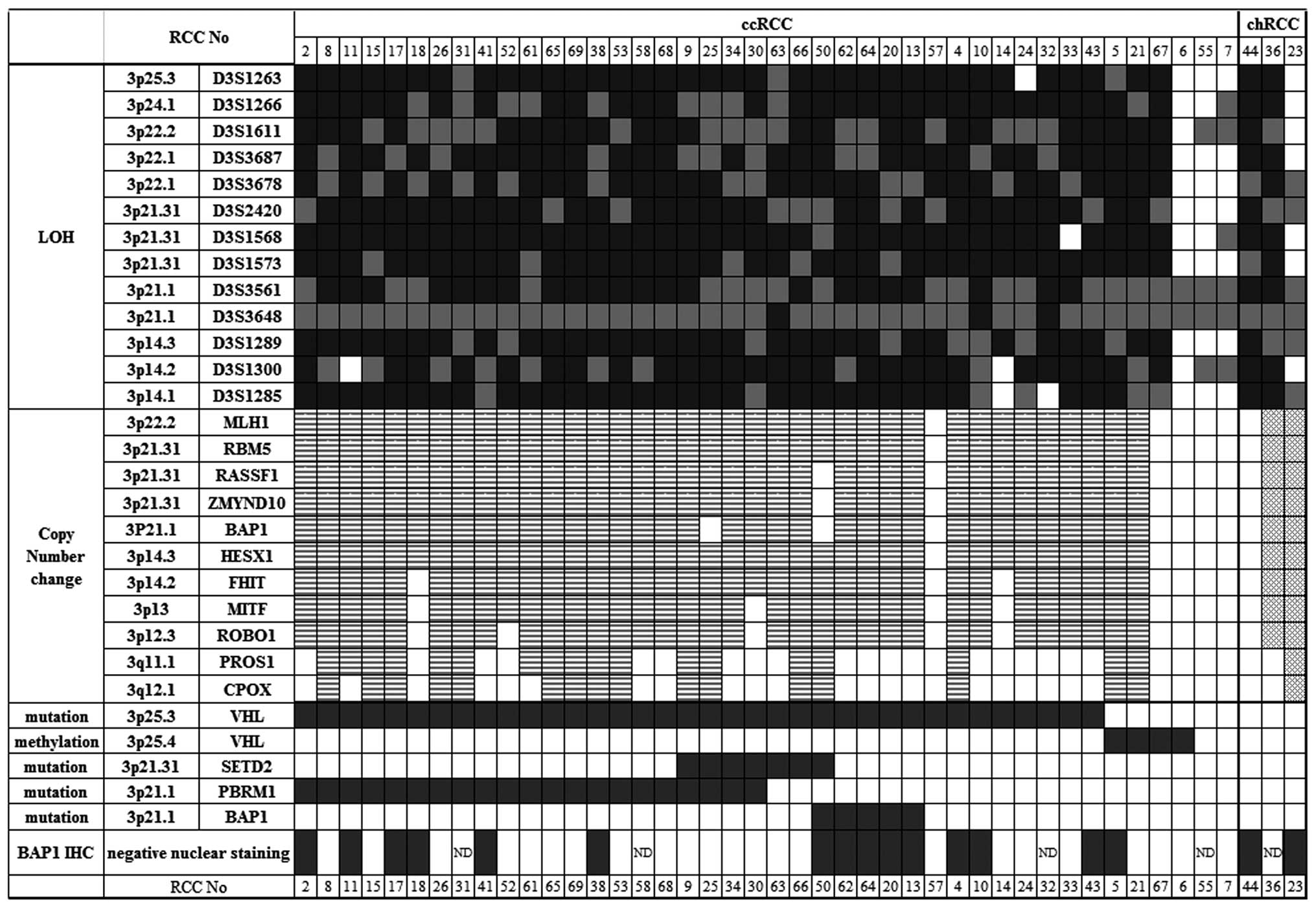
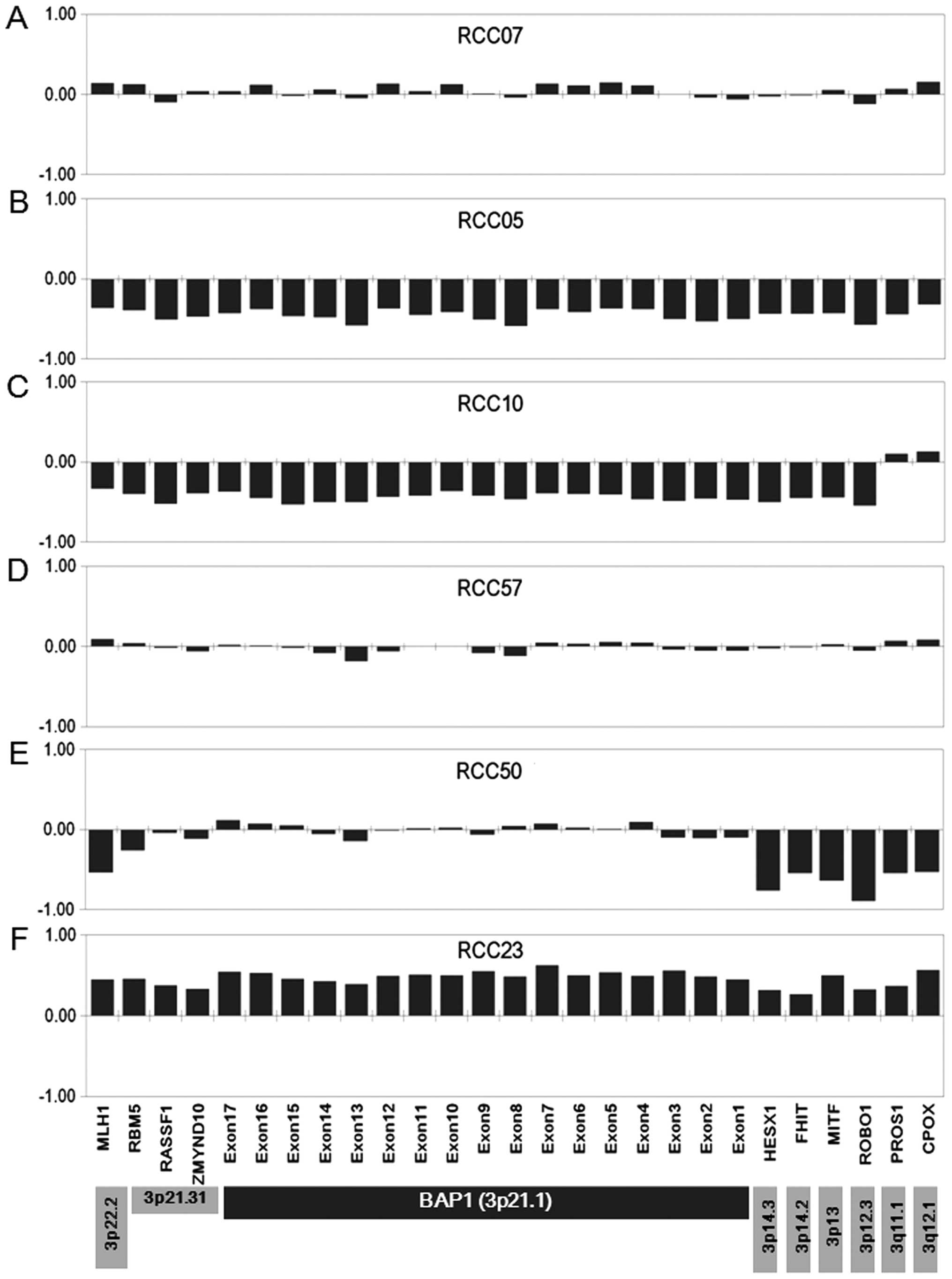
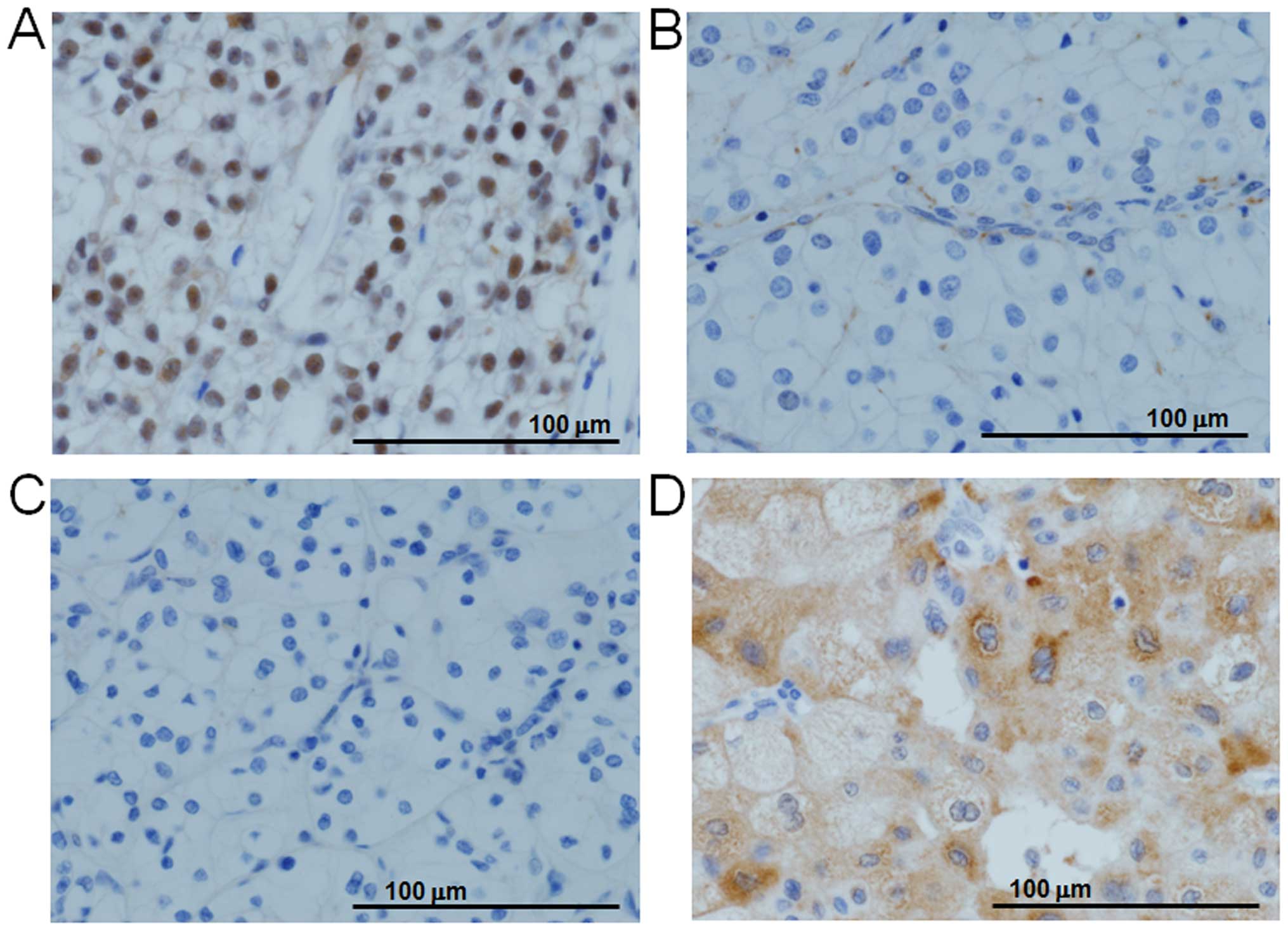
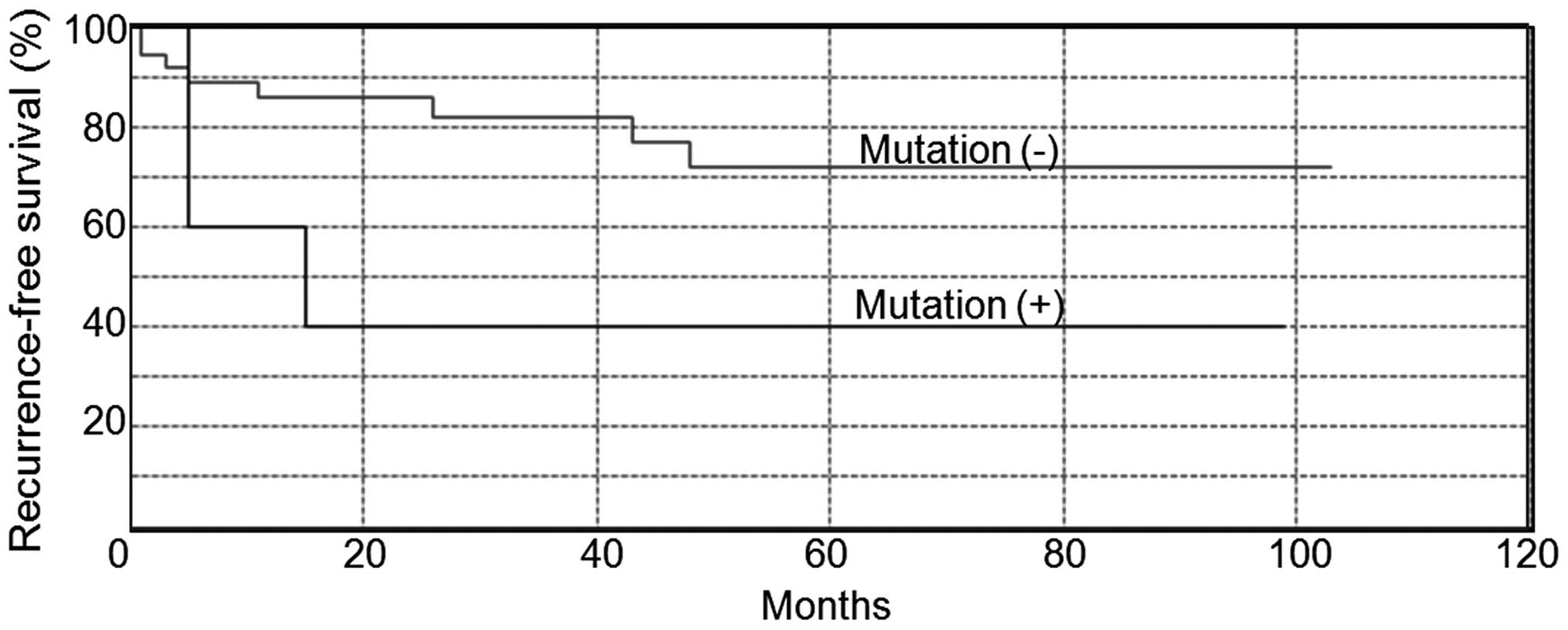
|
1
|
Angeloni D: Molecular analysis of
deletions in human chromosome 3p21 and the role of resident cancer
genes in disease. Brief Funct Genomics Proteomics. 6:19–39. 2007.
View Article : Google Scholar
|
|
2
|
Kok K, Naylor SL and Buys CH: Deletions of
the short arm of chromosome 3 in solid tumors and the search for
suppressor genes. Adv Cancer Res. 71:27–92. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gnarra JR, Tory K, Weng Y, Schmidt L, Wei
MH, Li H, Latif F, Liu S, Chen F, Duh FM, et al: Mutations of the
VHL tumour suppressor gene in renal carcinoma. Nat Genet. 7:85–90.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Clifford SC, Prowse AH, Affara NA, Buys CH
