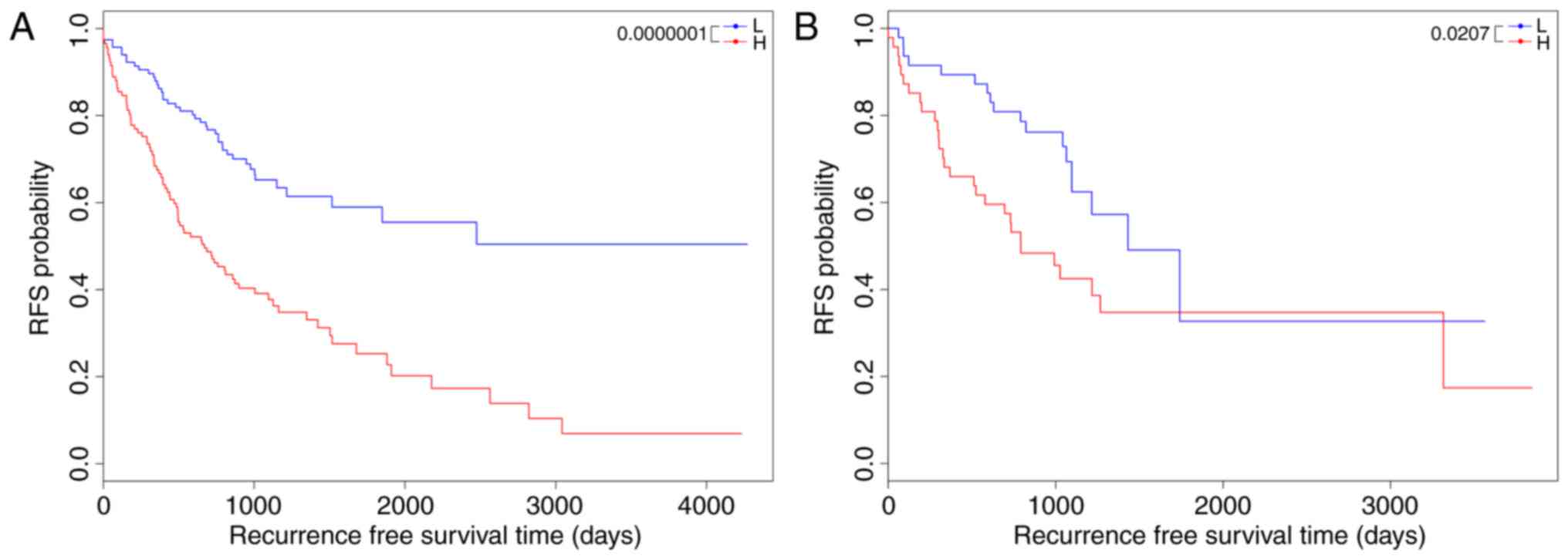
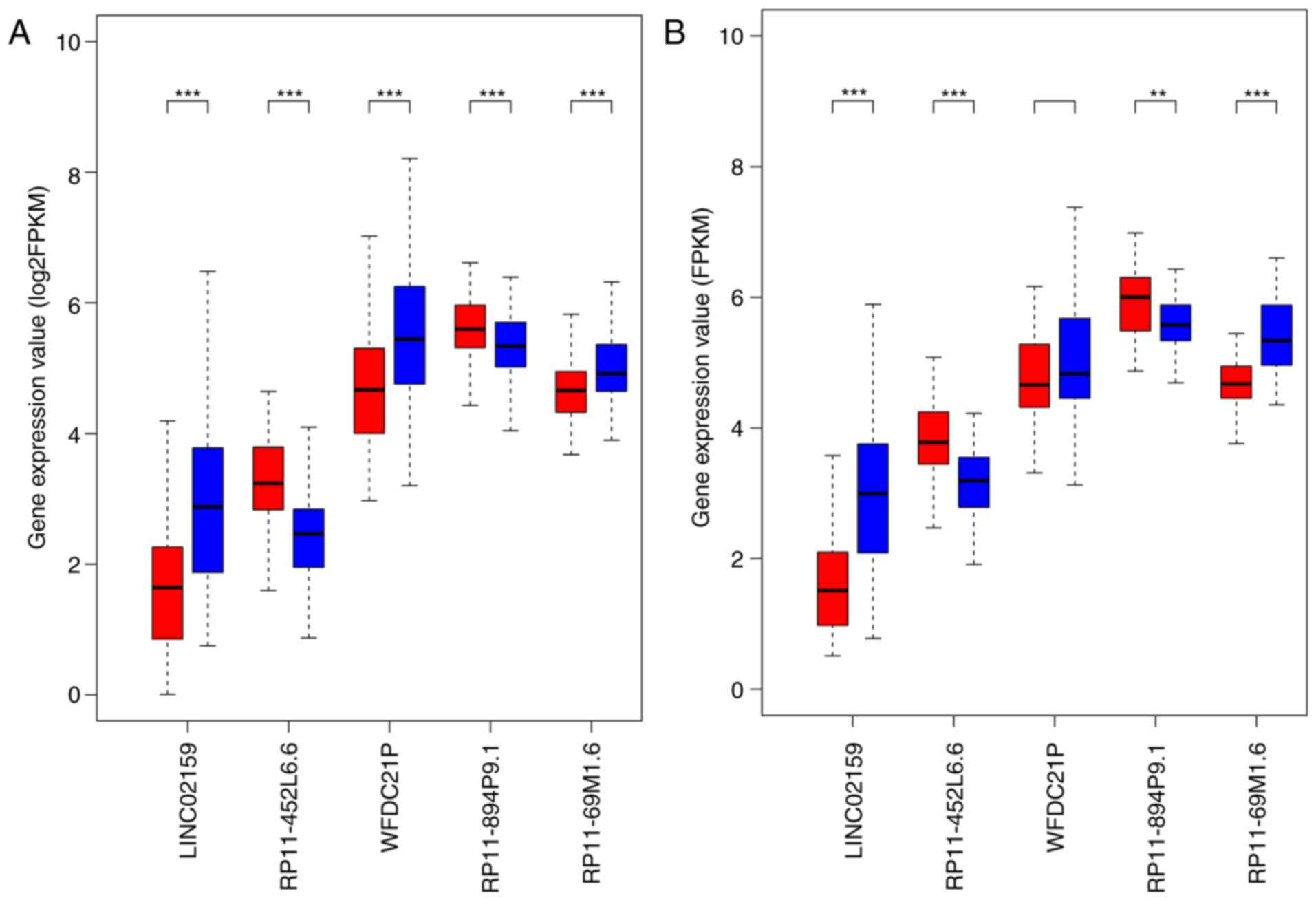
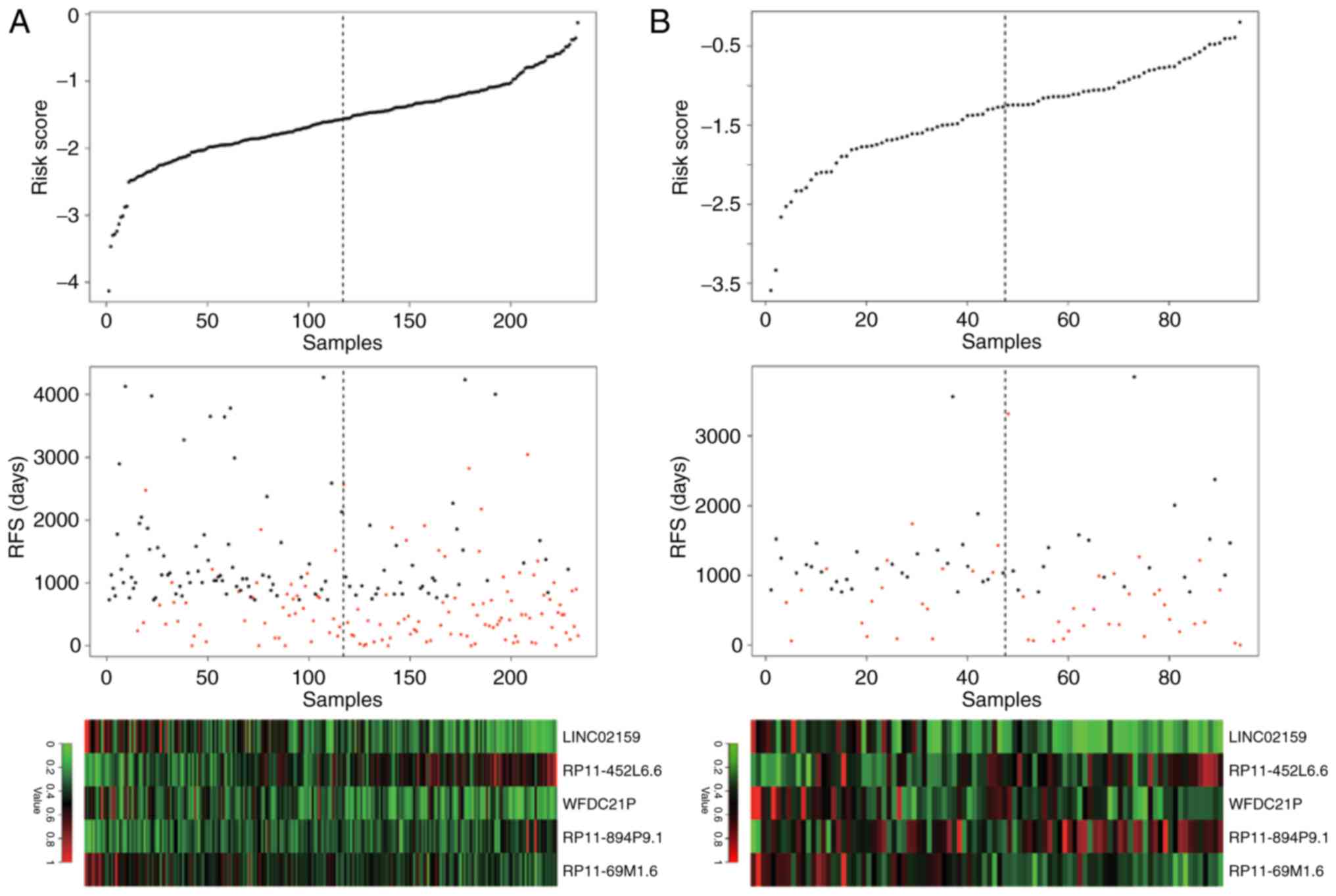
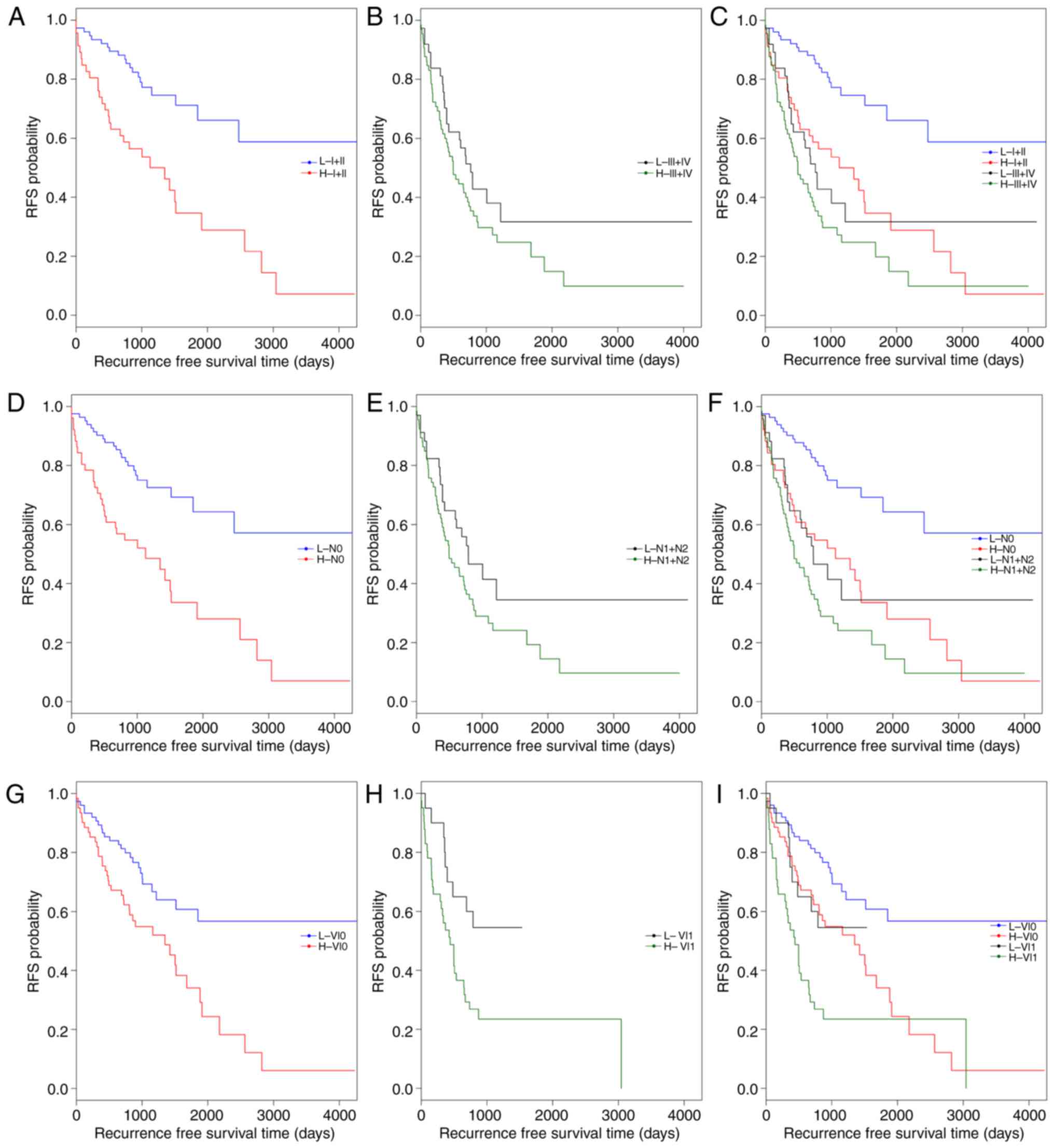
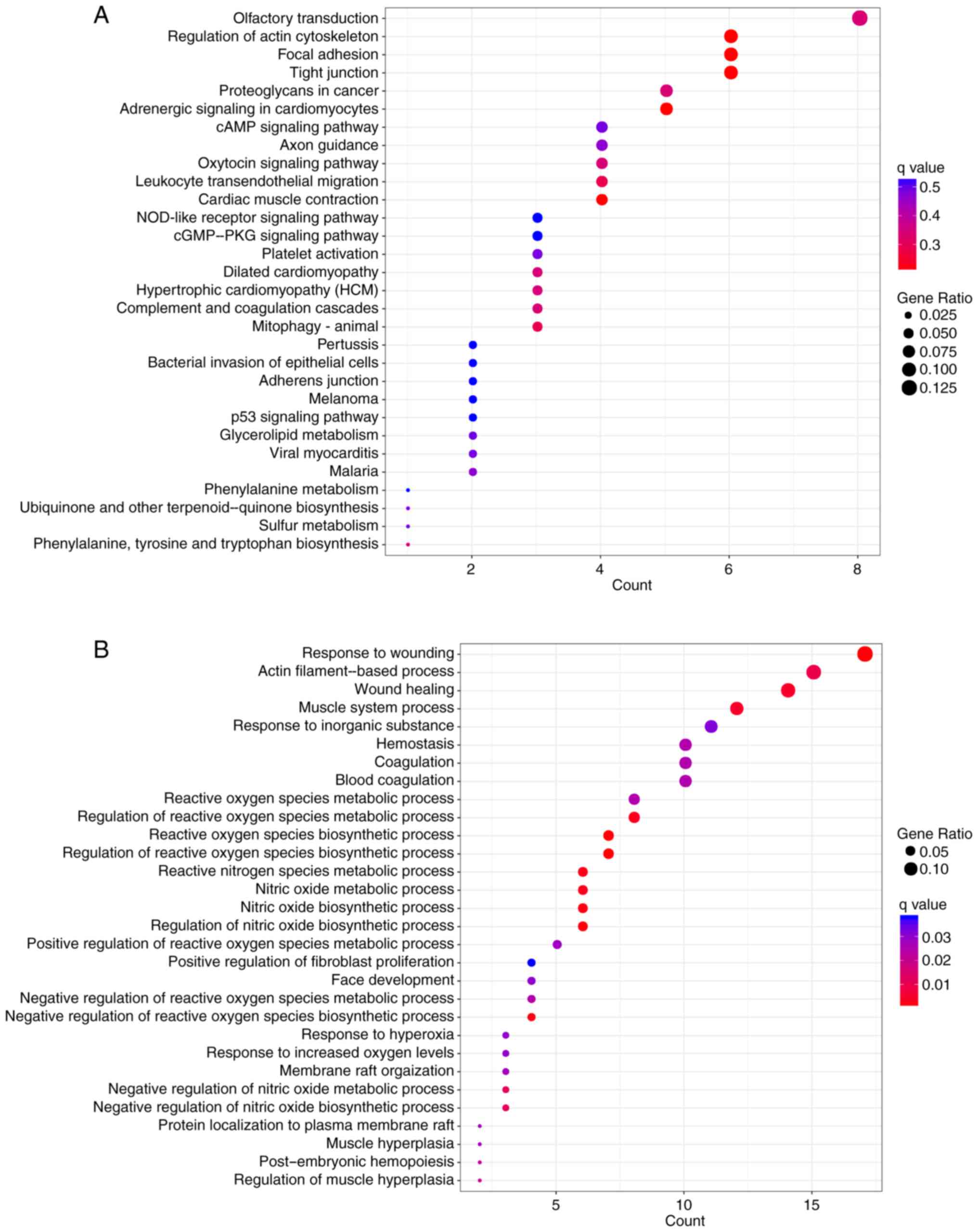
|
1
|
Miller KD, Siegel RL, Lin CC, Mariotto AB,
Kramer JL, Rowland JH, Stein KD, Alteri R and Jemal A: Cancer
treatment and survivorship statistics, 2016. CA Cancer J Clin.
66:271–289. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Brenner H, Kloor M and Pox CP: Colorectal
cancer. Lancet. 383:1490–1502. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
García Sánchez J: Colonoscopic polypectomy
and long-term prevention of colorectal cancer deaths. Rev Clin Esp.
212:4082012.(In Spanish).
|
|
4
|
Li X, Wu Z, Fu X and Han W: lncRNAs:
Insights into their function and mechanics in underlying disorders.
Mutat Res Rev Mutat Res. 762:1–21. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Schmitt AM and Chang HY: Long noncoding
RNAs in cancer pathways. Cancer Cell. 29:452–463. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Huarte M: The emerging role of lncRNAs in
cancer. Nat Med. 21:1253–1261. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zheng HT, Shi DB, Wang YW, Li XX, Xu Y,
Tripathi P, Gu WL, Cai GX and Cai SJ: High expression of lncRNA
MALAT1 suggests a biomarker of poor prognosis in colorectal cancer.
