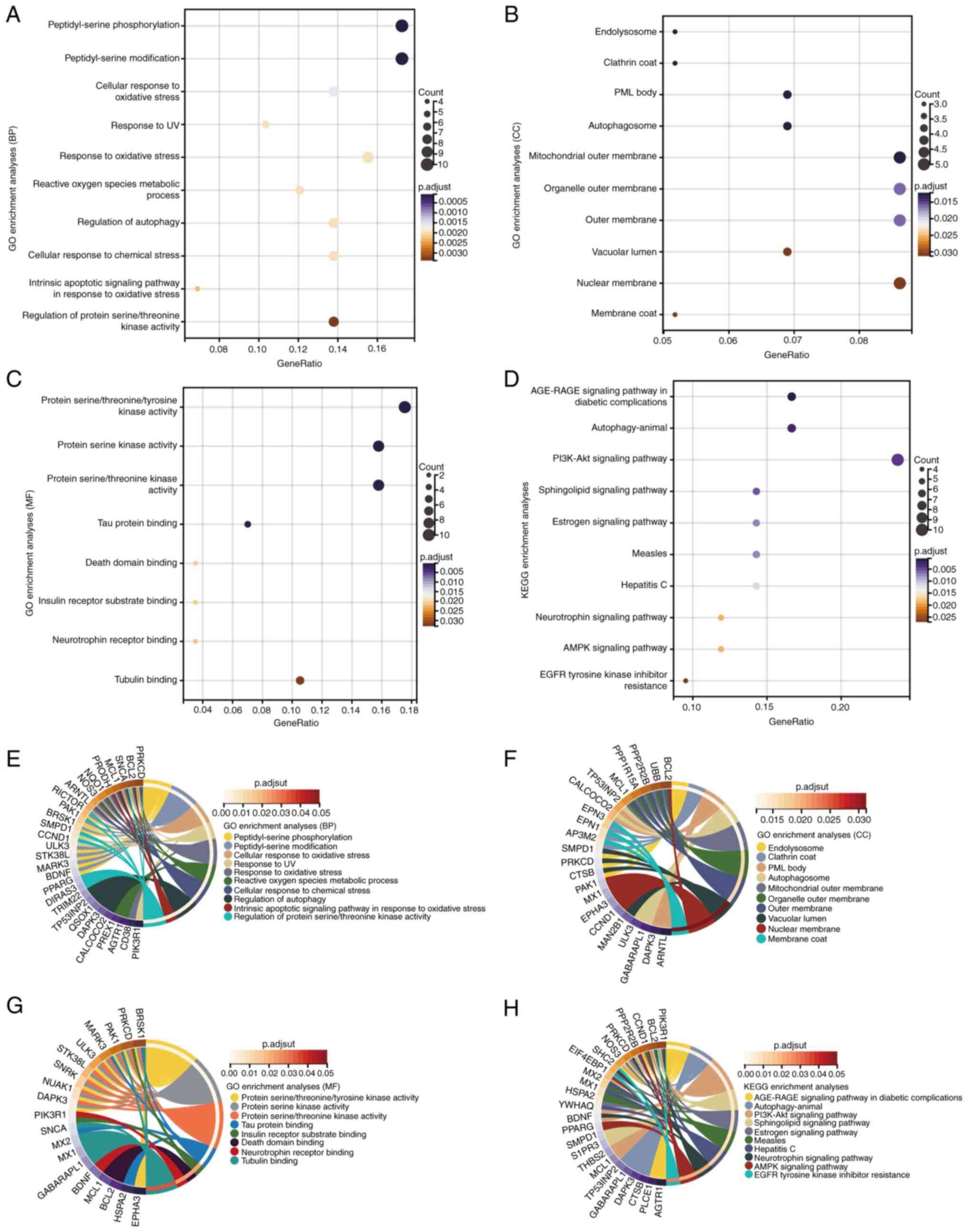 | Figure 2GO and KEGG enrichment analyses of
DEARGs. GO enrichment analysis of DEARGs in the (A) BP, (B) CC and
(C) MF categories. (D) KEGG enrichment analysis of DEARGs.
Crosstalk analysis between DEARGs and gene functions in (E) BP, (F)
CC and (G) MF categories, and (H) KEGG pathways. AGE-RAGE, advanced
glycation end product-receptor for advanced glycation endproducts;
AMPK, AMP-activated protein kinase; BP, biological process; CC,
cellular component; DEARG, differentially expressed
autophagy-related gene; GO, Gene Ontology; KEGG, Kyoto Encyclopedia
of Genes and Genomes; MF, molecular function; p.adjust, adjusted
P-value; PML, promyelocytic leukemia. |
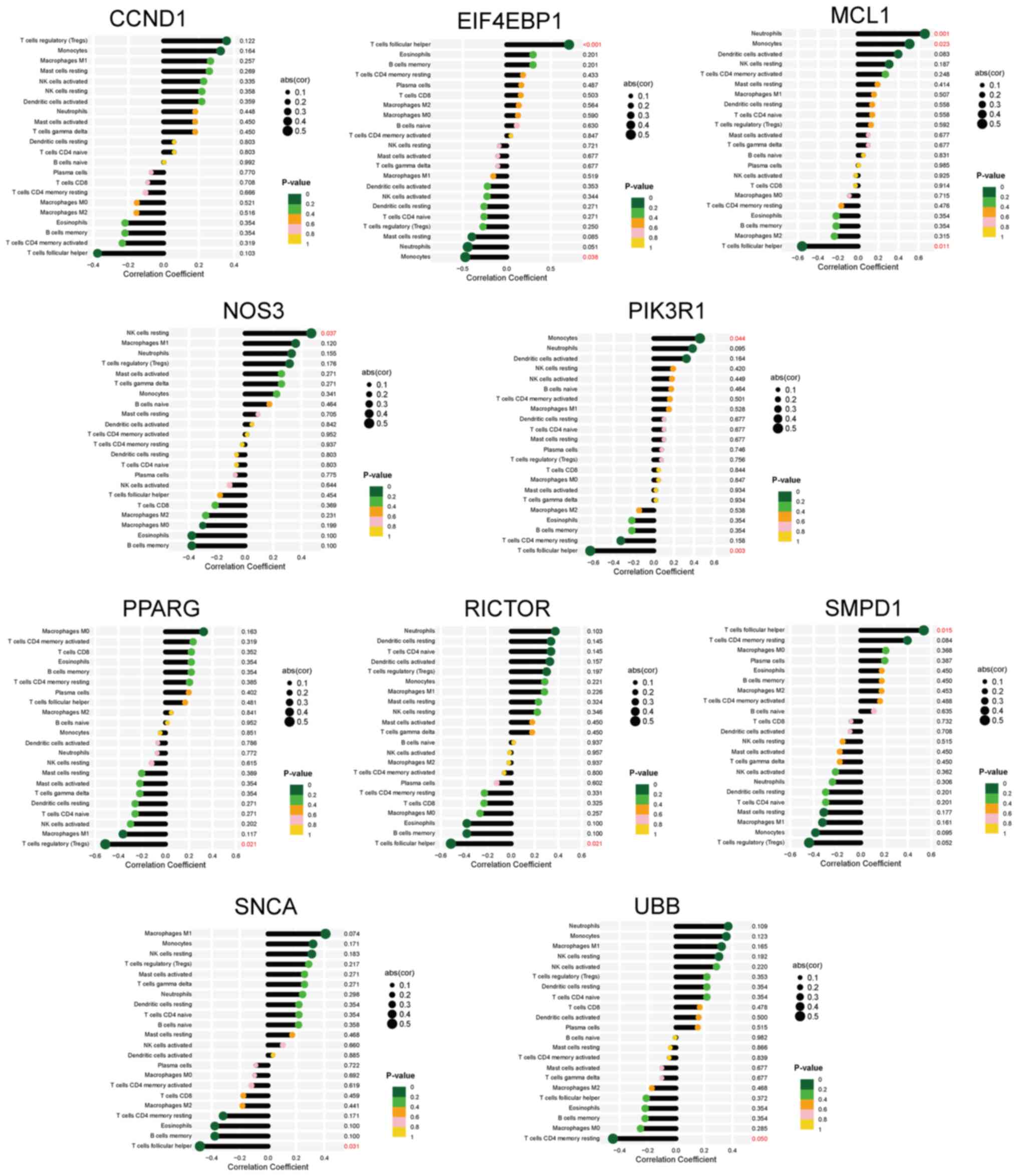
PPI network analysis, functional
module construction and hub gene identification
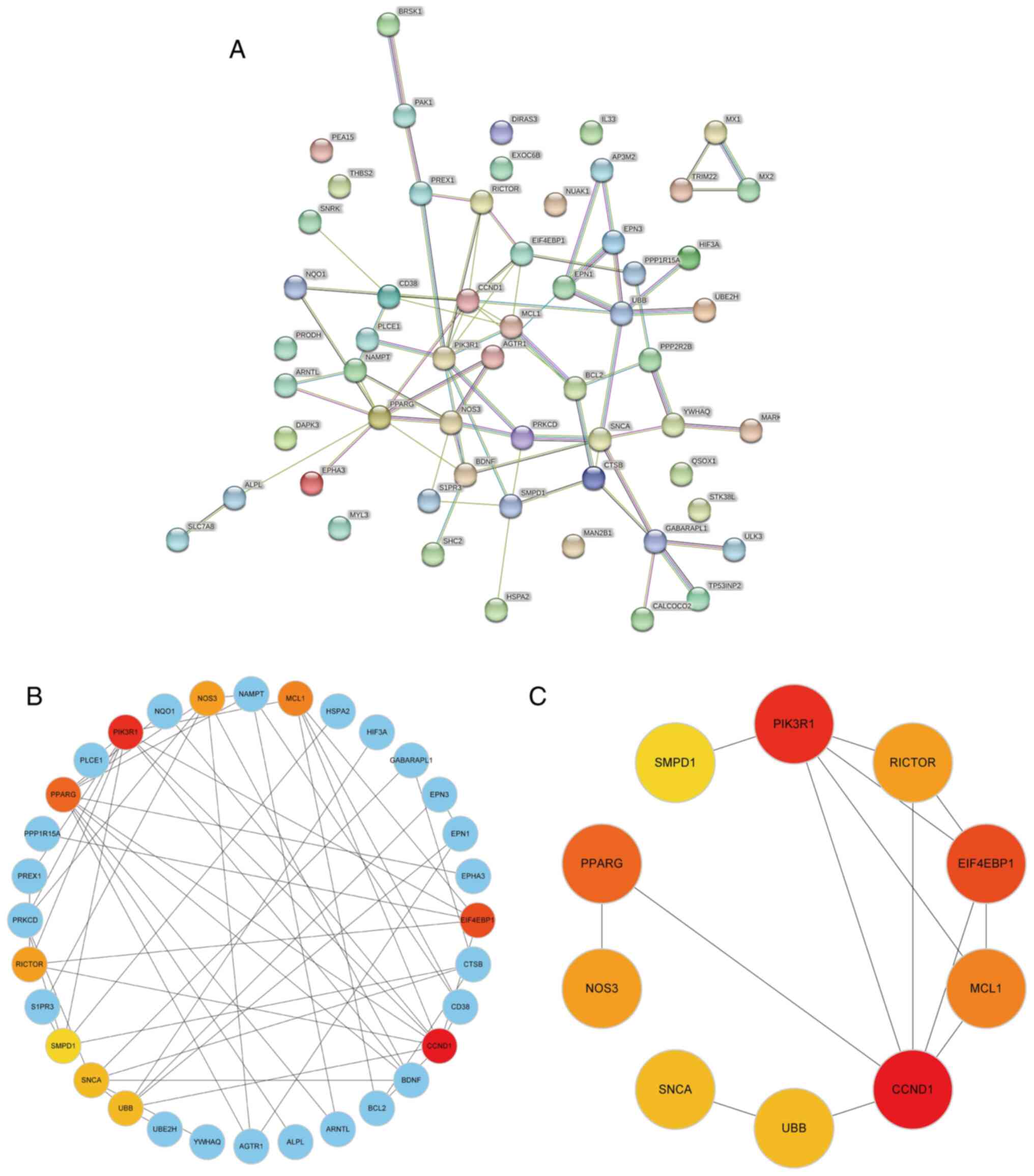
A PPI network of 58 DEARGs was constructed using the
STRING database (Fig. 3A). After
the network was imported into the Cytoscape software, the Maximal
Clique Centrality algorithm was used to identify a sub-network
comprising 32 hub genes. To filter these hub genes, the MCODE