and Maher ER: Inactivation of the von Hippel-Lindau (VHL) tumour
suppressor gene and allelic losses at chromosome arm 3p in primary
renal cell carcinoma: Evidence for a VHL-independent pathway in
clear cell renal tumourigenesis. Genes Chromosomes Cancer.
22:200–209. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sükösd F, Kuroda N, Beothe T, Kaur AP and
Kovacs G: Deletion of chromosome 3p14.2-p25 involving the VHL and
FHIT genes in conventional renal cell carcinoma. Cancer Res.
63:455–457. 2003.PubMed/NCBI
|
|
6
|
Singh RB and Amare Kadam PS: Investigation
of tumor suppressor genes apart from VHL on 3p by deletion mapping
in sporadic clear cell renal cell carcinoma (cRCC). Urol Oncol.
31:1333–1342. 2013. View Article : Google Scholar
|
|
7
|
Varela I, Tarpey P, Raine K, Huang D, Ong
CK, Stephens P, Davies H, Jones D, Lin ML, Teague J, et al: Exome
sequencing identifies frequent mutation of the SWI/SNF complex gene
PBRM1 in renal carcinoma. Nature. 469:539–542. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dalgliesh GL, Furge K, Greenman C, Chen L,
Bignell G, Butler A, Davies H, Edkins S, Hardy C, Latimer C, et al:
Systematic sequencing of renal carcinoma reveals inactivation of
histone modifying genes. Nature. 463:360–363. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Peña-Llopis S, Vega-Rubín-de-Celis S, Liao
A, Leng N, Pavía-Jiménez A, Wang S, Yamasaki T, Zhrebker L,
Sivanand S, Spence P, et al: BAP1 loss defines a new class of renal
cell carcinoma. Nat Genet. 44:751–759. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Guo G, Gui Y, Gao S, Tang A, Hu X, Huang
Y, Jia W, Li Z, He M, Sun L, et al: Frequent mutations of genes
encoding ubiquitin-mediated proteolysis pathway components in clear
cell renal cell carcinoma. Nat Genet. 44:17–19. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sato Y, Yoshizato T, Shiraishi Y, Maekawa
S, Okuno Y, Kamura T, Shimamura T, Sato-Otsubo A, Nagae G, Suzuki
H, et al: Integrated molecular analysis of clear-cell renal cell
carcinoma. Nat Genet. 45:860–867. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Testa JR, Cheung M, Pei J, Below JE, Tan
Y, Sementino E, Cox NJ, Dogan AU, Pass HI, Trusa S, et al: Germline
BAP1 mutations predispose to malignant mesothelioma. Nat Genet.