Int J Clin Exp Pathol. 7:3174–3181. 2014.PubMed/NCBI
|
|
8
|
Ge X, Chen Y, Liao X, Liu D, Li F, Ruan H
and Jia W: Overexpression of long noncoding RNA PCAT-1 is a novel
biomarker of poor prognosis in patients with colorectal cancer. Med
Oncol. 30:5882013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Qi P, Xu MD, Ni SJ, Huang D, Wei P, Tan C,
Zhou XY and Du X: Low expression of LOC285194 is associated with
poor prognosis in colorectal cancer. J Transl Med. 11:1222013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ozawa T, Matsuyama T, Toiyama Y, Takahashi
N, Ishikawa T, Uetake H, Yamada Y, Kusunoki M, Calin G and Goel A:
CCAT1 and CCAT2 long noncoding RNAs, located within the 8q.24.21
‘gene desert’, serve as important prognostic biomarkers in
colorectal cancer. Ann Oncol. 28:1882–1888. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Harrow J, Frankish A, Gonzalez JM,
Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa
A, Searle S, et al: GENCODE: the reference human genome annotation
for The ENCODE Project. Genome Res. 22:1760–1774. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sillar B and Plint CW: The price of a
false-negative result of mammography and an overenthusiastic lay
press. Med J Aust. 151:4181989.PubMed/NCBI
|
|
13
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
McCarthy DJ, Chen Y and Smyth GK:
Differential expression analysis of multifactor RNA-Seq experiments
with respect to biological variation. Nucleic Acids Res.
40:4288–4297. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15:5502014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang P, Wang Y, Bo H, Zou X and Mao JH: A
novel gene expression-based prognostic scoring system to predict
survival in gastric cancer. Oncotarget. 7:55343–55351.
2016.PubMed/NCBI
|
|
17
|
Zhou M, Wang X, Shi H, Cheng L, Wang Z,
Zhao H, Yang L and Sun J: Characterization of long non-coding
RNA-associated ceRNA network to reveal potential prognostic lncRNA
biomarkers in human ovarian cancer. Oncotarget. 7:12598–12611.
2016.PubMed/NCBI
|
|
18
|
Kolde R, Laur S, Adler P and Vilo J:
Robust rank aggregation for gene list integration and
meta-analysis. Bioinformatics. 28:573–580. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Adler P, Kolde R, Kull M, Tkachenko A,
Peterson H, Reimand J and Vilo J: Mining for coexpression across
hundreds of datasets using novel rank aggregation and visualization
methods. Genome Biol. 10:R1392009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45:D362–D368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xie C, Mao X, Huang J, Ding Y, Wu J, Dong
S, Kong L, Gao G, Li CY and Wei L: KOBAS 2.0: A web server for
annotation and identification of enriched pathways and diseases.
Nucleic Acids Res. 39:W316–W322. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xu MD, Qi P and Du X: Long non-coding RNAs
in colorectal cancer: Implications for pathogenesis and clinical
application. Mod Pathol. 27:1310–1320. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S,
Jiang Z, Xu J, Liu Q and Cao X: The STAT3-binding long noncoding
RNA lnc-DC controls human dendritic cell differentiation. Science.
344:310–313. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li Q and Liu Q: Noncoding RNAs in Cancer
Immunology. Adv Exp Med Biol. 927:2432016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wu GC, Li J, Leng RX, Li XP, Li XM, Wang
DG, Pan HF and Ye DQ: Identification of long non-coding RNAs GAS5,
linc0597 and lnc-DC in plasma as novel biomarkers for systemic
lupus erythematosus. Oncotarget. 8:23650–23663. 2017.PubMed/NCBI
|
|
28
|
Zhang W, Zhou Y and Ding Y: Lnc-DC
mediates the over-maturation of decidual dendritic cells and
induces the increase in Th1 cells in preeclampsia. Am J Reprod
Immunol. 77:2017. View Article : Google Scholar
|
|
29
|
Di Salvo TG, Guo Y, Su YR, Clark T,
Brittain E, Absi T, Maltais S and Hemnes A: Right ventricular long
noncoding RNA expression in human heart failure. Pulm Circ.