plug-in was used to identify important functional modules within
the PPI network. Notably, a key cluster in the network consisted of
a functional module with 10 nodes and 14 edges, including EIF4EBP1,
MCL1, PIK3R1, CCND1, PPARG, SMPD1, RICTOR, NOS3, SNCA and UBB.
Fig. 3B and C illustrate the interactions between
DEARGs and hub genes.
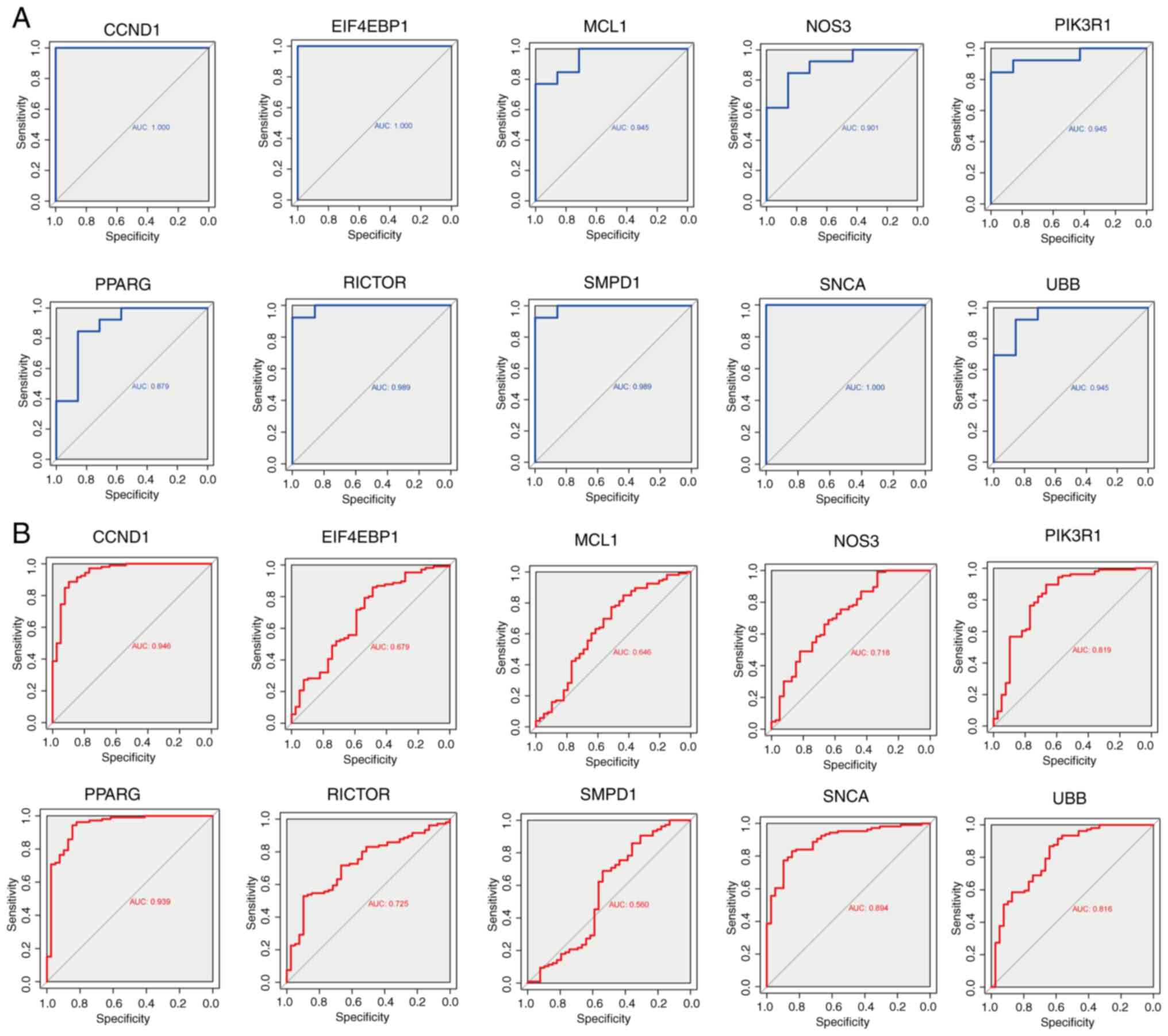
Diagnostic value of the hub genes
ROC curves were generated to evaluate the diagnostic
efficacy of the 10 hub genes (Fig.
4A). In the GSE180313 dataset, all hub genes exhibited AUC
values of >0.8, indicating a significant association with HCM
and promising diagnostic potential. An external dataset (GSE36961)
was used to validate these findings (Fig. 4B). Notably, discrepancies were
observed in the results of hub gene analysis between the two
datasets. During the validation of external datasets, it was
observed that the AUC values for EIF4EBP1, MCL1, and SMPD1 were
<0.7, indicating that these three hub genes exhibit limited
diagnostic performance for HCM. By contrast, most other hub genes
demonstrated robust diagnostic capabilities (AUC >0.7),
suggesting a potential association between autophagy and HCM.