43:1022–1025. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wiesner T, Obenauf AC, Murali R, Fried I,
Griewank KG, Ulz P, Windpassinger C, Wackernagel W, Loy S, Wolf I,
et al: Germline mutations in BAP1 predispose to melanocytic tumors.
Nat Genet. 43:1018–1021. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Abdel-Rahman MH, Pilarski R, Cebulla CM,
Massengill JB, Christopher BN, Boru G, Hovland P and Davidorf FH:
Germline BAP1 mutation predisposes to uveal melanoma, lung
adenocarcinoma, meningioma, and other cancers. J Med Genet.
48:856–859. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Carbone M, Ferris LK, Baumann F,
Napolitano A, Lum CA, Flores EG, Gaudino G, Powers A,
Bryant-Greenwood P, Krausz T, et al: BAP1 cancer syndrome:
Malignant mesothelioma, uveal and cutaneous melanoma, and MBAITs. J
Transl Med. 10:1792012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Popova T, Hebert L, Jacquemin V, Gad S,
Caux-Moncoutier V, Dubois-d'Enghien C, Richaudeau B, Renaudin X,
Sellers J, Nicolas A, et al: Germline BAP1 mutations predispose to
renal cell carcinomas. Am J Hum Genet. 92:974–980. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Farley MN, Schmidt LS, Mester JL,
Peña-Llopis S, Pavia-Jimenez A, Christie A, Vocke CD, Ricketts CJ,
Peterson J, Middelton L, et al: A novel germline mutation in BAP1
predisposes to familial clear-cell renal cell carcinoma. Mol Cancer
Res. 11:1061–1071. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liao L, Testa JR and Yang H: The roles of
chromatin-remodelers and epigenetic modifiers in kidney cancer.
Cancer Genet. 208:206–214. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang SS, Gu YF, Wolff N, Stefanius K,
Christie A, Dey A, Hammer RE, Xie XJ, Rakheja D, Pedrosa I, et al:
Bap1 is essential for kidney function and cooperates with Vhl in
renal tumorigenesis. Proc Natl Acad Sci USA. 111:16538–16543. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Peña-Llopis S, Christie A, Xie XJ and
Brugarolas J: Cooperation and antagonism among cancer genes: The
renal cancer paradigm. Cancer Res. 73:4173–4179. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Scheuermann JC, de Ayala Alonso AG, Oktaba
K, Ly-Hartig N, McGinty RK, Fraterman S, Wilm M, Muir TW and Müller
J: Histone H2A deubiquitinase activity of the Polycomb repressive
complex PR-DUB. Nature. 465:243–247. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Harbour JW, Onken MD, Roberson ED, Duan S,
Cao L, Worley LA, Council ML, Matatall KA, Helms C and Bowcock AM:
Frequent mutation of BAP1 in metastasizing uveal melanomas.
Science. 330:1410–1413. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ewens KG, Kanetsky PA, Richards-Yutz J,
Purrazzella J, Shields CL, Ganguly T and Ganguly A: Chromosome 3
status combined with BAP1 and EIF1AX mutation profiles are
associated with metastasis in uveal melanoma. Invest Ophthalmol Vis
Sci. 55:5160–5167. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yoshikawa Y, Sato A, Tsujimura T, Emi M,
Morinaga T, Fukuoka K, Yamada S, Murakami A, Kondo N, Matsumoto S,
et al: Frequent inactivation of the BAP1 gene in epithelioid-type
malignant mesothelioma. Cancer Sci. 103:868–874. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Emi M, Yoshikawa Y, Sato C, Sato A, Sato
H, Kato T, Tsujimura T, Hasegawa S, Nakano T and Hashimoto-Tamaoki
T: Frequent genomic rearrangements of BRCA1 associated protein-1
(BAP1) gene in Japanese malignant mesothelioma-characterization of
deletions at exon level. J Hum Genet. 60:647–649. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bott M, Brevet M, Taylor BS, Shimizu S,
Ito T, Wang L, Creaney J, Lake RA, Zakowski MF, Reva B, et al: The
nuclear deubiquitinase BAP1 is commonly inactivated by somatic
mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat
Genet. 43:668–672. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zauderer MG, Bott M, McMillan R, Sima CS,
Rusch V, Krug LM and Ladanyi M: Clinical characteristics of
patients with malignant pleural mesothelioma harboring somatic BAP1
mutations. J Thorac Oncol. 8:1430–1433. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Arzt L, Quehenberger F, Halbwedl I,
Mairinger T and Popper HH: BAP1 protein is a progression factor in
malignant pleural mesothelioma. Pathol Oncol Res. 20:145–151. 2014.
