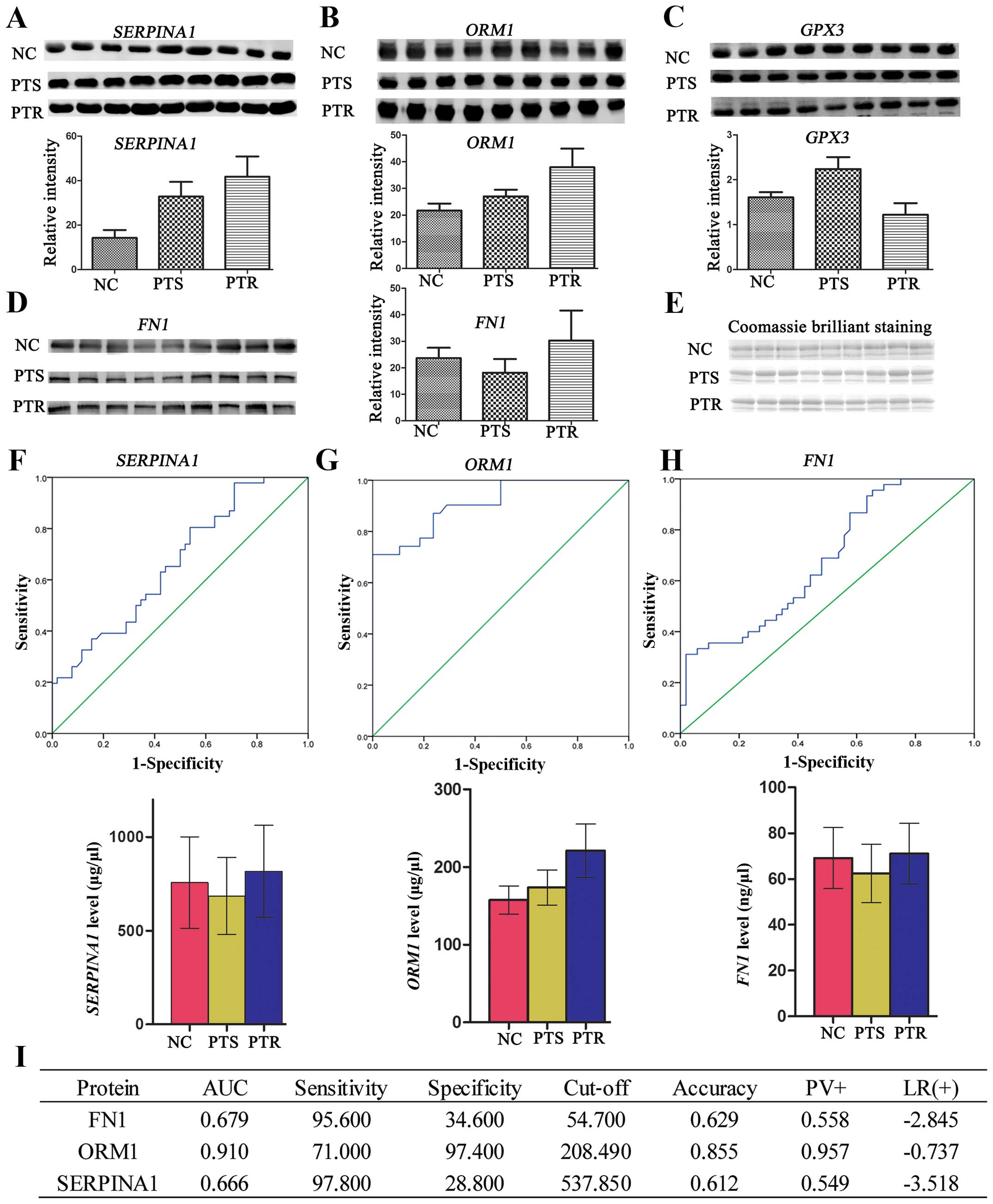
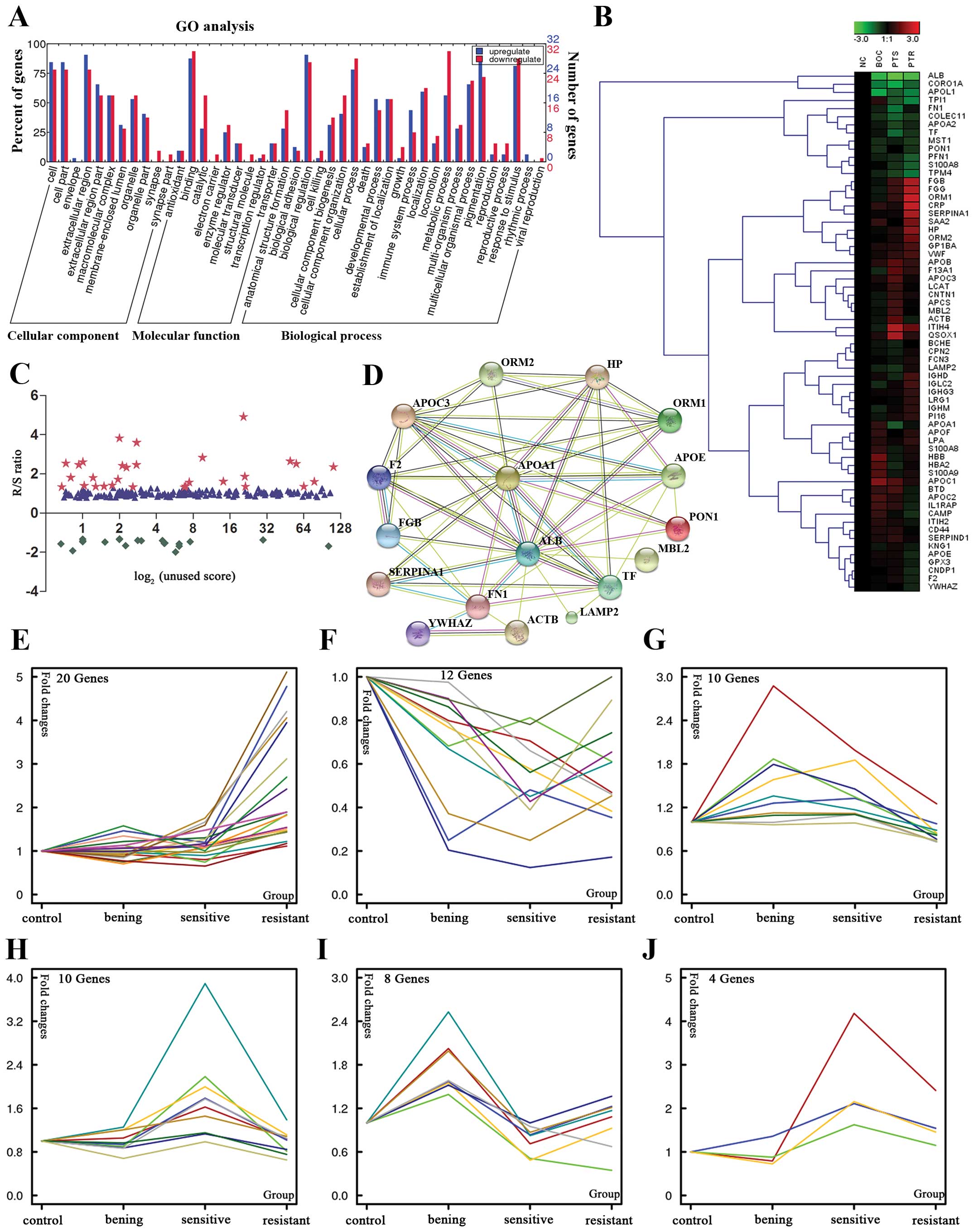
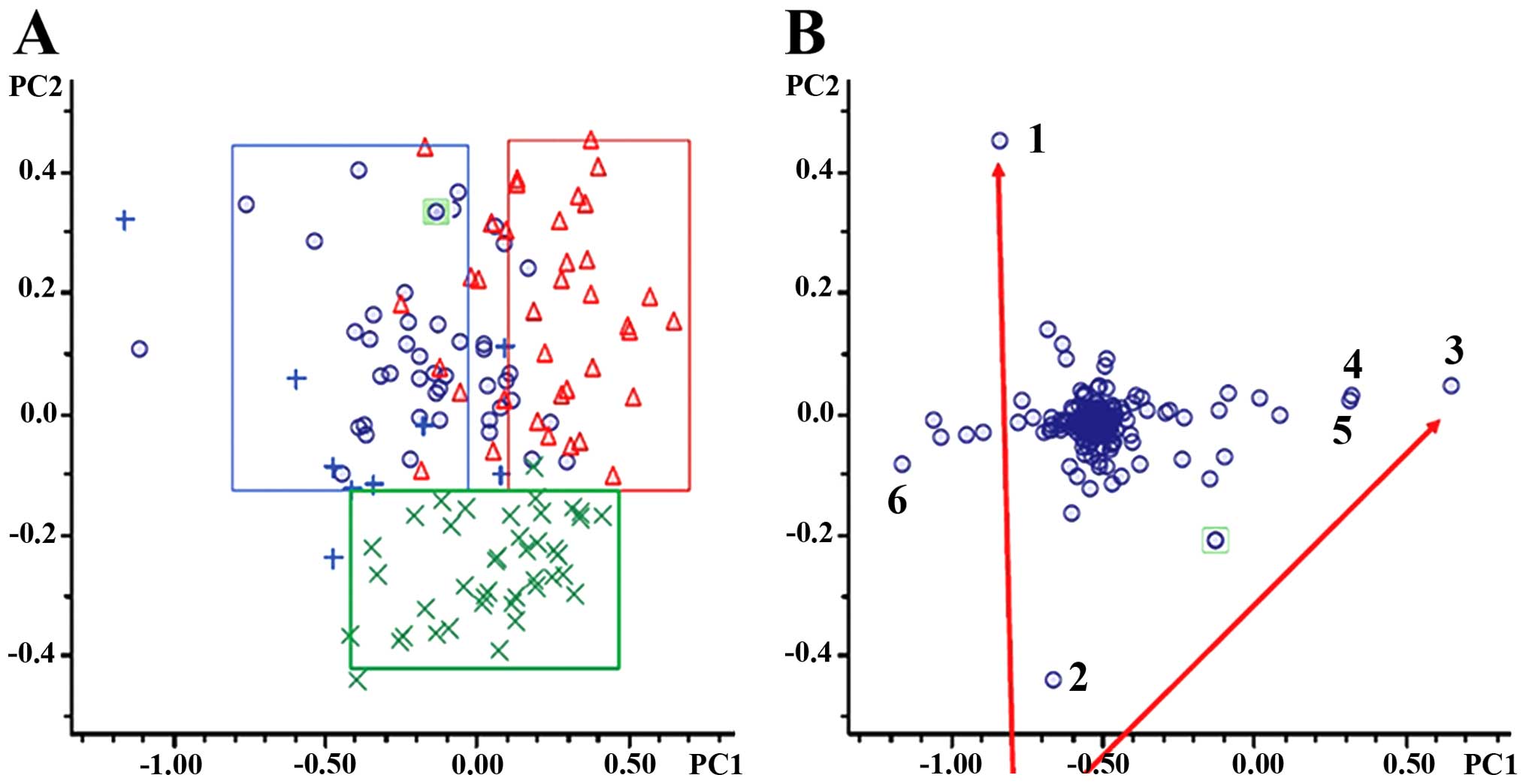
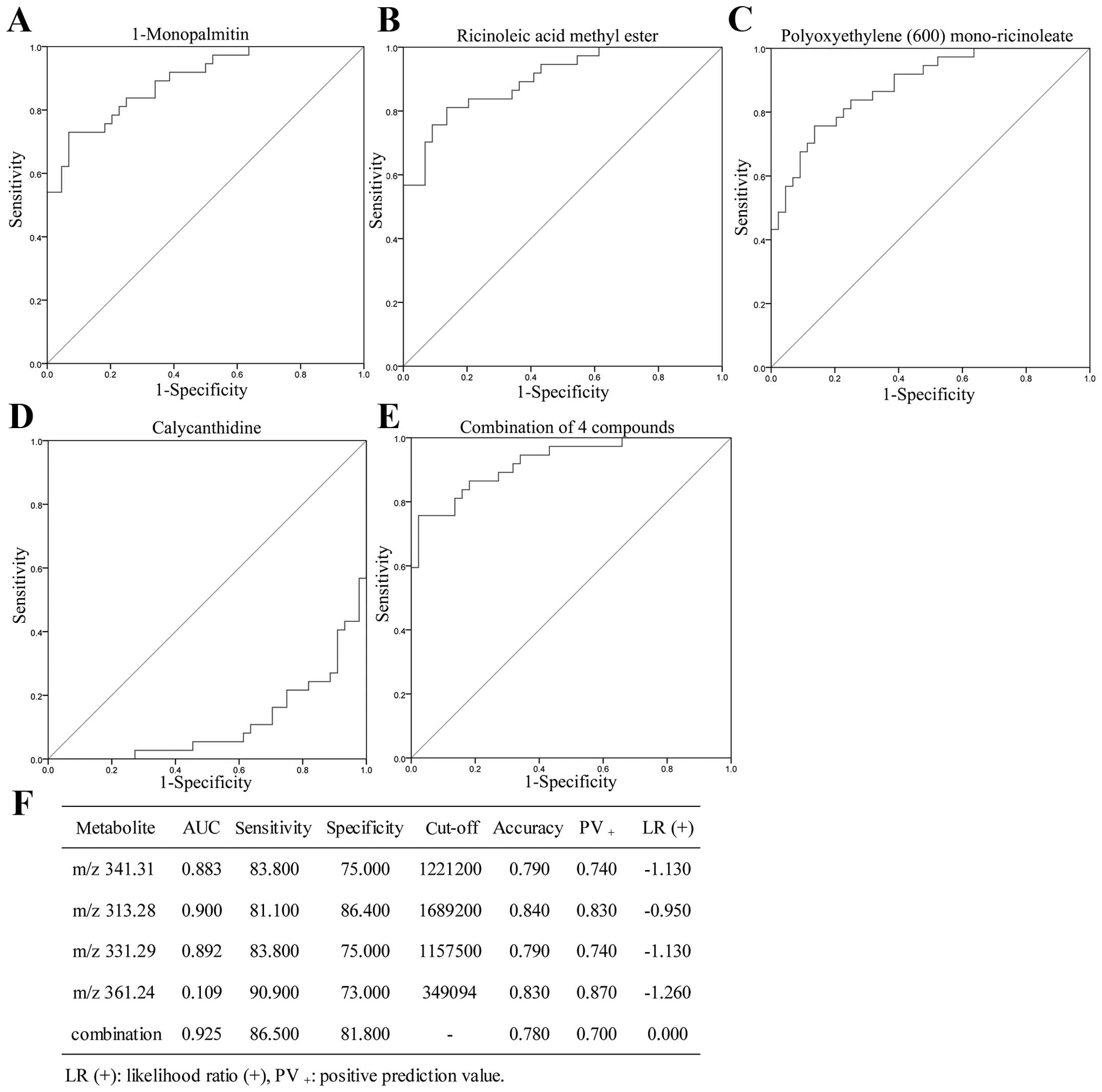
|
1
|
Leonessa F and Clarke R: ATP binding
cassette transporters and drug resistance in breast cancer. Endocr
Relat Cancer. 10:43–73. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gottesman MM, Fojo T and Bates SE:
Multidrug resistance in cancer: Role of ATP-dependent transporters.
Nat Rev Cancer. 2:48–58. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang D, Wu M, Nelson DE, Pasula R and
Martin WJ II: Alpha-1-antitrypsin expression in the lung is
increased by airway delivery of gene-transfected macrophages. Gene
Ther. 10:2148–2152. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hansson M, Jönsson S, Persson AM, Calafat
J, Tapper H and Olsson I: Targeting proteins to secretory lysosomes
of natural killer cells as a principle for immunoregulation. Mol
Immunol. 40:363–372. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cekmen M, Evereklioglu C, Er H, Inalöz HS,
Doganay S, Türköz Y and Ozerol IH: Vascular endothelial growth
factor levels are increased and associated with disease activity in
patients with Behçet’s syndrome. Int J Dermatol. 42:870–875. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lu Y, Liu P, Wen W, Grubbs CJ, Townsend
RR, Malone JP, Lubet RA and You M: Cross-species comparison of
orthologous gene expression in human bladder cancer and
carcinogen-induced rodent models. Am J Transl Res. 3:8–27.