Nonetheless, the significance of these hub genes warrants further
investigation in future studies
Prediction of drugs and molecular
docking simulations
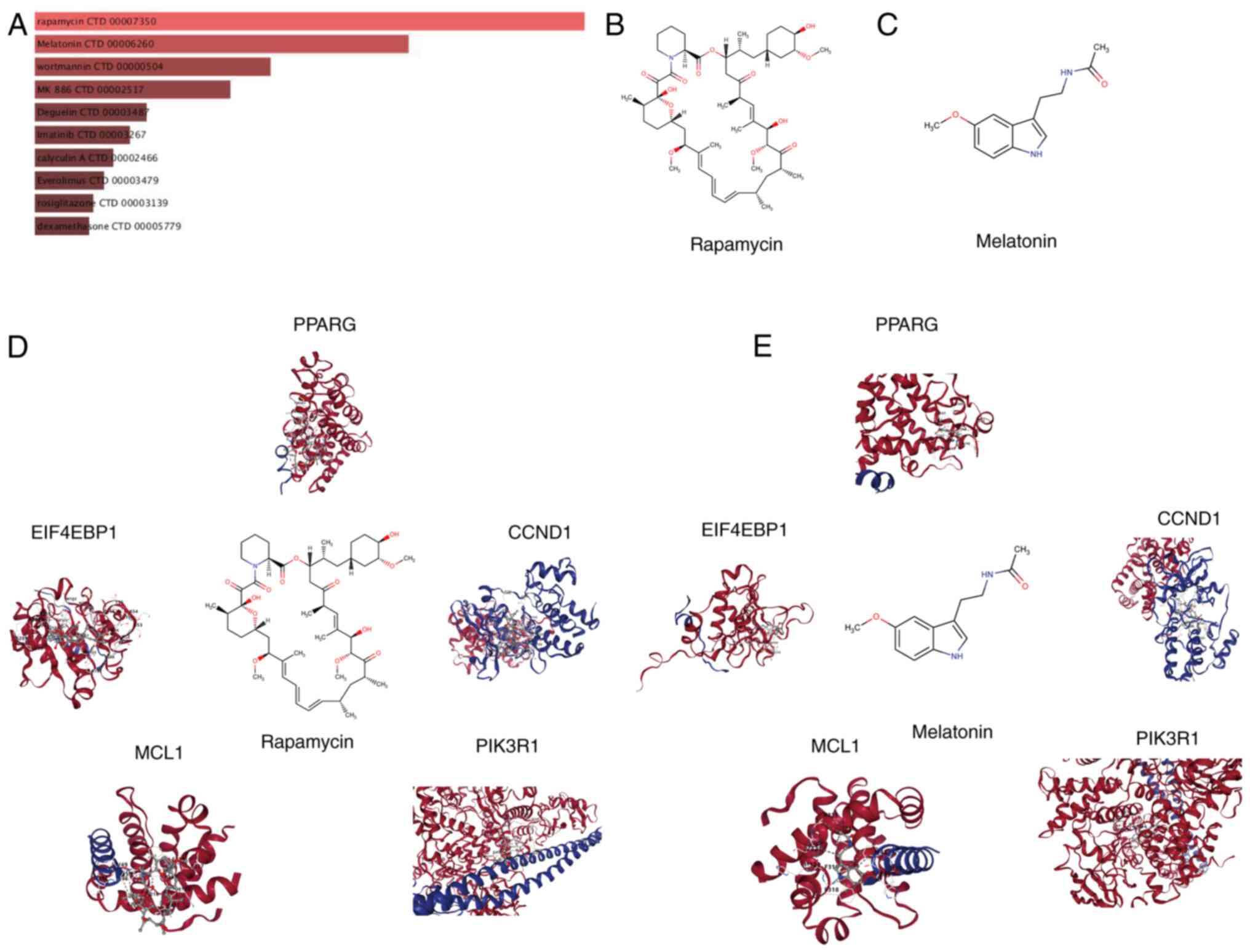
The DSigDB in the Enrichr platform was used to
identify small-molecule drugs targeting hub genes for the treatment
of HCM. A total of 1,332 drugs with potential therapeutic value
were identified based on the degree of gene-compound match and
median number, with the screening criteria being set as a false
discovery rate of <0.05 and composite scores of >5,000. The
top 10 small-molecule drugs with the most significant impact on the
expression of hub genes are shown in Fig. 5A. Among these, rapamycin and
melatonin are the top two candidates. Fig. 5B and C show the molecular structure of
rapamycin and melatonin (Mel; N-acetyl-5-methoxytryptamine).
Research has demonstrated that rapamycin exerts a notable effect on
the treatment and prevention of HCM (26,27).
Mel possesses antioxidant properties and exerts protective effects
against various cardiovascular diseases, including diabetic
cardiomyopathy and myocardial hypertrophy (28,29).
To gain insights into the binding between hub genes and predicted
drugs, molecular docking simulations were performed using rapamycin
and Mel as examples (Fig. 5D and
E).
Relationship between ARGs and immune
cell infiltration in HCM
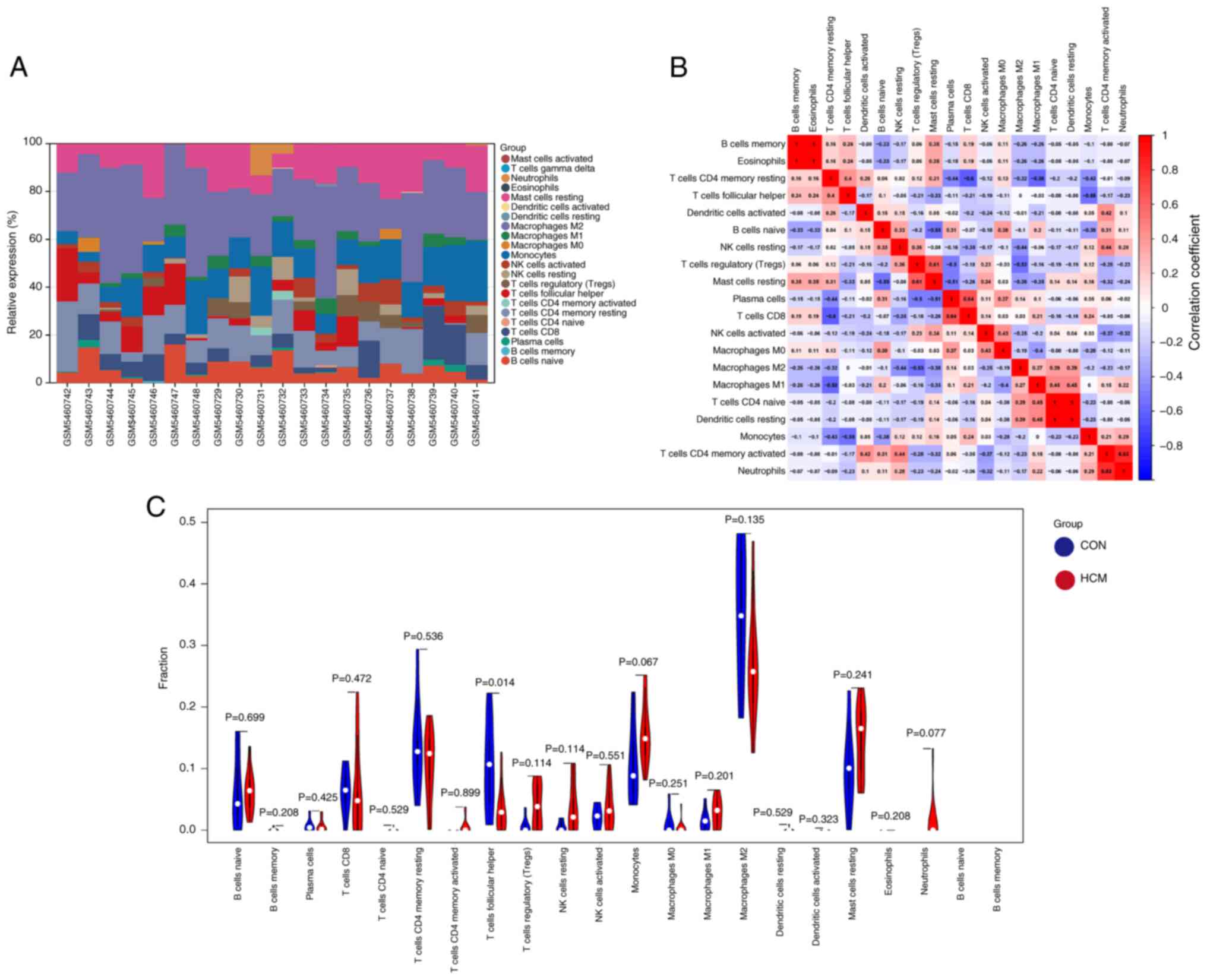
Cellular and humoral immune functions serve a
crucial role in the development of HCM (30). The relative proportions of
infiltrating immune cells in heart samples from the GSE180313
dataset were assessed and quantified using the CIBERSORT algorithm
(Fig. 6A). The resulting heatmap
illustrates the relationships between various infiltrating immune
cells (Fig. 6B). In addition,
violin plots were generated to visualize the expression profiles of
the 20 immune cell subtypes in the control and HCM groups (Fig. 6C). Only the infiltration levels of
T follicular helper (Tfh) cells were significantly different
between the HCM and control groups, with lower levels being
observed in the HCM group. Therefore, Tfh cells were identified as
differentially infiltrating immune cells. Furthermore, the
correlation between immune cells and the 10 hub DEARGs was examined
(Fig. 7). Nine hub genes, except
for CCND1, exhibited varying correlations with six types of immune
cells, namely Tfh cells, monocytes, neutrophils, regulatory T
cells, resting natural killer cells and T cells CD4 memory resting.
These findings suggested that the hub genes serve an important role
in the immune response to HCM.