View Article : Google Scholar
|
|
29
|
Nasu M, Emi M, Pastorino S, Tanji M,
Powers A, Luk H, Baumann F, Zhang YA, Gazdar A, et al: High
incidence of somatic BAP1 alterations in sporadic malignant
mesothelioma. J Thorac Oncol. 10:565–576. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mori T, Sumii M, Fujishima F, Ueno K, Emi
M, Nagasaki M, Ishioka C and Chiba N: Somatic alteration and
depleted nuclear expression of BAP1 in human esophageal squamous
cell carcinoma. Cancer Sci. 106:1118–1129. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yu H, Mashtalir N, Daou S, Hammond-Martel
I, Ross J, Sui G, Hart GW, Rauscher FJ III, Drobetsky E, Milot E,
et al: The ubiquitin carboxyl hydrolase BAP1 forms a ternary
complex with YY1 and HCF-1 and is a critical regulator of gene
expression. Mol Cell Biol. 30:5071–5085. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mashtalir N, Daou S, Barbour H, Sen NN,
Gagnon J, Hammond-Martel I, Dar HH, Therrien M and Affar B:
Autodeubiquitination protects the tumor suppressor BAP1 from
cytoplasmic sequestration mediated by the atypical ubiquitin ligase
UBE2O. Mol Cell. 54:392–406. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dietrich W, Katz H, Lincoln SE, Shin HS,
Friedman J, Dracopoli NC and Lander ES: A genetic map of the mouse
suitable for typing intraspecific crosses. Genetics. 131:423–447.
1992.PubMed/NCBI
|
|
34
|
Cingolani P, Platts A, Wang L, Coon M,
Nguyen T, Wang L, Land SJ, Lu X and Ruden DM: A program for
annotating and predicting the effects of single nucleotide
polymorphisms, SnpEff: SNPs in the genome of Drosophila
melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 6:80–92.
2012. View Article : Google Scholar
|
|
35
|
Liu X, Jian X and Boerwinkle E: dbNSFP
v2.0: A database of human non-synonymous SNVs and their functional
predictions and annotations. Hum Mutat. 34:E2393–E2402. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Herman JG, Graff JR, Myöhänen S, Nelkin BD
and Baylin SB: Methylation-specific PCR: A novel PCR assay for
methylation status of CpG islands. Proc Natl Acad Sci USA.
93:9821–9826. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kapur P, Christie A, Raman JD, Then MT,
Nuhn P, Buchner A, Bastian P, Seitz C, Shariat SF, Bensalah K, et
al: BAP1 immunohistochemistry predicts outcomes in a
multi-institutional cohort with clear cell renal cell carcinoma. J
Urol. 191:603–610. 2014. View Article : Google Scholar
|
|
38
|
Gerlinger M, Horswell S, Larkin J, Rowan
AJ, Salm MP, Varela I, Fisher R, McGranahan N, Matthews N, Santos
CR, et al: Genomic architecture and evolution of clear cell renal
cell carcinomas defined by multiregion sequencing. Nat Genet.
46:225–233. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ibragimova I, Maradeo ME, Dulaimi E and
Cairns P: Aberrant promoter hypermethylation of PBRM1, BAP1, SETD2,
KDM6A and other chromatin-modifying genes is absent or rare in
clear cell RCC. Epigenetics. 8:486–493. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hakimi AA, Ostrovnaya I, Reva B, Schultz
N, Chen YB, Gonen M, Liu H, Takeda S, Voss MH, Tickoo SK, et al;
ccRCC Cancer Genome Atlas (KIRC TCGA) Research Network
investigators. Adverse outcomes in clear cell renal cell carcinoma
with mutations of 3p21 epigenetic regulators BAP1 and SETD2: A
report by MSKCC and the KIRC TCGA research network. Clin Cancer
Res. 19:3259–3267. 2013. View Article : Google Scholar : PubMed/NCBI
|