2010.PubMed/NCBI
|
|
7
|
Normandin K, Péant B, Le Page C, de
Ladurantaye M, Ouellet V, Tonin PN, Provencher DM and Mes-Masson
AM: Protease inhibitor SERPINA1 expression in epithelial ovarian
cancer. Clin Exp Metastasis. 27:55–69. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
de la Fuente MT, Casanova B, Garcia-Gila
M, Silva A and Garcia-Pardo A: Fibronectin interaction with
alpha4beta1 integrin prevents apoptosis in B cell chronic
lymphocytic leukemia: Correlation with Bcl-2 and Bax. Leukemia.
13:266–274. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Schwarzbauer JE and DeSimone DW:
Fibronectins, their fibrillogenesis, and in vivo functions. Cold
Spring Harb Perspect Biol. 3:32011. View Article : Google Scholar
|
|
10
|
Tapper J, Kettunen E, El-Rifai W, Seppälä
M, Andersson LC and Knuutila S: Changes in gene expression during
progression of ovarian carcinoma. Cancer Genet Cytogenet. 128:1–6.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mutlu P, Ural AU and Gündüz U:
Differential gene expression analysis related to extracellular
matrix components in drug-resistant RPMI-8226 cell line. Biomed
Pharmacother. 66:228–231. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ahmed N, Riley C, Rice G and Quinn M: Role
of integrin receptors for fibronectin, collagen and laminin in the
regulation of ovarian carcinoma functions in response to a matrix
microenvironment. Clin Exp Metastasis. 22:391–402. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Akiyama SK, Olden K and Yamada KM:
Fibronectin and integrins in invasion and metastasis. Cancer
Metastasis Rev. 14:173–189. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shibata K, Kikkawa F, Nawa A, Suganuma N
and Hamaguchi M: Fibronectin secretion from human peritoneal tissue
induces Mr 92,000 type IV collagenase expression and invasion in
ovarian cancer cell lines. Cancer Res. 57:5416–5420.
1997.PubMed/NCBI
|
|
15
|
Lou X, Han X, Jin C, Tian W, Yu W, Ding D,
Cheng L, Huang B, Jiang H and Lin B: SOX2 targets fibronectin 1 to
promote cell migration and invasion in ovarian cancer: New
molecular leads for therapeutic intervention. OMICS. 17:510–518.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jinawath N, Vasoontara C, Jinawath A, Fang
X, Zhao K, Yap KL, Guo T, Lee CS, Wang W, Balgley BM, et al:
Oncoproteomic analysis reveals co-upregulation of RELA and STAT5 in
carboplatin resistant ovarian carcinoma. PLoS One. 5:e111982010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Qian P, Zuo Z, Wu Z, Meng X, Li G, Wu Z,
Zhang W, Tan S, Pandey V, Yao Y, et al: Pivotal role of reduced
let-7g expression in breast cancer invasion and metastasis. Cancer
Res. 71:6463–6474. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Waalkes S, Atschekzei F, Kramer MW,
Hennenlotter J, Vetter G, Becker JU, Stenzl A, Merseburger AS,
Schrader AJ, Kuczyk MA, et al: Fibronectin 1 mRNA expression
correlates with advanced disease in renal cancer. BMC Cancer.
10:5032010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yokomizo A, Takakura M, Kanai Y, et al:
Use of quantitative shotgun proteomics to identify fibronectin 1 as
a potential plasma biomarker for clear cell carcinoma of the
kidney. Cancer biomarkers: Section. Dis Markers. 10:175–183.
2011.
|
|
20
|
He X, Wang Y, Zhang W, Li H, Luo R, Zhou
Y, Li C, Liao M, Huang H, Lv X, et al: Screening differential
expression of serum proteins in AFP-negative HBV-related
hepatocellular carcinoma using iTRAQ -MALDI-MS/MS. Neoplasma.