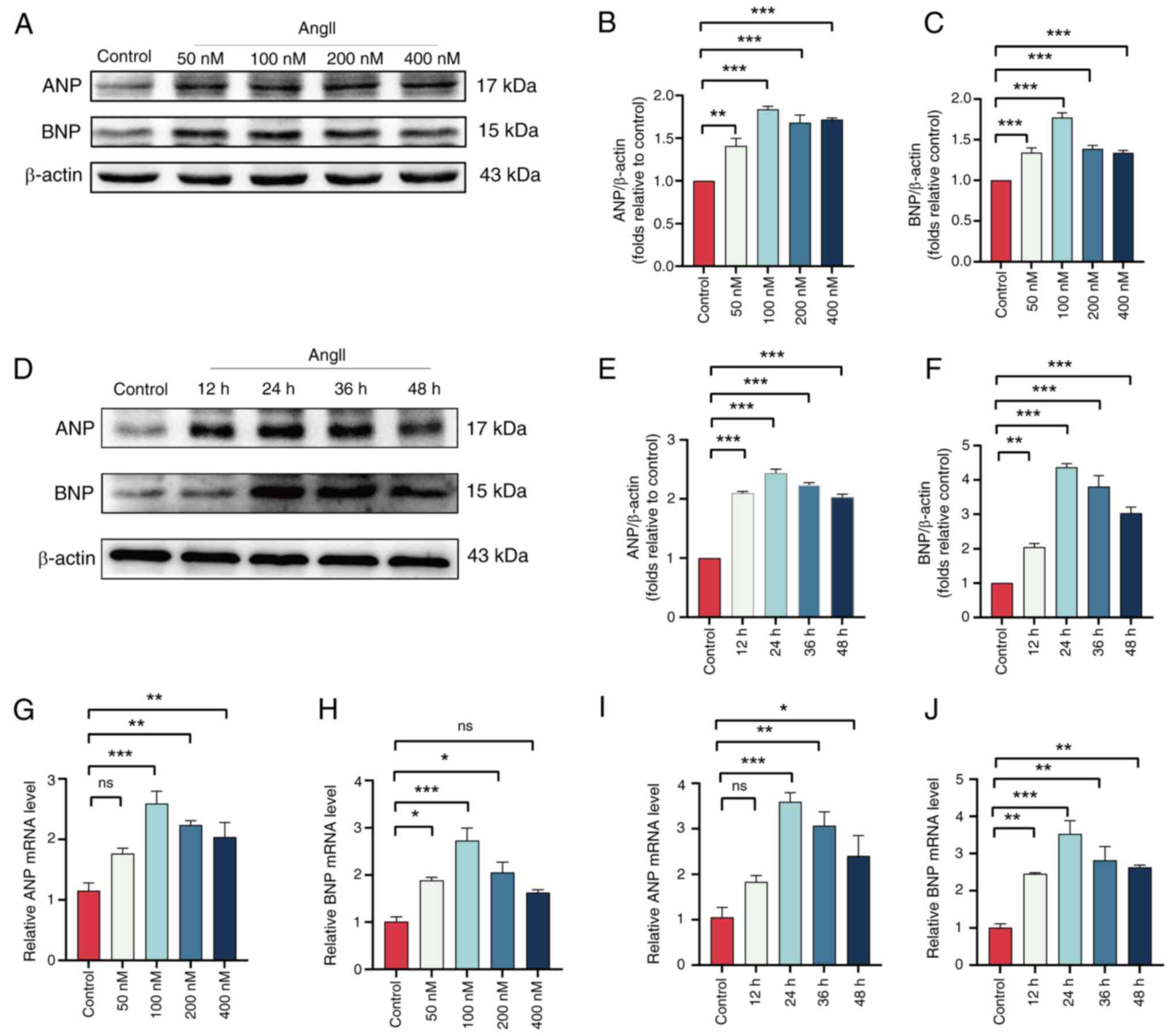
Validation of hub genes in an in vitro
model of HCM
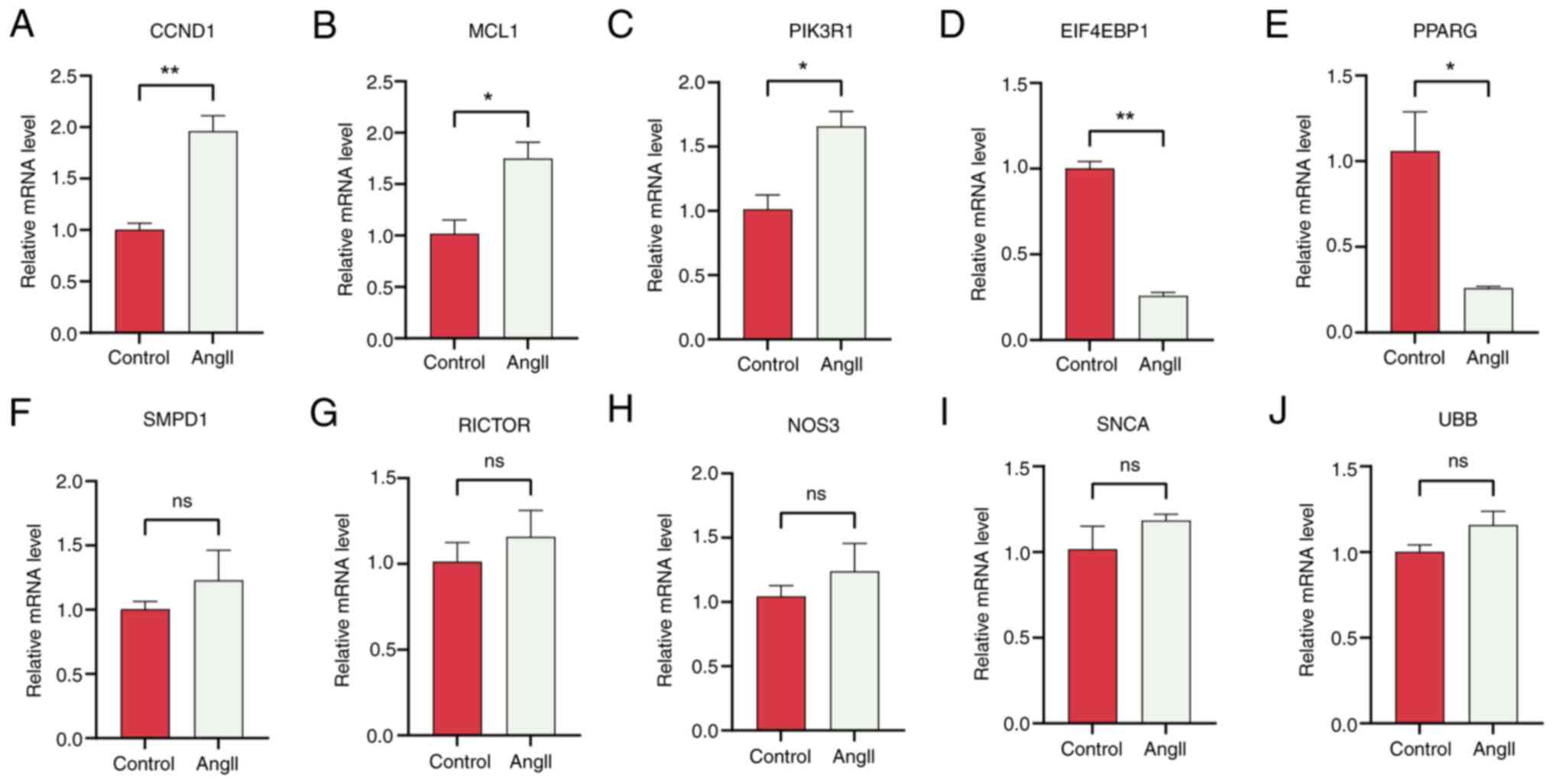
To validate the expression of the hub genes
EIF4EBP1, MCL1, PIK3R1, CCND1, PPARG, SMPD1, RICTOR, NOS3, SNCA and
UBB in vitro, H9c2 cells were stimulated with AngII to
induce HCM. Western blotting and RT-qPCR were used to determine the
optimal concentration and duration of AngII treatment. The results
demonstrated that when H9C2 cells were pretreated with AngII for
the same duration, the highest protein expression levels of ANP and
BNP were observed in the 100 nM AngII group (Fig. 8A-C). Additionally, when H9C2 cells
were pretreated with 100 nM AngII for different time periods, the
highest ANP and BNP protein expression levels were found at 24 h of
pretreatment (Fig. 8D-F).