5:135–161. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Roxburgh CS, Mcmillan DC, Richards CH,
Atwan M, Anderson JH, Harvey T, Horgan PG and Foulis AK: The
clinical utility of the combination of T stage and venous invasion
to predict survival in patients undergoing surgery for colorectal
cancer. Ann Surg. 259:1156–1165. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Berger AC, Sigurdson ER, Levoyer T, Hanlon
A, Mayer RJ, Macdonald JS, Catalano PJ and Haller DG: Colon cancer
survival is associated with decreasing ratio of metastatic to
examined lymph nodes. J Clin Oncol. 23:8706–8712. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
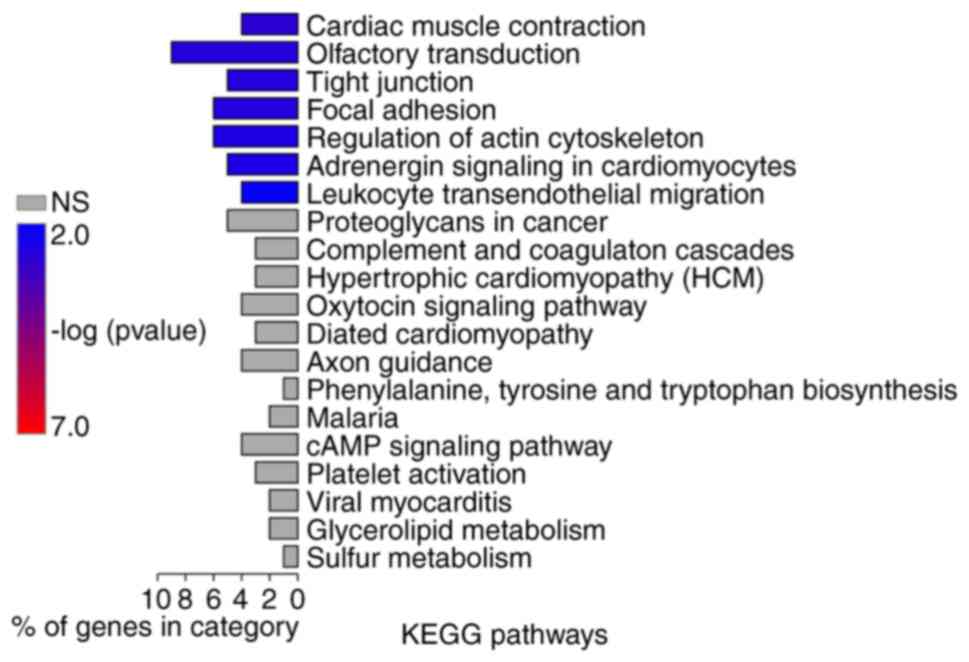
Martin TA: The role of tight junctions in
cancer metastasis. Semin Cell Dev Biol. 36:224–231. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Eke I and Cordes N: Focal adhesion
signaling and therapy resistance in cancer. Seminars in Cancer
Biol. 31:65–75. 2015. View Article : Google Scholar
|
|
34
|
Olson MF and Sahai E: The actin
cytoskeleton in cancer cell motility. Clin Exp Metastasis.
26:273–287. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Costa A, Scholerdahirel A and
Mechta-Grigoriou F: The role of reactive oxygen species and
metabolism on cancer cells and their microenvironment. Semin Cancer
Biol. 25:23–32. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Colin DJ, Limagne E, Ragot K, Lizard G,
Ghiringhelli F, Solary É, Chauffert B, Latruffe N and Delmas D: The
role of reactive oxygen species and subsequent DNA-damage response
in the emergence of resistance towards resveratrol in colon cancer
models. Cell Death Dis. 5:e15332014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chang CF, Diers AR and Hogg N: Cancer cell
metabolism and the modulating effects of nitric oxide. Free Radic
Biol Med. 79:324–336. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wink DA, Vodovotz Y, Laval J, Laval F,
Dewhirst MW and Mitchell JB: The multifaceted roles of nitric oxide
in cancer. Carcinogenesis. 19:711–721. 1998. View Article : Google Scholar : PubMed/NCBI
|