61:17–26. 2014. View Article : Google Scholar
|
|
21
|
Cochrane DR, Spoelstra NS, Howe EN,
Nordeen SK and Richer JK: MicroRNA-200c mitigates invasiveness and
restores sensitivity to microtubule-targeting chemotherapeutic
agents. Mol Cancer Ther. 8:1055–1066. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu L, Zou J, Wang Q, Yin FQ, Zhang W and
Li L: Novel microRNAs expression of patients with chemotherapy
drug-resistant and chemotherapy-sensitive epithelial ovarian
cancer. Tumour Biol. 35:7713–7717. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang X, Yang JJ, Kim YS, Kim KY, Ahn WS
and Yang S: An 8-gene signature, including methylated and
down-regulated glutathione peroxidase 3, of gastric cancer. Int J
Oncol. 36:405–414. 2010.PubMed/NCBI
|
|
24
|
Bottaro DP, Rubin JS, Faletto DL, Chan AM,
Kmiecik TE, Vande Woude GF and Aaronson SA: Identification of the
hepatocyte growth factor receptor as the c-met proto-oncogene
product. Science. 251:802–804. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Park M, Dean M, Kaul K, Braun MJ, Gonda MA
and Vande Woude G: Sequence of MET protooncogene cDNA has features
characteristic of the tyrosine kinase family of growth-factor
receptors. Proc Natl Acad Sci USA. 84:6379–6383. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Azad MB, Chen Y and Gibson SB: Regulation
of autophagy by reactive oxygen species (ROS): Implications for
cancer progression and treatment. Antioxid Redox Signal.
11:777–790. 2009. View Article : Google Scholar
|
|
27
|
Davis W Jr, Ronai Z and Tew KD: Cellular
thiols and reactive oxygen species in drug-induced apoptosis. J
Pharmacol Exp Ther. 296:1–6. 2001.
|
|
28
|
Lee HJ, Do JH, Bae S, Yang S, Zhang X, Lee
A, Choi YJ, Park DC and Ahn WS: Immunohistochemical evidence for
the over-expression of Glutathione peroxidase 3 in clear cell type
ovarian adenocarcinoma. Med Oncol. 28(Suppl 1): S522–S527. 2011.
View Article : Google Scholar
|
|
29
|
Saga Y, Ohwada M, Suzuki M, Konno R,
Kigawa J, Ueno S and Mano H: Glutathione peroxidase 3 is a
candidate mechanism of anticancer drug resistance of ovarian clear
cell adenocarcinoma. Oncol Rep. 20:1299–1303. 2008.PubMed/NCBI
|
|
30
|
Hochepied T, Berger FG, Baumann H and
Libert C: Alpha(1)-acid glycoprotein: An acute phase protein with
inflammatory and immunomodulating properties. Cytokine Growth
Factor Rev. 14:25–34. 2003. View Article : Google Scholar
|
|
31
|
Fournier T, Medjoubi-N N and Porquet D:
Alpha-1-acid glycoprotein. Biochim Biophys Acta. 1482:157–171.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fan C, Stendahl U, Stjernberg N and
Beckman L: Association between orosomucoid types and cancer.
Oncology. 52:498–500. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Duché JC, Urien S, Simon N, Malaurie E,
Monnet I and Barré J: Expression of the genetic variants of human
alpha-1-acid glycoprotein in cancer. Clin Biochem. 33:197–202.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tilg H, Vannier E, Vachino G, Dinarello CA
and Mier JW: Antiinflammatory properties of hepatic acute phase
proteins: Preferential induction of interleukin 1 (IL-1) receptor
antagonist over IL-1 beta synthesis by human peripheral blood
mononuclear cells. J Exp Med. 178:1629–1636. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Matsumoto K, Nishi K, Kikuchi M, Watanabe
H, Nakajou K, Komori H, Kadowaki D, Suenaga A, Maruyama T and
Otagiri M: Receptor-mediated uptake of human alpha1-acid
glycoprotein into liver parenchymal cells in mice. Drug Metab
Pharmacokinet. 25:101–107. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lee YS, Choi JW, Hwang I, Lee JW, Lee JH,
Kim AY, Huh JY, Koh YJ, Koh GY, Son HJ, et al: Adipocytokine
orosomucoid integrates inflammatory and metabolic signals to
preserve energy homeostasis by resolving immoderate inflammation. J
Biol Chem. 285:22174–22185. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gunnarsson P, Levander L, Påhlsson P and
Grenegård M: The acute-phase protein alpha 1-acid glycoprotein
(AGP) induces rises in cytosolic Ca2+ in neutrophil
granulocytes via sialic acid binding immunoglobulin-like lectins
(siglecs). FASEB J. 21:4059–4069. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Atemezem A, Mbemba E, Vassy R, Slimani H,
Saffar L and Gattegno L: Human alpha1-acid glycoprotein binds to
CCR5 expressed on the plasma membrane of human primary macrophages.