Consistent with these changes in protein expression, the mRNA
levels also corroborated this finding (Fig. 8G-J). Therefore, a pretreatment of
H9C2 cells with 100 nM AngII for 24 h was selected to induce
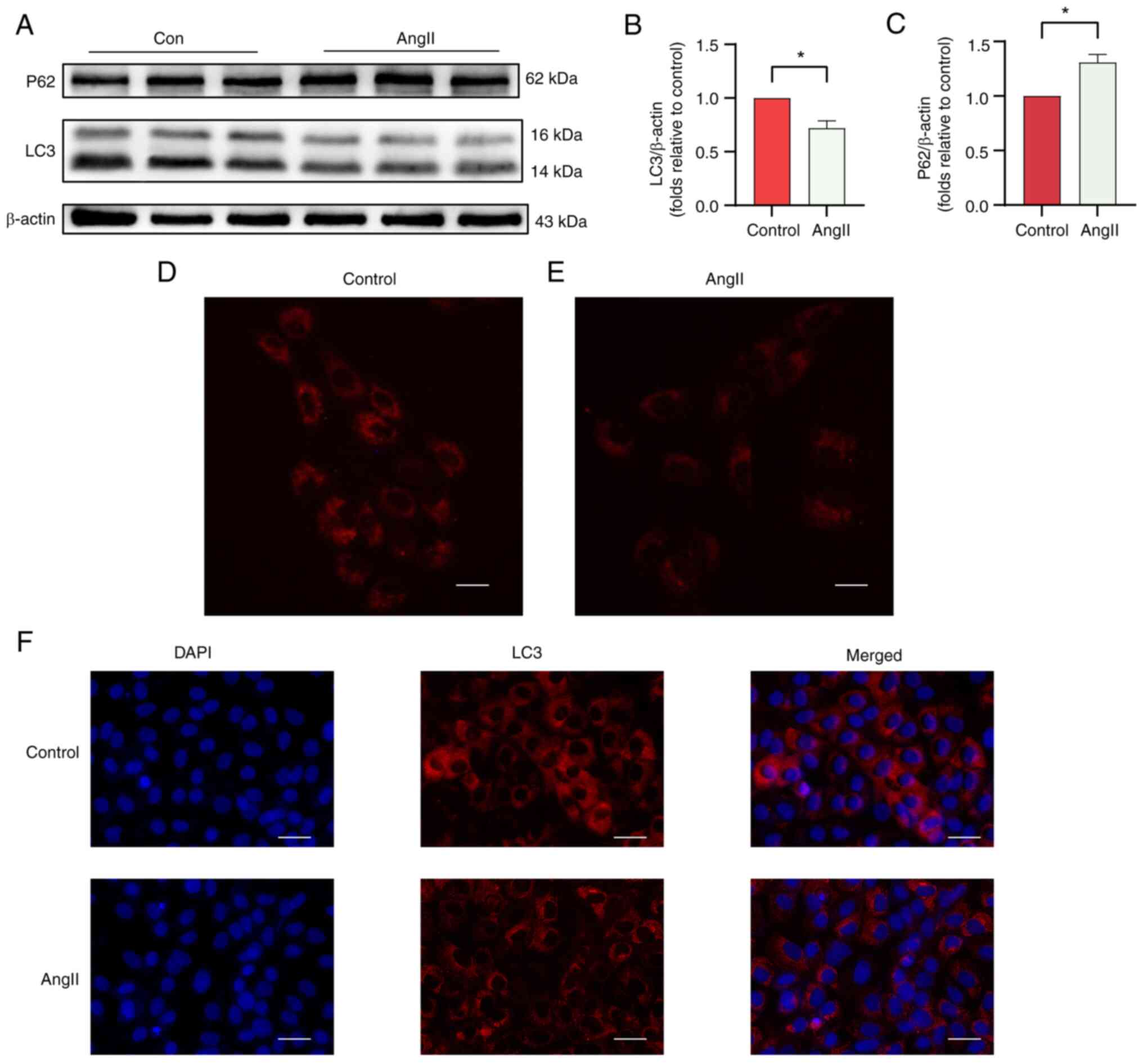
hypertrophy. Western blotting was used to evaluate the expression
levels of the AT-associated proteins LC3 and P62 in the AngII and
control groups (Fig. 9A). As shown
in Fig. 9B and C, the LC3/β-actin ratio was significantly
lower and the protein expression of P62 was higher in the AngII
group. Furthermore, autolysosome acidification was detected, and
immunofluorescence analysis was performed to assess the level of
AT. H9c2 cells stained with LysoTracker Red showed reduced
fluorescence intensity in the AngII group compared with that in the
control group (Fig. 9D).
Immunofluorescence analysis revealed a decrease in LC3 fluorescence
intensity in the AngII group compared with that in the control
group (Fig. 9F). The
aforementioned results indicate that autophagy is reduced in the
AngII-induced H9C2 cell hypertrophy model. To ensure the
reliability of the results, RT-qPCR was performed to evaluate the
expression levels of hub genes in both groups. Based on PPI network
analysis, EIF4EBP1, MCL1, PIK3R1, CCND1 and PPARG were identified
as the most significant hub ARGs associated with HCM. RT-qPCR
revealed that EIF4EBP1 and PPARG were downregulated, while MCLl,
PIK3R1 and CCND1 were upregulated after treatment with AngII
(Fig. 10). Therefore, we propose
that EIF4EBP1, MCL1, PIK3R1, CCND1 and PPARG play roles in the
regulation of autophagy in HCM.
Discussion
HCM is a prevalent hereditary cardiac disease that
predisposes individuals, especially young athletes, to sudden
death, also known as exercise-induced sudden cardiac death
(31,32). Although HCM does not progress
rapidly, its complications such as sudden arrhythmogenic death,
heart failure and atrial fibrillation can occur abruptly or worsen
under any circumstances, posing a severe threat to the life of
patients (33). According to the
2018 Epidemiological Survey statistics, HCM remains a major health
concern worldwide, affecting ~88% of the global population and
imposing a long-lasting socioeconomic burden (34). However, no precise and efficient
therapeutic strategy has been developed to date. AT serves an
essential role in the development and progression of cardiovascular
diseases such as myocardial infarction, aortic coarctation,
atherosclerosis and ischemic cardiomyopathy (35-37).
Therefore, targeting AT represents a promising strategy for the
treatment of HCM. However, the precise role of AT in the
pathogenesis of HCM warrants further investigation. In the present
study, bioinformatics analysis was used to examine the roles and
mechanisms of ARGs in the development of HCM to explore novel
avenues for effective treatment.
A total of 58 DEARGs associated with HCM were
identified through comprehensive analysis of a GEO dataset and an
ARG set. GSEA revealed a significant association between ARGs and
HCM, suggesting that AT serves a crucial role in the development of
HCM. Furthermore, GO functional annotation and KEGG pathway
enrichment analysis demonstrated that the DEARGs were closely
associated with various biological processes, cellular components
and molecular functions related to AT (Fig. 2). Notably, the findings indicated
that the pathogenesis of HCM involves not only AT but also other
classical pathways such as the ‘AGE-RAGE signaling pathway in
diabetic complications’ and the ‘PI3K-Akt signaling pathway’. AT
serves as a cytoprotective mechanism that maintains cellular
homeostasis by regulating intracellular and extracellular catabolic
and anabolic processes through the lysosomal degradation pathway
(38,39). Alterations in the levels of
cardiomyocyte AT have been reported to induce functional or
morphological changes, including apoptosis, atrophy, fibrosis or
hypertrophy (40-42).
A recent study revealed that excessive cardiomyocyte AT can result
in lysosomal storage disorders that lead to cellular damage and
cardiac dysfunction (16). These
findings highlight the extensive investigation of the role of AT in
HCM, while emphasizing the need for further exploration of other
molecules and pathways. In the present study, PPI network analysis
revealed 10 hub genes (EIF4EBP1, MCL1, PIK3R1, CCND1, PPARG, SMPD1,
RICTOR, NOS3, SNCA and UBB) associated with the development of HCM.
The diagnostic value of these hub genes was assessed through ROC
analysis and validated through cellular experiments. All hub genes
except for SMPD1 exhibited significant diagnostic potential.
Notably, EIF4EBP1, MCL1, PIK3R1, CCND1 and PPARG were
differentially expressed between control and AngII-treated H9c2
cells. In particular, PIK3R1, MCL-1 and CCND1 were significantly
upregulated in AngII-treated cells. These results suggested that
the aforementioned five ARGs serve as promising therapeutic targets
for HCM.
EIF4EBP1 functions as a regulatory protein in cell
signaling pathways and is involved in the initiation and
progression of various diseases (43-46).
Upregulation of EIF4EBP1 has been shown to delay the progression of
systemic lupus erythematosus through B-cell AT (43). Additionally, EIF4EBP1 acts as a
tumor suppressor gene. Upregulation of EIF4EBP1 promotes the
development and metastasis of breast cancer, whereas downregulation
of EIF4EBP1 in breast cancer impedes the proliferation of pituitary
tumor cells (45,46). Several studies have indicated that
EIF4EBP1 serves as a biomarker for evaluating the prognosis of
tumors (44). MCL1, an
anti-apoptotic gene, serves a crucial role in cell survival,
metabolism, apoptosis, immunity and tumor formation (47-49).
It has attracted attention in research on hematological
malignancies (48). Inhibition of
MCL1 can promote tumor cell apoptosis and enhance the cytotoxicity
or antitumor immune efficacy of drugs in acute myeloid leukemia
(AML) (50). Dysregulation or
inhibition of MCL1 is an essential factor contributing to drug
resistance in various cancer types (51-54),
as MCL1 is a major regulatory protein of the intrinsic apoptosis
pathway. Given that the PIK3R1/Akt/mTOR signaling pathway is
regulated by AT, targeting PIK3R1 can reduce cellular oxidative
stress and apoptosis, thereby regulating cardiomyocyte apoptosis in
the treatment of heart diseases (55). In addition, PIK3R1 is positively
associated with immune activation and serves an essential role in
regulating the tumor microenvironment, inflammation and drug
sensitivity or resistance (56-59).
CCND1, located on chromosome 11q, is a member of the
cell cycle protein D family and serves a crucial role in anti-aging
signaling pathways (60). Liu
et al (61) found that
downregulation of CCND1 activated anti-aging signaling pathways,
enhanced the expression of antioxidant genes, suppressed the
production of reactive oxygen species and prevented the osteogenic
differentiation of valve interstitial cells in heart valve disease.
CCND1 has been revealed to regulate the viability, proliferation
and cell cycle of oral squamous cell carcinoma cells through
microRNA-519d-3p (62).
Furthermore, detection of CCND1 rearrangements holds diagnostic
value for blood disorders (63).
PPARG, a member of the peroxisome proliferator-activated receptor
subfamily, serves a crucial role in regulating various signaling
pathways involved in the pathophysiological mechanisms of various
diseases and states, including inflammation, lipid metabolism, AT,
apoptosis and cell cycle progression (64,65).
Notably, PPARG has been closely associated with chemosensitivity in
gastric cancer, AML, colorectal cancer and breast cancer (66), highlighting its important role in
modulating the response of tumor cells to chemotherapeutic agents.
Mechanistically, PPARG influences chemosensitivity by regulating
cell cycle progression and AT, and participating in inflammatory
responses (67). In the present
study, DSigDB was utilized to predict potential drugs targeting the
identified hub genes. The results indicated that rapamycin and Mel
are promising drugs targeting ARGs and pathways in HCM. Previous
studies have demonstrated the therapeutic efficacy of rapamycin and
Melatonin in HCM, which is consistent with the findings of the
present study (28,68). However, the therapeutic efficacy of
other drugs warrants further investigation and validation.
The precise role of the immune system in the
development of HCM remains elusive. Previous studies have
demonstrated that AT serves an essential role in immunity,
primarily through its involvement in pathogen clearance and
inflammation regulation (69-71).
Therefore, in the current study, the relationship between ARGs and
immune cell infiltration in HCM was investigated. Only the
infiltration levels of Tfh cells were significantly different
between the HCM and control groups. Tfh cells represent an
independent subset of CD4(+) T effector cells involved in humoral
immunity and activation of other immune cells (72). The infiltration levels of Tfh cells
were positively correlated with EIF4EBP1 but negatively correlated
with MCL1 and PIK3R1. Furthermore, the infiltration levels of
monocytes and neutrophils were positively correlated with MCL1, and
those of regulatory T cells were negatively correlated with PPARG.
However, CCND1 did not exhibit a significant correlation with the
22 immune cell types examined in the present study. These results
provided valuable insights into how ARGs influence the development
of HCM by regulating immunity. However, the present study did not
validate the relationship between immune infiltrating cells and
core ARGs through specific experiments. Further research and
evidence in this area are required in the future.
The level of autophagic activity reported in studies
on HCM is inconsistent possibly due to differences in study
participants, experimental design and sample size (73,74).
Upregulation of certain ARGs is also observed in HCM, which may be
attributed to the following reasons: Firstly, it can be considered
as a compensatory mechanism wherein cells attempt to restore
autophagic function by increasing the expression of specific ARGs.
This upregulation aims to compensate for the reduced autophagic
activity either by enhancing particular steps of AT or by
augmenting the number of autophagosomes (73,75,76).
Secondly, due to the complex nature of HCM as a disease, involving
alterations in multiple genes and signaling pathways during its
pathological progression, certain factors within this process might
contribute to the upregulation of select ARGs (77-79).
The objective of the present study was to investigate the function
and expression of ARGs in HCM through bioinformatics analysis and
cellular experiments in order to predict potential drugs for HCM. A
total of 10 drugs targeting hub genes were identified, including
rapamycin and Mel. Studies have validated the therapeutic or
preventive effects of rapamycin in HCM (26,27,80).
Activation of the mTOR signaling pathway serves a crucial role in
regulating cell proliferation and protein activation. Rapamycin
inhibits mTOR, directly influencing metabolic disorders, fibrosis
and myocardial hypertrophy. It attenuates myocardial hypertrophy
and fibrosis, while reversing ventricular remodeling and restoring
cardiac function (68,73,81,82).
Notably, mTOR signaling has been investigated in studies on heart
diseases (83). Furthermore, Mel
possesses antioxidant properties and exerts protective effects
against various cardiovascular diseases, including diabetic
cardiomyopathy and myocardial hypertrophy (28,29).
Additionally, studies have demonstrated that activation of
macrophage stimulating 1/nuclear factor erythroid 2-related factor
2 signaling and MICU1 could effectively reduce oxidative stress,
alleviating myocardial hypertrophy (84,85).
In addition to rapamycin and Mel, other ARG-targeted drugs
predicted in the present study include wortmannin, deguelin,
imatinib, everolimus and rosiglitazone. However, the mechanisms of
action of these drugs warrant further investigation.
The present study emphasized the important role of
AT in HCM. ARGs associated with the development of HCM were
analyzed using bioinformatics tools, and the findings were
validated using an external dataset and a cell model of HCM. The
present study provides novel insights into the pathological
mechanisms of HCM and offers promising avenues for developing
therapeutic strategies targeting ARGs.
Despite its important findings, the current study
had some limitations that should be acknowledged. First, the sample
size should be increased to enhance the reliability of the results,
and a more comprehensive prospective study is warranted to validate
the results of the present study. Second, clinical samples and
animal models are required to verify the functional roles of the
identified hub ARGs. Third, validation of the protein expression
levels of core ARGs should be added. Lastly, further investigation
is required to verify the therapeutic efficacy of the predicted
drugs and elucidate the specific mechanisms through which the hub
ARGs regulate the development of HCM.
In conclusion, the present study demonstrated that
the expression of ARGs was significantly altered in HCM. In
particular, EIF4EBP1, MCL1, PIK3R1, CCND1 and PPARG were identified
as key ARGs that serve as potential diagnostic markers and
therapeutic targets for HCM. Additionally, 10 drugs targeting the
key ARGs were identified, which may be used in the ARG-targeted
treatment of HCM. In conclusion, the current study improved the
understanding of the pathogenesis of HCM and highlighted the
potential diagnostic and therapeutic value of ARGs in HCM,
providing a crucial theoretical foundation for the development of
personalized therapies.
Supplementary Material
Overall protocol of the present study.
ARG, autophagy-related gene; DEARG, differentially expressed ARG;
DEGs, differentially expressed genes; DIIC, differentially
infiltrating immune cell; DSigDB, Drug Signatures Database; GO,
Gene Ontology; GSEA, Gene Set Enrichment Analysis; HADb, Human
Autophagy Database; KEGG, Kyoto Encyclopedia of Genes and Genomes;
PPI, protein-protein interaction.
Sequences of primers used for reverse
transcription-quantitative PCR.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the Natural Science
Foundation of Jiangxi (grant nos. 20212ACB206011, 20224ACB206002
and 20232BAB206009) and the National Natural Science Foundation of
China (grant nos. 82160073 and 81860082).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
RBQ conducted the cell experiments, and analyzed and
mapped the experimental data. STZ contributed to the experimental
design and performed data analysis for the bioinformatics analysis.
ZWL drafted the article, provided software support and analyzed the
data. RYZ, ZCQ, and HZP confirm the authenticity of all the raw
data and contributed to data interpretation. LFZ and ZQX
contributed to the cell experiments. SQL and LW designed the
experiments and provided financial support. All authors read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
He J, Gu D, Wu X, Reynolds K, Duan X, Yao
C, Wang J, Chen CS, Chen J, Wildman RP, et al: Major causes of
death among men and women in China. N Engl J Med. 353:1124–1134.
2005.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Abbas MT, Baba Ali N, Farina JM, Mahmoud
AK, Pereyra M, Scalia IG, Kamel MA, Barry T, Lester SJ, Cannan CR,
et al: Role of genetics in diagnosis and management of hypertrophic
cardiomyopathy: A glimpse into the future. Biomedicines.
12(682)2024.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Wang Z, Xia Q, Su W, Cao M, Sun Y, Zhang
M, Chen W and Jiang T: Exploring the communal pathogenesis,
ferroptosis mechanism, and potential therapeutic targets of dilated
cardiomyopathy and hypertrophic cardiomyopathy via a microarray
data analysis. Front Cardiovasc Med. 9(824756)2022.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Chase Cole J, Benvie SF and DeLosSantos M:
Mavacamten: A novel agent for hypertrophic cardiomyopathy. Clin
Ther. 46:368–373. 2024.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Zampieri M, Argirò A, Marchi A, Berteotti
M, Targetti M, Fornaro A, Tomberli A, Stefàno P, Marchionni N and
Olivotto I: Mavacamten, a novel therapeutic strategy for
obstructive hypertrophic cardiomyopathy. Curr Cardiol Rep.
23(79)2021.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Ottaviani A, Mansour D, Molinari LV,
Galanti K, Mantini C, Khanji MY, Chahal AA, Zimarino M, Renda G,
Sciarra L, et al: Revisiting diagnosis and treatment of
hypertrophic cardiomyopathy: Current practice and novel
perspectives. J Clin Med. 12(5710)2023.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Bakalakos A, Monda E and Elliott PM: The
diagnostic and therapeutic implications of phenocopies and mimics
of hypertrophic cardiomyopathy. Can J Cardiol. 40:754–765.
2024.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Pu L, Li J, Qi W, Zhang J, Chen H, Tang Z,
Han Y, Wang J and Chen Y: Current perspectives of sudden cardiac
death management in hypertrophic cardiomyopathy. Heart Fail Rev.
29:395–404. 2024.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Faisaluddin M, Balasubramanian S, Ahmed A,
Hussain K, Nso N, Gaznabi S, Erwin JP III, Pursnani A and Ricciardi
M: Temporal trends and procedural safety of transcatheter mitral
valve repair with mitraclip in patients with hypertrophic
cardiomyopathy: Insights from the national inpatient sample. Curr
Probl Cardiol. 49(102354)2024.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Yacoub MS, El-Nakhal T, Hasabo EA, Shehata
N, Wilson K, Ismail KH, Bakr MS, Mohsen M, Mohamed A, Abdelazim E,
et al: A systematic review and meta-analysis of the efficacy and
safety of Mavacamten therapy in international cohort of 524
patients with hypertrophic cardiomyopathy. Heart Fail Rev.
29:479–496. 2024.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Chen X, Tsvetkov AS, Shen HM, Isidoro C,
Ktistakis NT, Linkermann A, Koopman WJH, Simon HU, Galluzzi L, Luo
S, et al: International consensus guidelines for the definition,
detection, and interpretation of autophagy-dependent ferroptosis.