Biochem J. 356:121–128. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ligresti G, Aplin AC, Dunn BE, Morishita A
and Nicosia RF: The acute phase reactant orosomucoid-1 is a bimodal
regulator of angiogenesis with time- and context-dependent
inhibitory and stimulatory properties. PLoS One. 7:e413872012.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Irmak S, Oliveira-Ferrer L, Singer BB,
Ergün S and Tilki D: Pro-angiogenic properties of orosomucoid
(ORM). Exp Cell Res. 315:3201–3209. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Goh BC, Lee SC, Wang LZ, Fan L, Guo JY,
Lamba J, Schuetz E, Lim R, Lim HL, Ong AB, et al: Explaining
interindividual variability of docetaxel pharmacokinetics and
pharmacodynamics in Asians through phenotyping and genotyping
strategies. J Clin Oncol. 20:3683–3690. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Delbaldo C, Chatelut E, Ré M, Deroussent
A, Séronie-Vivien S, Jambu A, Berthaud P, Le Cesne A, Blay JY and
Vassal G: Pharmacokinetic-pharmacodynamic relationships of imatinib
and its main metabolite in patients with advanced gastrointestinal
stromal tumors. Clin Cancer Res. 12:6073–6078. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Bruno R, Olivares R, Berille J, Chaikin P,
Vivier N, Hammershaimb L, Rhodes GR and Rigas JR: Alpha-1-acid
glycoprotein as an independent predictor for treatment effects and
a prognostic factor of survival in patients with non-small cell
lung cancer treated with docetaxel. Clin Cancer Res. 9:1077–1082.
2003.PubMed/NCBI
|
|
44
|
Katori N, Sai K, Saito Y, Fukushima-Uesaka
H, Kurose K, Yomota C, Kawanishi T, Nishimaki-Mogami T, Naito M,
Sawada J, et al: Genetic variations of orosomucoid genes associated
with serum alpha-1-acid glycoprotein level and the pharmacokinetics
of paclitaxel in Japanese cancer patients. J Pharm Sci.
100:4546–4559. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Pandey PR, Liu W, Xing F, Fukuda K and
Watabe K: Anti-cancer drugs targeting fatty acid synthase (FAS).
Recent Patents Anticancer Drug Discov. 7:185–197. 2012. View Article : Google Scholar
|
|
46
|
Flavin R, Peluso S, Nguyen PL and Loda M:
Fatty acid synthase as a potential therapeutic target in cancer.
Future Oncol. 6:551–562. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chen Y, Ma Z, Li A, Li H, Wang B, Zhong J,
Min L and Dai L: Metabolomic profiling of human serum in lung
cancer patients using liquid chromatography/hybrid quadrupole
time-of-flight mass spectrometry and gas chromatography/mass
spectrometry. J Cancer Res Clin Oncol. 141:705–718. 2015.
View Article : Google Scholar
|
|
48
|
Liu H, Liu Y and Zhang JT: A new mechanism
of drug resistance in breast cancer cells: Fatty acid synthase
overexpression-mediated palmitate overproduction. Mol Cancer Ther.
7:263–270. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Meena AS, Sharma A, Kumari R, Mohammad N,
Singh SV and Bhat MK: Inherent and acquired resistance to
paclitaxel in hepatocellular carcinoma: Molecular events involved.
PLoS One. 8:e615242013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Roodhart JM, Daenen LG, Stigter EC, Prins
HJ, Gerrits J, Houthuijzen JM, Gerritsen MG, Schipper HS, Backer
MJ, van Amersfoort M, et al: Mesenchymal stem cells induce
resistance to chemotherapy through the release of platinum-induced
fatty acids. Cancer Cell. 20:370–383. 2011. View Article : Google Scholar : PubMed/NCBI
|