Autophagy. 24:1213–1246. 2024.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Kaplan JL, Rivas VN and Connolly DJ:
Advancing treatments for feline hypertrophic cardiomyopathy: The
role of animal models and targeted therapeutics. Vet Clin North Am
Small Anim Pract. 53:1293–1308. 2023.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Rivas VN, Kaplan JL, Kennedy SA,
Fitzgerald S, Crofton AE, Farrell A, Grubb L, Jauregui CE,
Grigorean G, Choi E, et al: Multi-omic, histopathologic, and
clinicopathologic effects of once-weekly oral rapamycin in a
naturally occurring feline model of hypertrophic cardiomyopathy: A
pilot study. Animals (Basel). 13(3184)2023.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Dang JY, Zhang W, Chu Y, Chen JH, Ji ZL
and Feng P: Downregulation of salusins alleviates hypertrophic
cardiomyopathy via attenuating oxidative stress and autophagy. Eur
J Med Res. 29(109)2024.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Huang X, Zhang J, Wang W, Huang Z and Han
P: Vps4a regulates autophagic flux to prevent hypertrophic
cardiomyopathy. Int J Mol Sci. 24(10800)2023.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Rabinovich-Nikitin I and Kirshenbaum LA:
YAP/TFEB pathway promotes autophagic cell death and hypertrophic
cardiomyopathy in lysosomal storage diseases. J Clin Invest.
131(e146821)2021.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Zhang Y, Zhao J, Jin Q and Zhuang L:
Transcriptomic analyses and experimental validation identified
immune-related lncRNA-mRNA Pair MIR210HG-BPIFC regulating the
progression of hypertrophic cardiomyopathy. Int J Mol Sci.
25(2816)2024.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43(e47)2015.PubMed/NCBI View Article : Google Scholar
|
|
19
|
R Core Team: A Language and Environment
for Statistical Computing. R Foundation for Statistical Computing,
Vienna, 2020. Available from: https://www.R-project.org/.
|
|
20
|
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT and
Lin CY: cytoHubba: Identifying hub objects and sub-networks from
complex interactome. BMC Syst Biol. 8 (Suppl 4)(S11)2014.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z,
Feng T, Zhou L, Tang W, Zhan L, et al: clusterProfiler 4.0: A
universal enrichment tool for interpreting omics data. Innovation
(Camb). 2(100141)2021.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Gong J, Shi B, Yang P, Khan A, Xiong T and
Li Z: Unveiling immune infiltration characterizing genes in
hypertrophic cardiomyopathy through transcriptomics and
bioinformatics. J Inflamm Res. 17:3079–3092. 2024.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Li N, Wang W, Zhou H, Wu Q, Duan M, Liu C,
Wu H, Deng W, Shen D and Tang Q: Ferritinophagy-mediated
ferroptosis is involved in sepsis-induced cardiac injury. Free
Radic Biol Med. 160:303–318. 2020.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expres-sion data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Gao XM, Wong G, Wang B, Kiriazis H, Moore
XL, Su YD, Dart A and Du XJ: Inhibition of mTOR reduces chronic
pressure-overload cardiac hypertrophy and fibrosis. J Hypertens.
24:1663–1670. 2006.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Völkers M, Konstandin MH, Doroudgar S,
Toko H, Quijada P, Din S, Joyo A, Ornelas L, Samse K, Thuerauf DJ,
et al: Mechanistic target of rapamycin complex 2 protects the heart
from ischemic damage. Circulation. 128:2132–2144. 2013.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Pei HF, Hou JN, Wei FP, Xue Q, Zhang F,
Peng CF, Yang Y, Tian Y, Feng J, Du J, et al: Melatonin attenuates
postmyocardial infarction injury via increasing Tom70 expression. J
Pineal Res. 62:2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Reiter RJ, Mayo JC, Tan DX, Sainz RM,
Alatorre-Jimenez M and Qin L: Melatonin as an antioxidant: Under
promises but over delivers. J Pineal Res. 61:253–278.
2016.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Dai H, Liu Y, Zhu M, Tao S, Hu C, Luo P,
Jiang A and Zhang G: Machine learning and experimental validation
of novel biomarkers for hypertrophic cardiomyopathy and cancers. J
Cell Mol Med. 28(e70034)2024.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Abbasi M, Ong KC, Newman DB, Dearani JA,
Schaff HV and Geske JB: Obstruction in hypertrophic cardiomyopathy:
Many faces. J Am Soc Echocardiogr. 37:613–625. 2024.PubMed/NCBI View Article : Google Scholar
|
|
32
|
McKinney J, Isserow M, Wong J, Isserow S
and Moulson N: New insights and recommendations for athletes with
hypertrophic cardiomyopathy. Can J Cardiol. 40:921–933.
2024.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Schaff HV and Wei X: Contemporary surgical
management of hypertrophic cardiomyopathy. Ann Thorac Surg.
117:271–281. 2024.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Maron BJ, Rowin EJ and Maron MS: Global
burden of hypertrophic cardiomyopathy. JACC Heart Fail. 6:376–378.
2018.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Ding X, Zhu C, Wang W, Li M, Ma C and Gao
B: SIRT1 is a regulator of autophagy: Implications for the
progression and treatment of myocardial ischemia-reperfusion.
Pharmacol Res. 199(106957)2024.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Rabinovich-Nikitin I, Kirshenbaum E and
Kirshenbaum LA: Autophagy, clock genes, and cardiovascular disease.
Can J Cardiol. 39:1772–1780. 2023.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Zhao J, Liu GW and Tao C: Hotspots and
future trends of autophagy in traditional chinese medicine: A
bibliometric analysis. Heliyon. 9(e20142)2023.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Sazonova EV, Petrichuk SV, Kopeina GS and
Zhivotovsky B: A link between mitotic defects and mitotic
catastrophe: Detection and cell fate. Biol Direct.
16(25)2021.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Byrnes K, Blessinger S, Bailey NT, Scaife
R, Liu G and Khambu B: Therapeutic regulation of autophagy in
hepatic metabolism. Acta Pharm Sin B. 12:33–49. 2022.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Xiong R, Li N, Chen L, Wang W, Wang B,
Jiang W and Geng Q: STING protects against cardiac dysfunction and
remodelling by blocking autophagy. Cell Commun Signal.
19(109)2021.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Ikeda S, Zablocki D and Sadoshima J: The
role of autophagy in death of cardiomyocytes. J Mol Cell Cardiol.
165:1–8. 2022.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Chen B, Yang Y, Wu J, Song J and Lu J:
microRNA-17-5p downregulation inhibits autophagy and myocardial
remodelling after myocardial infarction by targeting STAT3.
Autoimmunity. 55:43–51. 2022.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Zhu QH, Zhou YL, Yang M, Yang BB, Cao WT,
Yuan LM and Deng DQ: Reduced miR-99a-3p levels in systemic lupus
erythematosus may promote B cell proliferation via NCAPG and the
PI3K/AKT signaling pathway. Lupus. 33:365–374. 2024.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Voeltzke K, Scharov K, Funk CM, Kahler A,
Picard D, Hauffe L, Orth MF, Remke M, Esposito I, Kirchner T, et
al: EIF4EBP1 is transcriptionally upregulated by MYCN and
associates with poor prognosis in neuroblastoma. Cell Death Discov.
8(157)2022.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Wu ZR, Yan L, Liu YT, Cao L, Guo YH, Zhang
Y, Yao H, Cai L, Shang HB, Rui WW, et al: Inhibition of mTORC1 by
lncRNA H19 via disrupting 4E-BP1/Raptor interaction in pituitary
tumours. Nat Commun. 9(4624)2018.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Nelson ED, Benesch MG, Wu R, Ishikawa T
and Takabe K: High EIF4EBP1 expression reflects mTOR pathway
activity and cancer cell proliferation and is a biomarker for poor
breast cancer prognosis. Am J Cancer Res. 14:227–242.
2024.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Montalban-Bravo G, Thongon N,
Rodriguez-Sevilla JJ, Ma F, Ganan-Gomez I, Yang H, Kim YJ, Adema V,
Wildeman B, Tanaka T, et al: Targeting MCL1-driven anti-apoptotic
pathways overcomes blast progression after hypomethylating agent
failure in chronic myelomonocytic leukemia. Cell Rep Med.
5(101585)2024.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Mukherjee N, Katsnelson E, Brunetti TM,
Michel K, Couts KL, Lambert KA, Robinson WA, McCarter MD, Norris
DA, Tobin RP and Shellman YG: MCL1 inhibition targets myeloid
derived suppressors cells, promotes antitumor immunity and enhances
the efficacy of immune checkpoint blockade. Cell Death Dis.
15(198)2024.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Clerbaux LA, Cordier P, Desboeufs N, Unger
K, Leary P, Semere G, Boege Y, Chan LK, Desdouets C, Lopes M and
Weber A: Mcl-1 deficiency in murine livers leads to nuclear
polyploidisation and mitotic errors: Implications for
hepatocellular carcinoma. JHEP Rep. 5(100838)2023.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Chiou JT and Chang LS: Synergistic
cytotoxicity of decitabine and YM155 in leukemia cells through
upregulation of SLC35F2 and suppression of MCL1 and survivin
expression. Apoptosis. 29:503–520. 2024.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Boët E and Sarry JE: Targeting metabolic
dependencies fueling the TCA cycle to circumvent therapy resistance
in acute myeloid leukemia. Cancer Res. 84:950–952. 2024.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Mukherjee N, Schwan JV, Fujita M, Norris
DA and Shellman YG: Alternative treatments for melanoma: Targeting
BCL-2 family members to de-bulk and kill cancer stem cells. J
Invest Dermatol. 135:2155–2161. 2015.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Kapoor I, Bodo J, Hill BT, Hsi ED and
Almasan A: Targeting BCL-2 in B-cell malignancies and overcoming
therapeutic resistance. Cell Death Dis. 11(941)2020.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Neophytou CM, Trougakos IP, Erin N and
Papageorgis P: Apoptosis deregulation and the development of cancer
multi-drug resistance. Cancers (Basel). 13(4363)2021.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Zhan H, Huang F, Niu Q, Jiao M, Han X,
Zhang K, Ma W, Mi S, Guo S and Zhao Z: Downregulation of miR-128
ameliorates Ang II-induced cardiac remodeling via SIRT1/PIK3R1
multiple targets. Oxid Med Cell Longev.
2021(8889195)2021.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Dsouza NR, Cottrell CE, Davies OMT,
Tollefson MM, Frieden IJ, Basel D, Urrutia R, Drolet BA and
Zimmermann MT: Structural and dynamic analyses of pathogenic
variants in PIK3R1 reveal a shared mechanism associated among
cancer, undergrowth, and overgrowth syndromes. Life (Basel).
14(297)2024.PubMed/NCBI View Article : Google Scholar
|
|
57
|
De Bortoli M, Queisser A, Pham VC,
Dompmartin A, Helaers R, Boutry S, Claus C, De Roo AK, Hammer F,
Brouillard P, et al: Somatic loss-of-function PIK3R1 and activating
non-hotspot PIK3CA mutations associated with capillary malformation
with dilated veins (CMDV). J Invest Dermatol. 144:2066–2077.
2024.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Yu X, Xu C, Zou Y, Liu W, Xie Y and Wu C:
A prognostic metabolism-related gene signature associated with the
tumor immune microenvironment in neuroblastoma. Am J Cancer Res.
14:253–273. 2024.PubMed/NCBI View Article : Google Scholar
|
|
59
|
He B, Quan L, Li C, Yan W, Zhang Z, Zhou
L, Wei Q, Li Z, Mo J, Zhang Z, et al: Targeting ERBB2 and PIK3R1 as
a therapeutic strategy for dilated cardiomyopathy: A single-cell
sequencing and mendelian randomization analysis. Heliyon.
10(e25572)2024.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Maura F and Bergsagel PL: Molecular
pathogenesis of multiple myeloma: Clinical implications. Hematol
Oncol Clin North Am. 38:267–279. 2024.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Liu Z, Wang K, Jiang C, Chen Y, Liu F, Xie
M, Yim WY, Yao D, Qian X, Chen S, et al: Morusin alleviates aortic
valve calcification by inhibiting valve interstitial cell
senescence through Ccnd1/Trim25/Nrf2 axis. Adv Sci (Weinh).
19(e2307319)2024.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Zhang W and Hong W: Upregulation of
miR-519d-3p inhibits viability, proliferation, and G1/S cell cycle
transition of oral squamous cell carcinoma cells through targeting
CCND1. Cancer Biother Radiopharm. 39:153–163. 2024.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Quesada AE, Hu S, Li S, Toruner GA, Wei Q,
Loghavi S, Ok CY, Jain P, Thakral B, Nwogbo OV, et al: Optical
genomic mapping is a helpful tool for detecting CCND1
rearrangements in CD5-negative small B-cell lymphoma: Two cases of
leukemic non-nodal mantle cell lymphoma. Hum Pathol. 144:71–76.
2024.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Han B, Chen J, Chen S, Shen X, Hou L, Fang
J and Lian M: PPARG and the PTEN-PI3K/AKT signaling axis may
cofunction in promoting chemosensitivity in hypopharyngeal squamous
cell carcinoma. PPAR Res. 2024(2271214)2024.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Qin Y, Ashrafizadeh M, Mongiardini V,
Grimaldi B, Crea F, Rietdorf K, Győrffy B, Klionsky DJ, Ren J,
Zhang W and Zhang X: Autophagy and cancer drug resistance in
dialogue: Pre-clinical and clinical evidence. Cancer Lett.
570(216307)2023.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Jia Q, Li B, Wang X, Ma Y and Li G:
Comprehensive analysis of peroxisome proliferator-activated
receptors to predict the drug resistance, immune microenvironment,
and prognosis in stomach adenocarcinomas. PeerJ.
12(e17082)2024.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Sun Y, Ma J, Lin J, Sun D, Song P, Shi L,
Li H, Wang R, Wang Z and Liu S: Circular RNA circ_ASAP2 regulates
drug sensitivity and functional behaviors of cisplatin-resistant
gastric cancer cells by the miR-330-3p/NT5E axis. Anticancer Drugs.
32:950–961. 2021.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Choi JC, Muchir A, Wu W, Iwata S, Homma S,
Morrow JP and Worman HJ: Temsirolimus activates autophagy and
ameliorates cardiomyopathy caused by lamin A/C gene mutation. Sci
Transl Med. 4(144ra102)2012.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Gan T, Qu S, Zhang H and Zhou XJ:
Modulation of the immunity and inflammation by autophagy. MedComm
(2020). 4(e311)2023.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Herb M, Gluschko A and Schramm M:
LC3-associated phagocytosis-the highway to hell for phagocytosed
microbes. Semin Cell Dev Biol. 101:68–76. 2020.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Castillo EF, Dekonenko A, Arko-Mensah J,
Mandell MA, Dupont N, Jiang S, Delgado-Vargas M, Timmins GS,
Bhattacharya D, Yang H, et al: Autophagy protects against active
tuberculosis by suppressing bacterial burden and inflammation. Proc
Natl Acad Sci USA. 109:E3168–E3176. 2012.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Ma CS: Human T follicular helper cells in
primary immunodeficiency: Quality just as important as quantity. J
Clin Immunol. 36 (Suppl 1):S40–S47. 2016.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Singh SR, Zech ATL, Geertz B,
Reischmann-Düsener S, Osinska H, Prondzynski M, Krämer E, Meng Q,
Redwood C, van der Velden J, et al: Activation of autophagy
ameliorates cardiomyopathy in mybpc3-targeted knockin mice. Circ
Heart Fail. 10(e004140)2017.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Hassoun R, Budde H, Zhazykbayeva S, Herwig
M, Sieme M, Delalat S, Mostafi N, Gömöri K, Tangos M, Jarkas M, et
al: Stress activated signalling impaired protein quality control
pathways in human hypertrophic cardiomyopathy. Int J Cardiol.
344:160–169. 2021.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Simpson JE and Gammoh N: Autophagy
cooperates with PDGFRA to support oncogenic growth signaling.
Autophagy. 20:1901–1902. 2024.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Ravikumar B, Sarkar S, Davies JE, Futter
M, Garcia-Arencibia M, Green-Thompson ZW, Jimenez-Sanchez M,
Korolchuk VI, Lichtenberg M, Luo S, et al: Regulation of Mammalian
autophagy in physiology and pathophysiology. Physiol Rev.
90:1383–1435. 2010.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Marian AJ and Braunwald E: Hypertrophic
cardiomyopathy: Genetics, pathogenesis, clinical manifestations,
diagnosis, and therapy. Circ Res. 121:749–770. 2017.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Tannous P, Zhu H, Johnstone JL, Shelton
JM, Rajasekaran NS, Benjamin IJ, Nguyen L, Gerard RD, Levine B,
Rothermel BA and Hill JA: Autophagy is an adaptive response in
desmin-related cardiomyopathy. Proc Natl Acad Sci USA.
105:9745–9750. 2008.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Sheng SY, Li JM, Hu XY and Wang Y:
Regulated cell death pathways in cardiomyopathy. Acta Pharmacol
Sin. 44:1521–1535. 2023.PubMed/NCBI View Article : Google Scholar
|
|
80
|
Buss SJ, Muenz S, Riffel JH, Malekar P,
Hagenmueller M, Weiss CS, Bea F, Bekeredjian R, Schinke-Braun M,
Izumo S, et al: Beneficial effects of Mammalian target of rapamycin
inhibition on left ventricular remodeling after myocardial
infarction. J Am Coll Cardiol. 54:2435–2446. 2009.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Sciarretta S, Volpe M and Sadoshima J:
Mammalian target of rapamycin signaling in cardiac physiology and
disease. Circ Res. 114:549–564. 2014.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Marin TM, Keith K, Davies B, Conner DA,
Guha P, Kalaitzidis D, Wu X, Lauriol J, Wang B, Bauer M, et al:
Rapamycin reverses hypertrophic cardiomyopathy in a mouse model of
LEOPARD syndrome-associated PTPN11 mutation. J Clin Invest.
121:1026–1043. 2011.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Sciarretta S, Forte M, Frati G and
Sadoshima J: New insights into the role of mTOR signaling in the
cardiovascular system. Circ Res. 122:489–505. 2018.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Yang Y, Du J, Xu R, Shen Y, Yang D, Li D,
Hu H, Pei H and Yang Y: Melatonin alleviates angiotensin-II-induced
cardiac hypertrophy via activating MICU1 pathway. Aging (Albany
NY). 13:493–515. 2020.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Chen S, Sun P, Li Y, Shen W, Wang C, Zhao
P, Cui H, Xue JY and Du GQ: Melatonin activates the Mst1-Nrf2
signaling to alleviate cardiac hypertrophy in pulmonary arterial
hypertension. Eur J Pharmacol. 933(175262)2022.PubMed/NCBI View Article : Google Scholar
|